RNAi-Mediated Knockdown of Tssk1 and Tektin1 Genes Impair Male Fertility in Bactrocera dorsalis
Abstract
1. Introduction
2. Materials and Methods
2.1. Flies Rearing
2.2. Selection of Target Genes for RNAi
2.3. Expression Profile Analysis
2.4. Preparation of Double Stranded RNAs
2.5. Feeding of dsRNA
2.6. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.7. Reproduction Bioassays
2.8. Spermatozoa Counts and Sperm Viability Assays
2.9. Data Analyses
3. Results
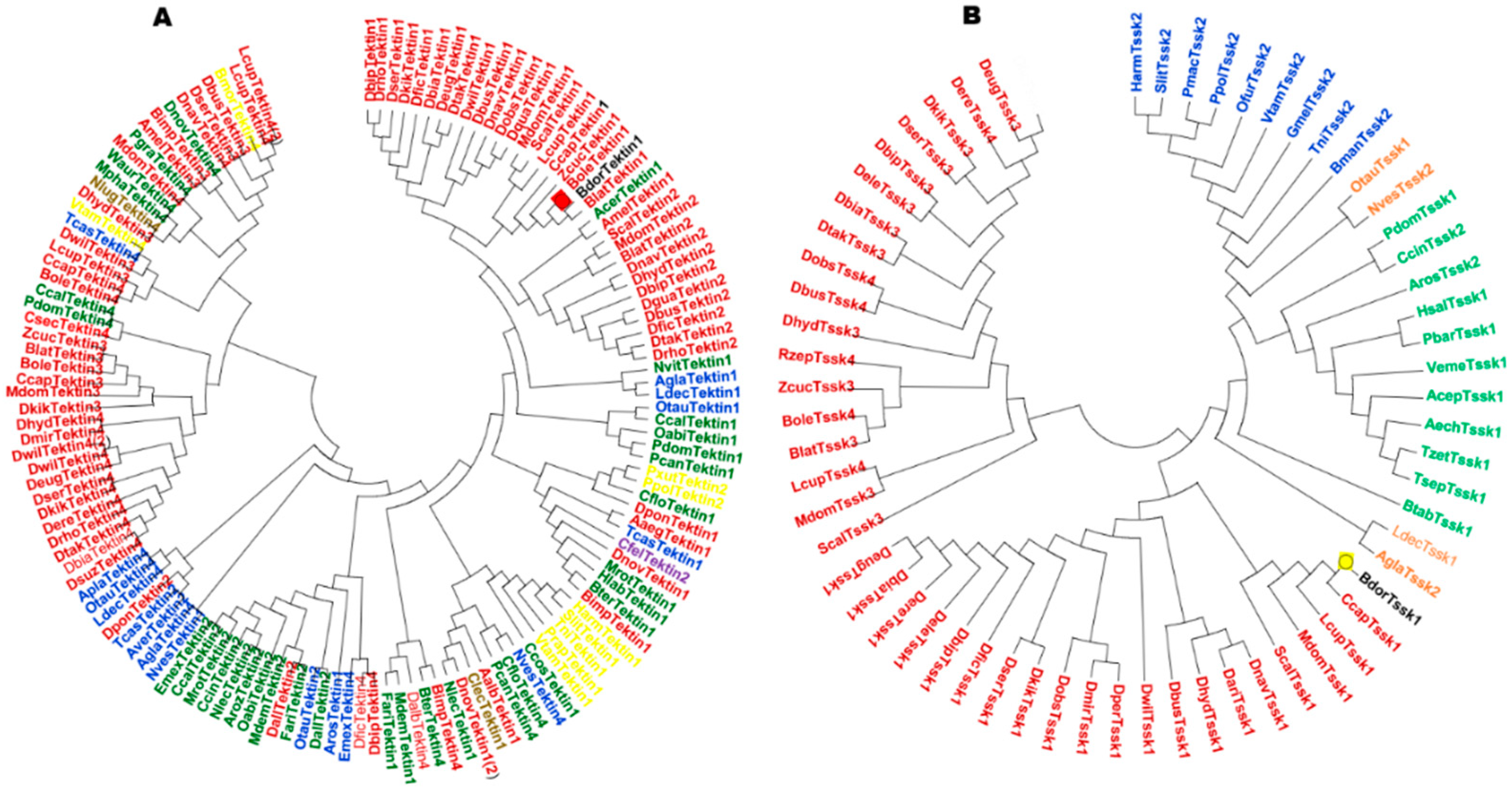
3.1. Selection of Genes Related to Spermatogenesis
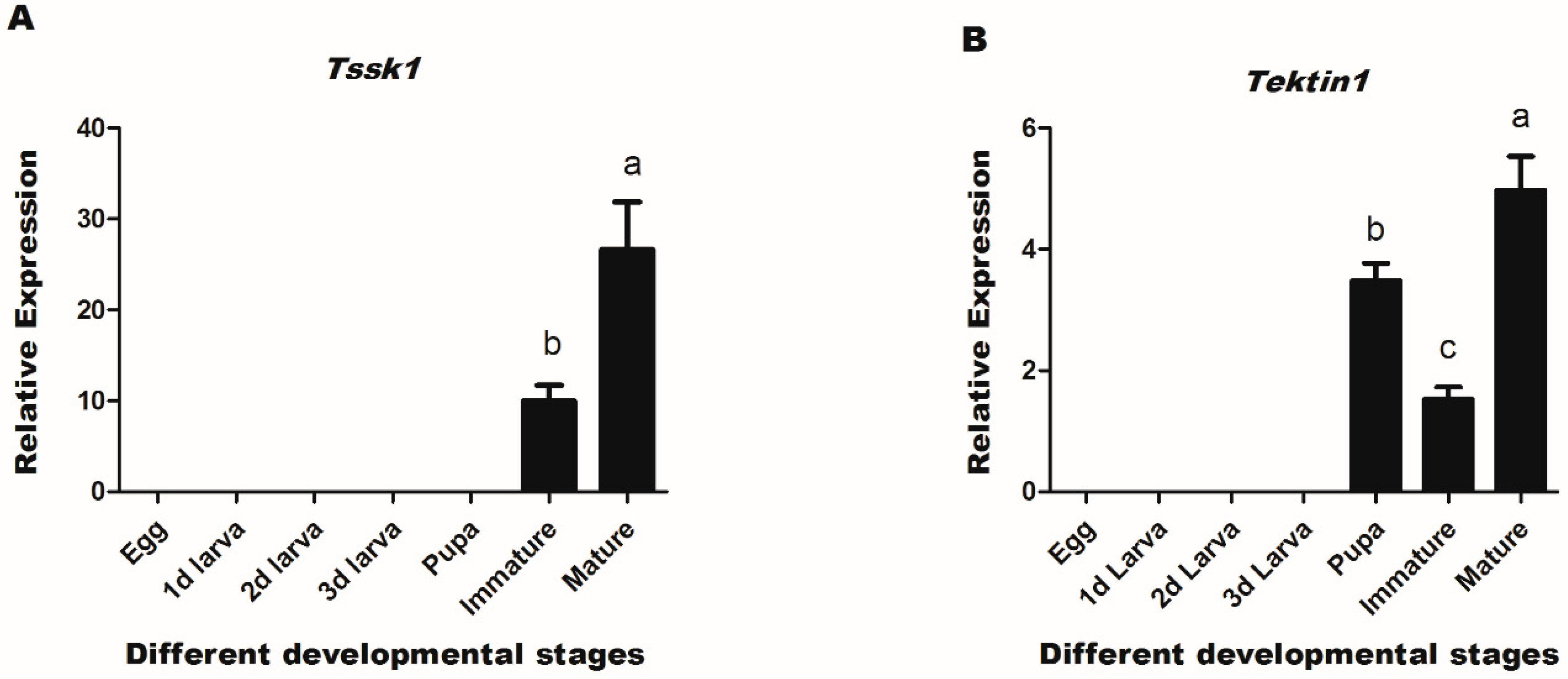
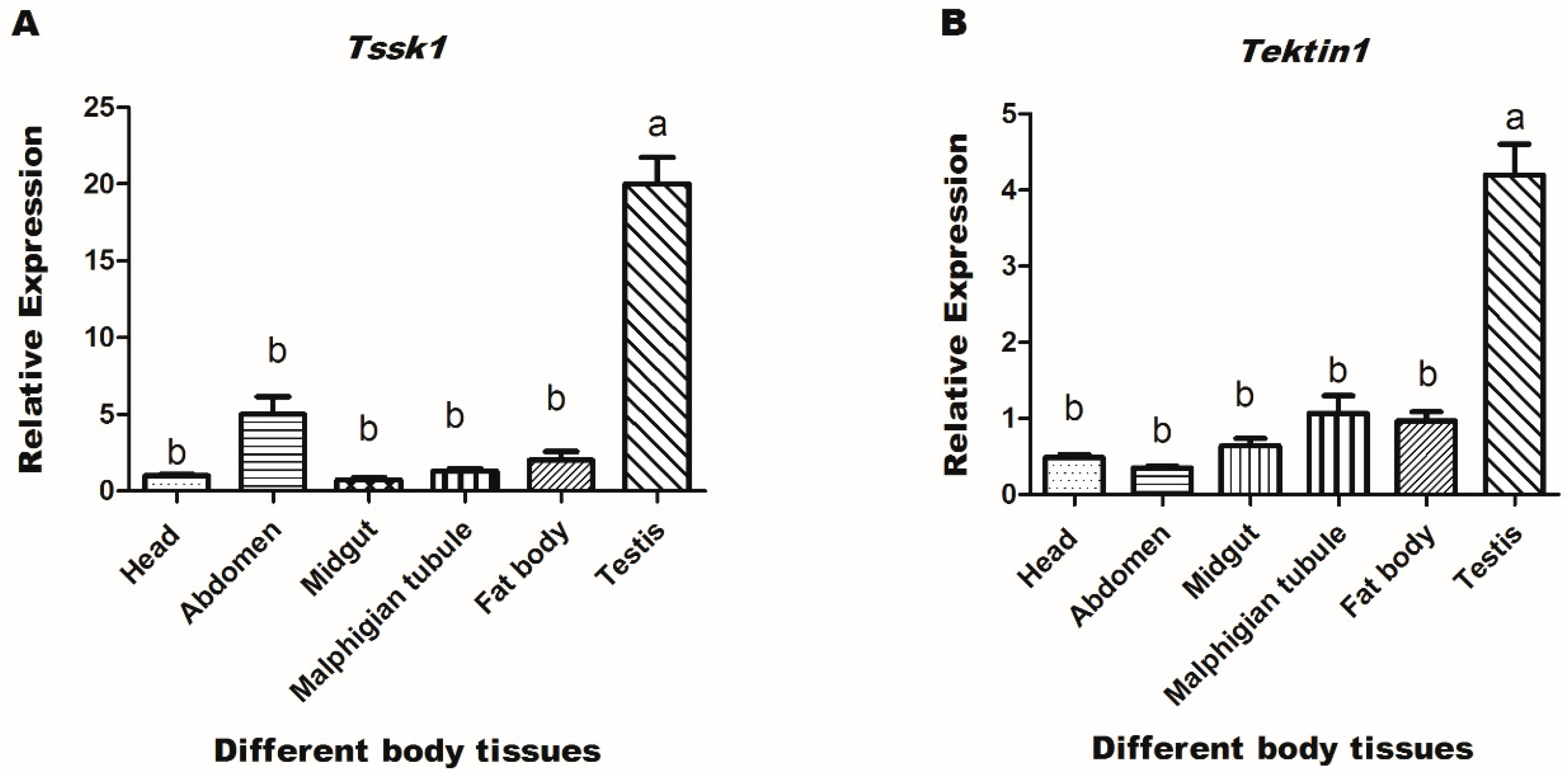
3.2. Expression Profiles of Target Genes
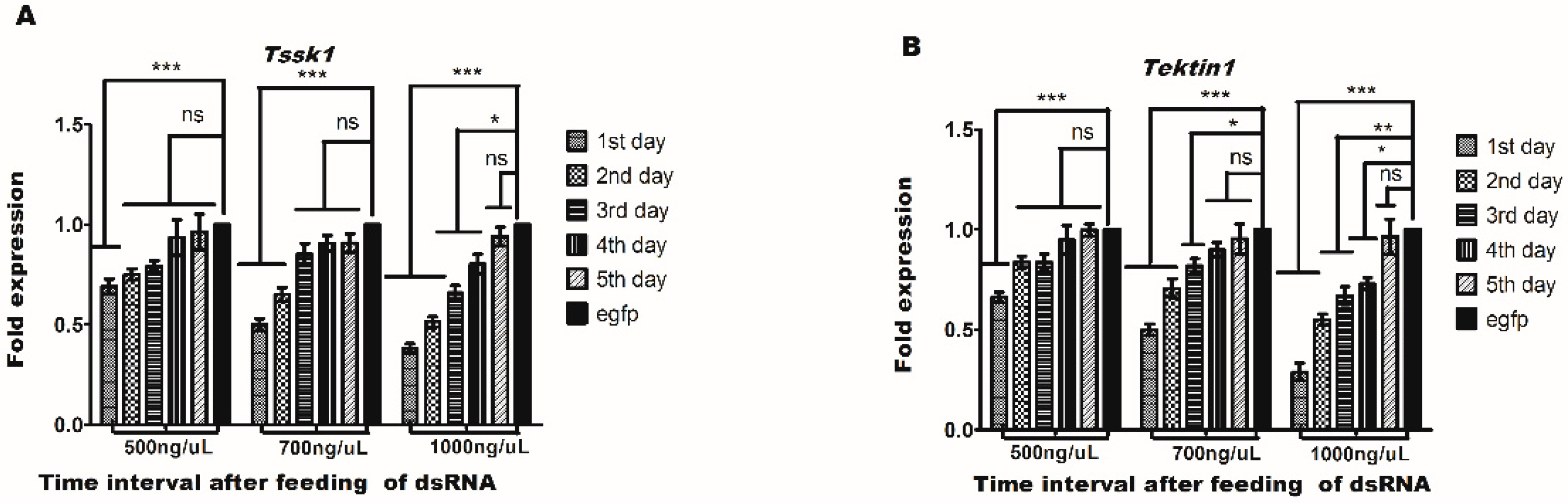
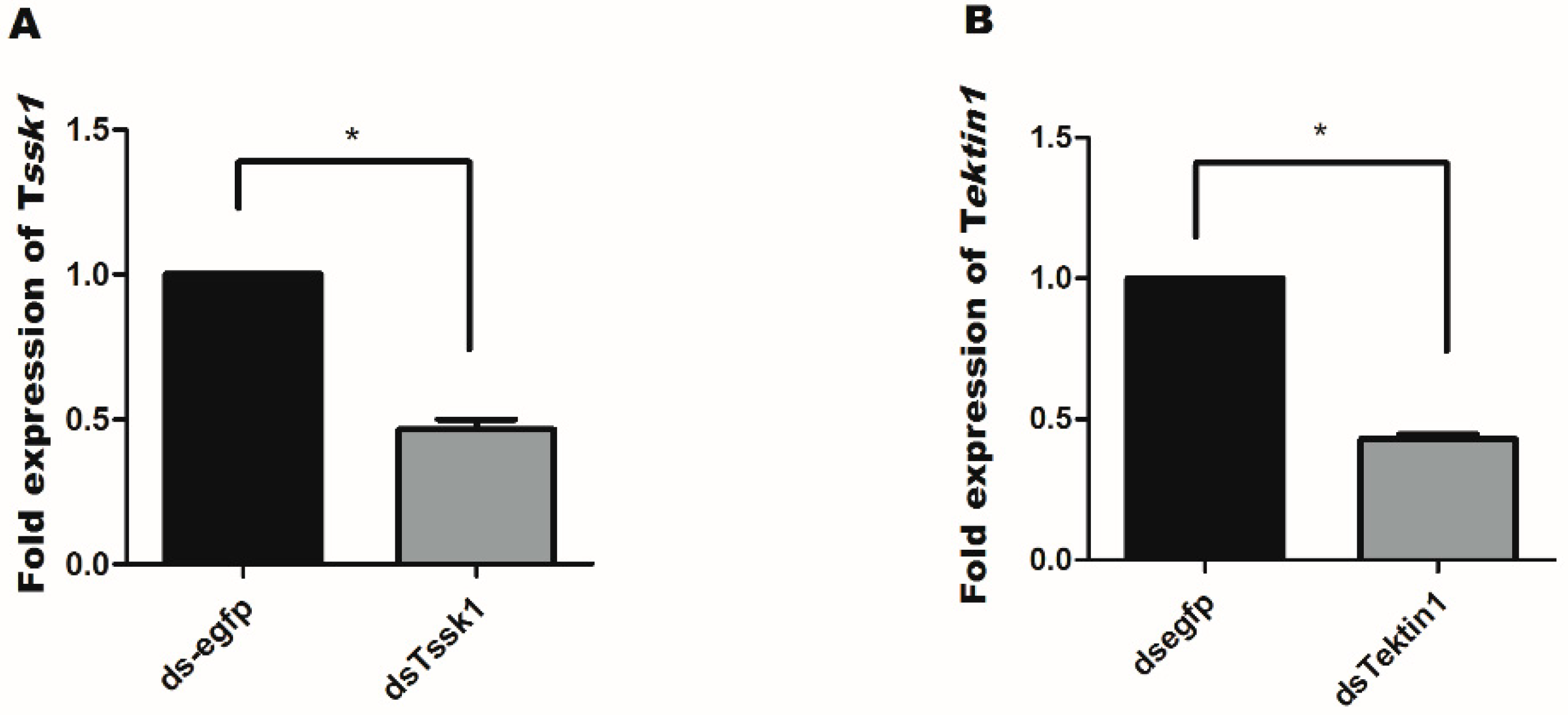
3.3. Effect of dsRNA Feeding on Tssk1 and Tektin1 Expression
3.4. Effect of Genes Silencing on the Reproductive Capacity of Males
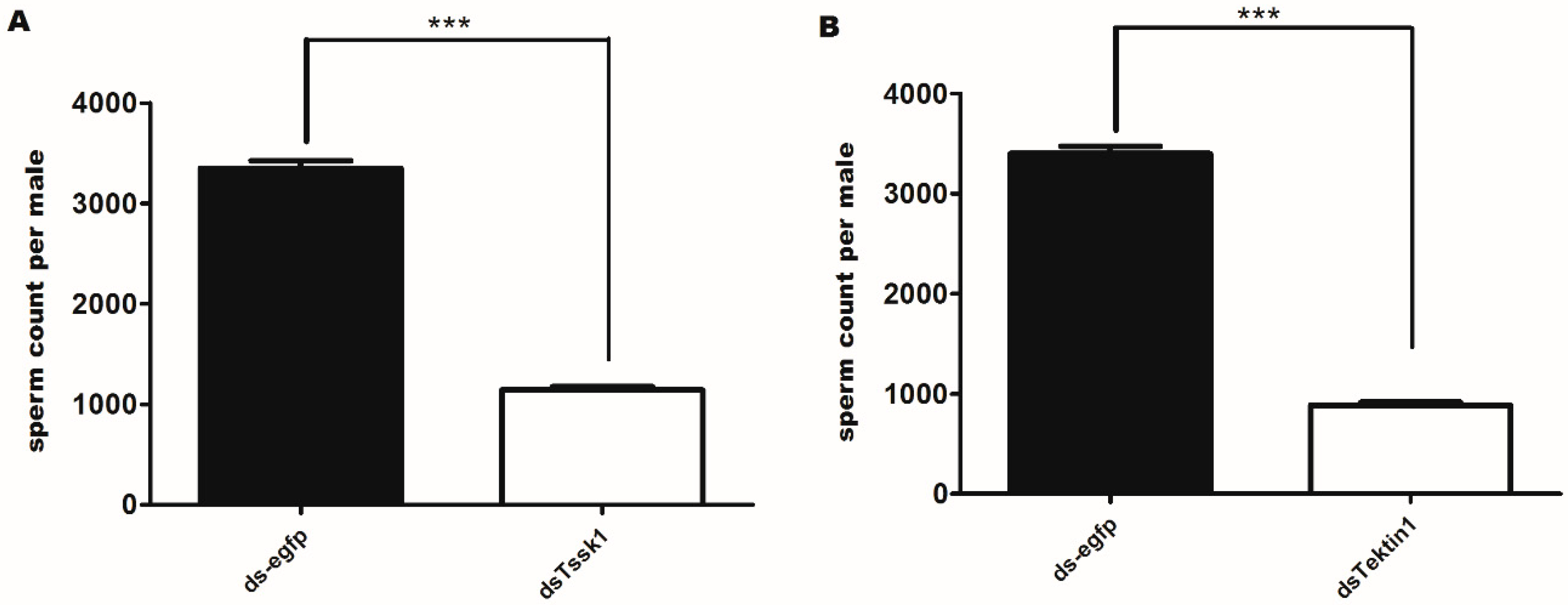
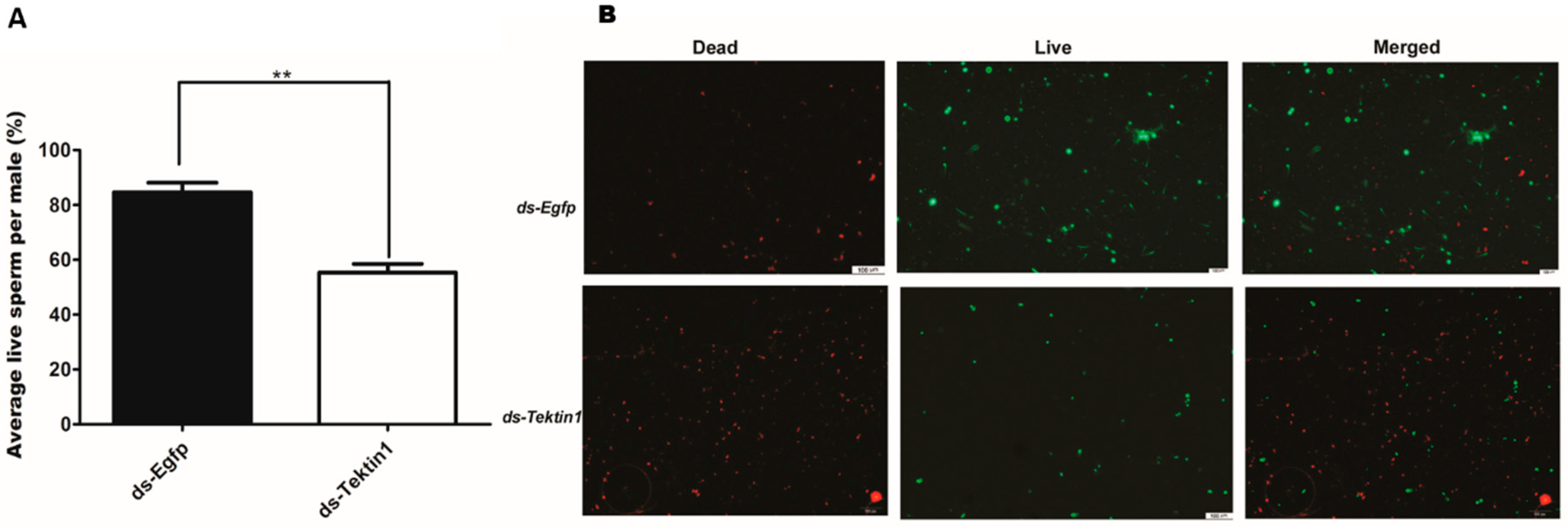
3.5. Effects of Gene Silencing on the Quantity and Quality of Spermatozoa
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Clarke, A.R.; Armstrong, K.F.; Carmichael, A.E.; Milne, J.R.; Raghu, S.; Roderick, G.K.; Yeates, D.K. Invasive Yeates. phytophagous pests arising through a recent tropical evolutionary radiation: The Bactrocera dorsalis complex of fruit flies. Annu. Rev. Entomol. 2005, 50, 293–319. [Google Scholar] [CrossRef]
- Chen, A.; Zheng, W.; Zheng, W.; Zhang, H. The effects of RNA interference targeting Bactrocera dorsalis ds-Bdrpl19 on the gene expression of rpl19 in non-target insects. Ecotoxicology 2015, 24, 595–603. [Google Scholar] [CrossRef]
- Li, X.; Zhang, M.; Zhang, H. RNA interference of four genes in adult Bactrocera dorsalis by feeding their dsRNAs. PLoS ONE 2011, 6, e17788. [Google Scholar] [CrossRef]
- Zhang, H.Y; Li, H.Y. Photographic Guide to Key Control Techniques for Citrus Disease and Insect Pests; Chinese Agricultural Press: Beijing, China, 2012. [Google Scholar]
- Jin, T.; Zeng, L.; Lin, Y.; Lu, Y.; Liang, G. Insecticide resistance of the oriental fruit fly, Bactrocera dorsalis (Hendel) (Diptera: Tephritidae), in mainland China. Pest Manag. Sci. 2011, 67, 370–376. [Google Scholar] [CrossRef]
- Lu, X.-P.; Wang, L.-L.; Huang, Y.; Dou, W.; Chen, C.-T.; Wei, D.; Wang, J.-J. The epsilon glutathione S-transferases contribute to the malathion resistance in the oriental fruit fly, Bactrocera dorsalis (Hendel). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2016, 180, 40–48. [Google Scholar] [CrossRef]
- Colborn, T.; Vom Saal, F.S.; Soto, A.M. Developmental effects of endocrine-disrupting chemicals in wildlife and humans. Environ. Health Perspect. 1993, 101, 378–384. [Google Scholar] [CrossRef]
- Leftwich, P.T.; Bolton, M.; Chapman, T. Evolutionary biology and genetic technique for insect control. Evol. Appl. 2016, 9, 212–230. [Google Scholar] [CrossRef]
- Lees, R.S.; Gilles, J.R.; Hendrichs, J.; Vreysen, M.J.; Bourtzis, K. Back to the future: The sterile insect technique against mosquito disease vectors. Curr. Opin. Insect. Sci. 2015, 10, 156–162. [Google Scholar] [CrossRef]
- Lance, D.; McInnis, D. Biological Basis of the Sterile Insect Technique, in Sterile Insect Technique; Springer: Dordrecht, The Netherlands, 2005; pp. 69–94. [Google Scholar]
- Fuller, M.T. Spermatogenesis. In The Development of Drosophila melanogaster; Martinez-Arias, M., Bate, M., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1993; pp. 71–147. [Google Scholar]
- Lindsley, D.L.; Tokuyasu, K. The Genetics and Biology of Drosophila; Academic Press: New York, NY, USA, 1980; pp. 225–294. [Google Scholar]
- Hake, L.E.; Alcivar, A.A.; Hecht, N.B. Changes in mRNA length accompany translational regulation of the somatic and testis-specific cytochrome c genes during spermatogenesis in the mouse. Development 1990, 110, 249–257. [Google Scholar]
- Tariq, K.; Metzendorf, C.; Peng, W.; Sohail, S.; Zhang, H. miR-8-3p regulates mitoferrin in the testes of Bactrocera dorsalis to ensure normal spermatogenesis. Sci. Rep. 2016, 6, 22565. [Google Scholar] [CrossRef]
- Li, Y.; Sosnik, J.; Brassard, L.; Reese, M.; Spiridonov, N.A.; Bates, T.C.; Salicioni, A.M. Expression and localization of five members of the testis-specific serine kinase (Tssk) family in mouse and human sperm and testis. Mol. Hum. Reprod. 2010, 17, 42–56. [Google Scholar] [CrossRef]
- Li, H.H.; Kong, L.F.; Yu, R.; Yu, H.; Li, Q. Characterization, expression, and functional analysis of testis-specific serine/threonine kinase 1 (Tssk1) in the pen shell Atrina pectinata. Invertebr. Reprod. Dev. 2016, 60, 118–125. [Google Scholar] [CrossRef]
- Zuercher, G.; Rohrbach, V.; Andres, A.-C.; Ziemiecki, A. A novel member of the testis specific serine kinase family, tssk-3, expressed in the Leydig cells of sexually mature mice. Mech. Dev. 2000, 93, 175–177. [Google Scholar] [CrossRef]
- Chen, X.; Lin, G.; Wei, Y.; Hexige, S.; Niu, Y.; Liu, L.; Yu, L. TSSK5, a novel member of the testis-specific serine/threonine kinase family, phosphorylates CREB at Ser-133, and stimulates the CRE/CREB responsive pathway. Biochem. Biophys. Res. Commun. 2005, 333, 742–749. [Google Scholar] [CrossRef]
- Spiridonov, N.A.; Wong, L.; Zerfas, P.M.; Starost, M.F.; Pack, S.D.; Paweletz, C.P.; Johnson, G.R. Identification and characterization of SSTK, a serine/threonine protein kinase essential for male fertility. Mol. Cell. Biol. 2005, 25, 4250–4261. [Google Scholar] [CrossRef]
- Bielke, W.; Blaschke, R.; Miescher, G.; Zürcher, G.; Andres, A.-C.; Ziemiecki, A. Characterization of a novel murine testis-specific serine/threonine kinase. Gene 1994, 139, 235–239. [Google Scholar] [CrossRef]
- Kueng, P.; Nikolova, Z.; Djonov, V.; Hemphill, A.; Rohrbach, V.; Boehlen, D.; Ziemiecki, A. A novel family of serine/threonine kinases participating in spermiogenesis. J. Cell. Biol. 1997, 139, 1851–1859. [Google Scholar] [CrossRef]
- Xu, B.; Hao, Z.; Jha, K.N.; Zhang, Z.; Urekar, C.; Digilio, L.; Herr, J.C. Targeted deletion of Tssk1 and 2 causes male infertility due to haploinsufficiency. Dev. Biol. 2008, 319, 211–222. [Google Scholar] [CrossRef]
- Cruz, C.; Tayler, A.; Whyard, S. RNA Interference-Mediated Knockdown of Male Fertility Genes in the Queensland Fruit Fly Bactrocera tryoni (Diptera: Tephritidae). Insects 2018, 9, 96. [Google Scholar] [CrossRef]
- Larsson, M.; Norrander, J.; Gräslund, S.; Brundell, E.; Linck, R.; Ståhl, S.; Höög, C. The spatial and temporal expression of Tekt1, a mouse tektin C homologue, during spermatogenesis suggest that it is involved in the development of the sperm tail basal body and axoneme. Eur. J. Cell Biol. 2000, 79, 718–725. [Google Scholar] [CrossRef]
- Linck, R.; Amos, L.; Amos, W. Localization of tektin filaments in microtubules of sea urchin sperm flagella by immunoelectron microscopy. J. Cell Biol. 1985, 100, 126–135. [Google Scholar] [CrossRef]
- Norrander, J.; Larsson, M.; Ståhl, S.; Höög, C.; Linck, R. Expression of ciliary tektins in brain and sensory development. J. Neurosci. 1998, 18, 8912–8918. [Google Scholar] [CrossRef]
- Steffen, W.; Linck, R. Evidence for tektins in centrioles and axonemal microtubules. Proc. Natl. Acad. Sci. USA 1988, 85, 2643–2647. [Google Scholar] [CrossRef]
- Ota, A.; Kusakabe, T.; Sugimoto, Y.; Takahashi, M.; Nakajima, Y.; Kawaguchi, Y.; Koga, K. Cloning and characterization of testis-specific tektin in Bombyx mori. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2002, 133, 371–382. [Google Scholar] [CrossRef]
- Oiki, S.; Hiyama, E.; Gotoh, T.; Iida, H. Localization of Tektin 1 at both acrosome and flagella of mouse and bull spermatozoa. Zool. Sci. 2014, 31, 101–108. [Google Scholar] [CrossRef]
- Iguchi, N.; Tanaka, H.; Nakamura, Y.; Nozaki, M.; Fujiwara, T.; Nishimune, Y. Cloning and characterization of the human Tektin-T gene. Mol. Hum. Reprod. 2002, 8, 525–530. [Google Scholar] [CrossRef]
- Linck, R.; Fu, X.; Lin, J.; Ouch, C.; Schefter, A.; Steffen, W.; Nicastro, D. Insights into the Structure and Function of Ciliary and Flagellar Doublet Microtubules TEKTINS, Ca2+-BINDING PROTEINS, AND STABLE PROTOFILAMENTS. J. Biol. Chem. 2014, 289, 17427–17444. [Google Scholar] [CrossRef]
- Norrander, J.M.; Perrone, C.A.; Amos, L.A.; Linck, R.W. Structural comparison of tektins and evidence for their determination of complex spacings in flagellar microtubules. J. Mol. Biol. 1996, 257, 385–397. [Google Scholar] [CrossRef]
- Xu, M.; Zhou, Z.; Cheng, C.; Zhao, W.; Tang, R.; Huang, Y.; Xie, Y. Cloning and characterization of a novel human TEKTIN1 gene. Int. J. Biochem. Cell Biol. 2001, 33, 1172–1182. [Google Scholar] [CrossRef]
- Tanaka, H.; Iguchi, N.; Toyama, Y.; Kitamura, K.; Takahashi, T.; Kaseda, K.; Nishimune, Y. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol. Cell. Biol. 2004, 24, 7958–7964. [Google Scholar] [CrossRef]
- Ali, M.W.; Zheng, W.; Sohail, S.; Li, Q.; Zheng, W.; Zhang, H. A genetically enhanced sterile insect technique against the fruit fly, Bactrocera dorsalis (Hendel) by feeding adult double stranded RNAs. Sci. Rep. 2017, 7, 4063. [Google Scholar] [CrossRef]
- Ant, T.; Koukidou, M.; Rempoulakis, P.; Gong, H.-F.; Economopoulos, A.; Vontas, J.; Alphey, L. Control of the olive fruit fly using genetics-enhanced sterile insect technique. BMC Biol. 2012, 10, 51. [Google Scholar] [CrossRef]
- Whyard, S.; Erdelyan, C.N.; Partridge, A.L.; Singh, A.D.; Beebe, N.W.; Capina, R. Silencing the buzz: A new approach to population suppression of mosquitoes by feeding larvae double-stranded RNAs. Parasit. Vectors 2015, 8, 96. [Google Scholar] [CrossRef]
- Wei, D.; Li, H.M.; Yang, W.J.; Wei, D.D.; Dou, W.; Huang, Y.; Wang, J.J. Transcriptome profiling of the testis reveals genes involved in spermatogenesis and marker discovery in the oriental fruit fly, Bactrocera dorsalis. Insect Mol. Biol. 2015, 24, 41–57. [Google Scholar] [CrossRef]
- Peng, W.; Zheng, W.; Handler, A.M.; Zhang, H. The role of the transformer gene in sex determination and reproduction in the tephritid fruit fly, Bactrocera dorsalis (Hendel). Genetica 2015, 143, 717–727. [Google Scholar] [CrossRef]
- Liu, G.; Wu, Q.; Li, J.; Zhang, G.; Wan, F. RNAi-mediated knock-down of transformer and transformer 2 to generate male-only progeny in the oriental fruit fly, Bactrocera dorsalis (Hendel). PLoS ONE 2015, 10, 0128892. [Google Scholar] [CrossRef]
- Yao, Z.; Wang, A.; Li, Y.; Cai, Z.; Lemaitre, B.; Zhang, H. The dual oxidase gene BdDuox regulates the intestinal bacterial community homeostasis of Bactrocera dorsalis. ISME J. 2016, 10, 1037. [Google Scholar] [CrossRef]
- Find Percentage with Percent Increase Online Calculator. Available online: https://www.marshu.com/articles/calculate-percentage-increase-decrease-percent-calculator.php (accessed on 31 January 2018).
- Bressac, C.; Chevrier, C. Offspring and sex ratio are independent of sperm management in Eupelmus orientalis females. J. Insect. Physiol. 1998, 44, 351–359. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Orankanok, W.; Chinvinijkul, S.; Thanaphum, S.; Sitilob, P.; Enkerlin, W. Area-wide integrated control of oriental fruit fly Bactrocera dorsalis and guava fruit fly Bactrocera correcta in Thailand. In Area-Wide Control of Insect Pests; Springer: Dordrecht, The Netherlands, 2007; pp. 517–526. [Google Scholar]
- Zheng, Z.; Guan, M.; Jia, Y.; Wang, D.; Pang, R.; Lv, F.; Xue, Y. The coordinated roles of miR-26a and miR-30c in regulating TGFβ1-induced epithelial-to-mesenchymal transition in diabetic nephropathy. Sci. Rep. 2016, 6, 37492. [Google Scholar] [CrossRef]
- Kupferschmidt, K. A lethal dose of RNA. Science 2013, 341, 732–733. [Google Scholar] [CrossRef]
- Kamath, R.S.; Martinez-Campos, M.; Zipperlen, P.; Fraser, A.G.; Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2000, 2, research0002.1. [Google Scholar] [CrossRef]
- Xie, Q.; Zhang, R.J. Study advance on biology and ecology of Bactrocera dorsalis (Hendel) and its control. Ecol. Sci. 2005, 24, 52–56. [Google Scholar]
- Perezgasga, L.; Jiang, J.; Bolival, B.J.; Hiller, M.; Benson, E.; Fuller, M.T.; White-Cooper, H. Regulation of transcription of meiotic cell cycle and terminal differentiation genes by the testis-specific Zn-finger protein matotopetli. Development 2004, 131, 1691–1702. [Google Scholar] [CrossRef]

| Primer Name | Primer Sequences for qRT-PCR |
| Tssk1-F | CTCCAATCGCCAACTGAATA |
| TSSK1-R | ATTTGTGTACGAAATCCGAG |
| Tektin1-F | TGTGGATGAAACCAAAGACA |
| Tektin1-R | CCAACGATTTCTCCTTCAGA |
| 16S-F | CTCGTCCAACCGTTCATACC |
| 16S-R | CTGACCTGCCCACTGAAGTT |
| Primer Name | Primer Sequence for dsRNA Synthesis |
| dstssk1-F | GGATCCTAATACGACTCACTATAGGATCACCCAAACATCATACAGA |
| dstssk1-R | GGATCCTAATACGACTCACTATAGGCAATTTCGGATCGTATGGC |
| dstektin1-F | GGATCCTAATACGACTCACTATAGGAGACATGCAAAATCAAACGG |
| dstektin1-R | GGATCCTAATACGACTCACTATAGGCGTGTAGCAAATAGCGTAAC |
| dsGFP-F | GGATCCTAATACGACTCACTATAGGATACGGCGTGCAGTGCT |
| dsGFP-R | GGATCCTAATACGACTCACTATAGGATGATCGCGCTTCTCG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohail, S.; Tariq, K.; Zheng, W.; Ali, M.W.; Peng, W.; Raza, M.F.; Zhang, H. RNAi-Mediated Knockdown of Tssk1 and Tektin1 Genes Impair Male Fertility in Bactrocera dorsalis. Insects 2019, 10, 164. https://doi.org/10.3390/insects10060164
Sohail S, Tariq K, Zheng W, Ali MW, Peng W, Raza MF, Zhang H. RNAi-Mediated Knockdown of Tssk1 and Tektin1 Genes Impair Male Fertility in Bactrocera dorsalis. Insects. 2019; 10(6):164. https://doi.org/10.3390/insects10060164
Chicago/Turabian StyleSohail, Summar, Kaleem Tariq, Weiwei Zheng, Muhammad Waqar Ali, Wei Peng, Muhammad Fahim Raza, and Hongyu Zhang. 2019. "RNAi-Mediated Knockdown of Tssk1 and Tektin1 Genes Impair Male Fertility in Bactrocera dorsalis" Insects 10, no. 6: 164. https://doi.org/10.3390/insects10060164
APA StyleSohail, S., Tariq, K., Zheng, W., Ali, M. W., Peng, W., Raza, M. F., & Zhang, H. (2019). RNAi-Mediated Knockdown of Tssk1 and Tektin1 Genes Impair Male Fertility in Bactrocera dorsalis. Insects, 10(6), 164. https://doi.org/10.3390/insects10060164






