Increases in Genistein in Medicago sativa Confer Resistance against the Pisum Host Race of Acyrthosiphon pisum
Abstract
1. Introduction
2. Materials and Methods
2.1. Aphids and Host Plants
2.2. Aphid Population Abundance
2.3. Aphid Feeding Behavior
2.4. Plant Material Sampling and Extraction of Phenolics
2.5. Quantification of Plant Secondary Metabolites by HPLC
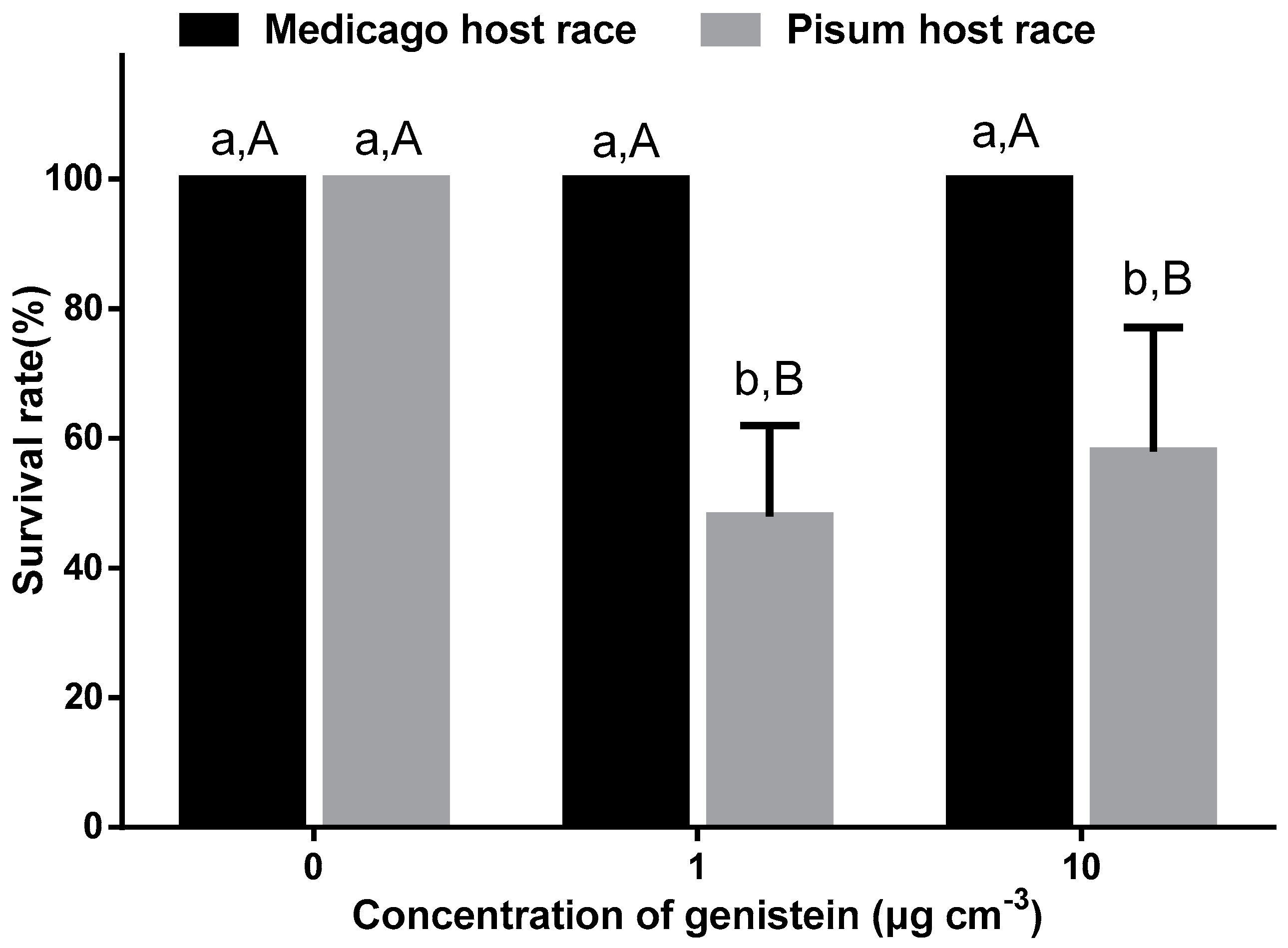
2.6. Bioassay with Pure Compound
2.7. Transcriptomics Analyses
2.8. Quantification of Gene Expression
2.9. Statistical Analysis
3. Result
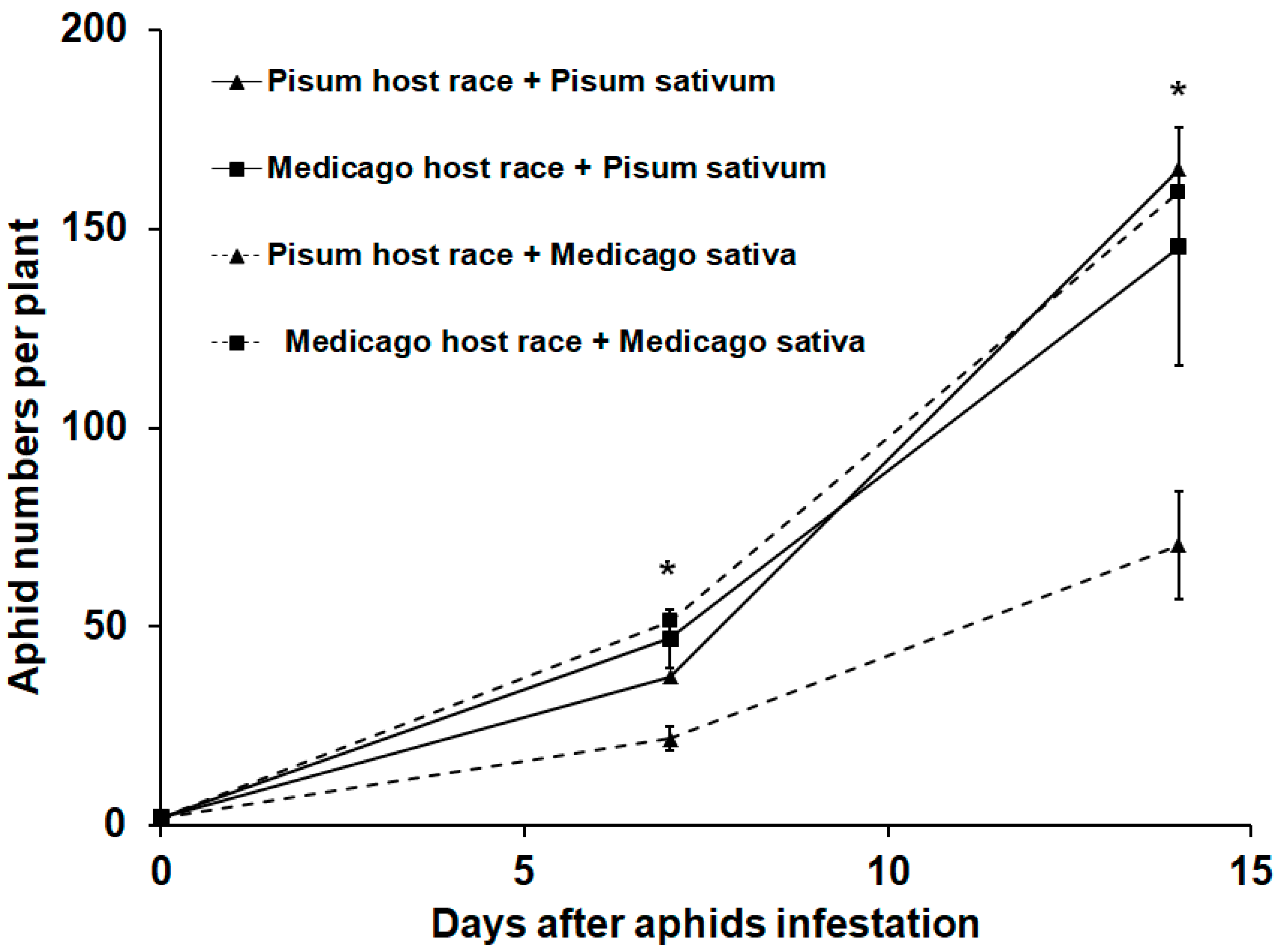
3.1. Aphid Performance
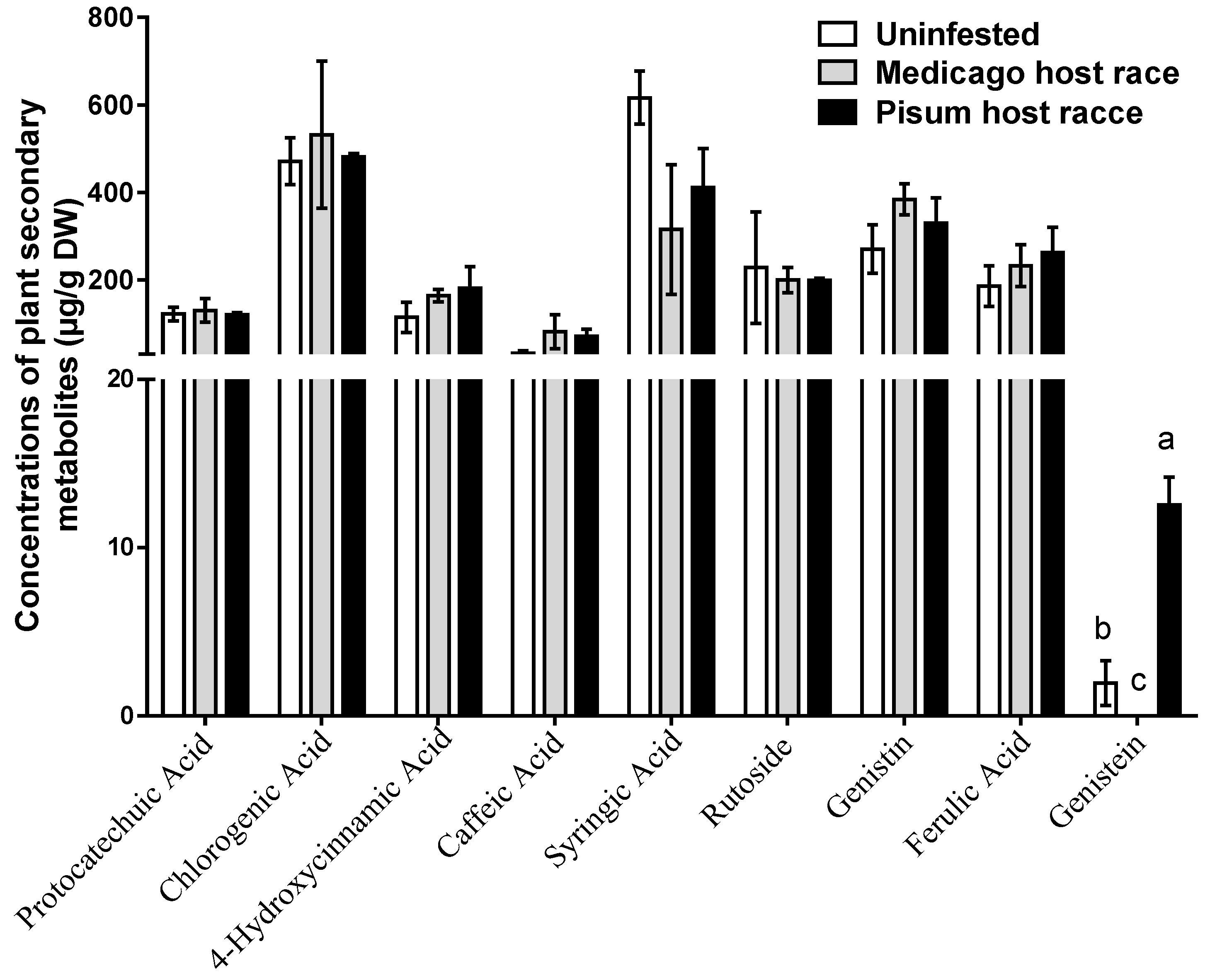
3.2. Concentrations of Foliar Phenolics in M. sativa
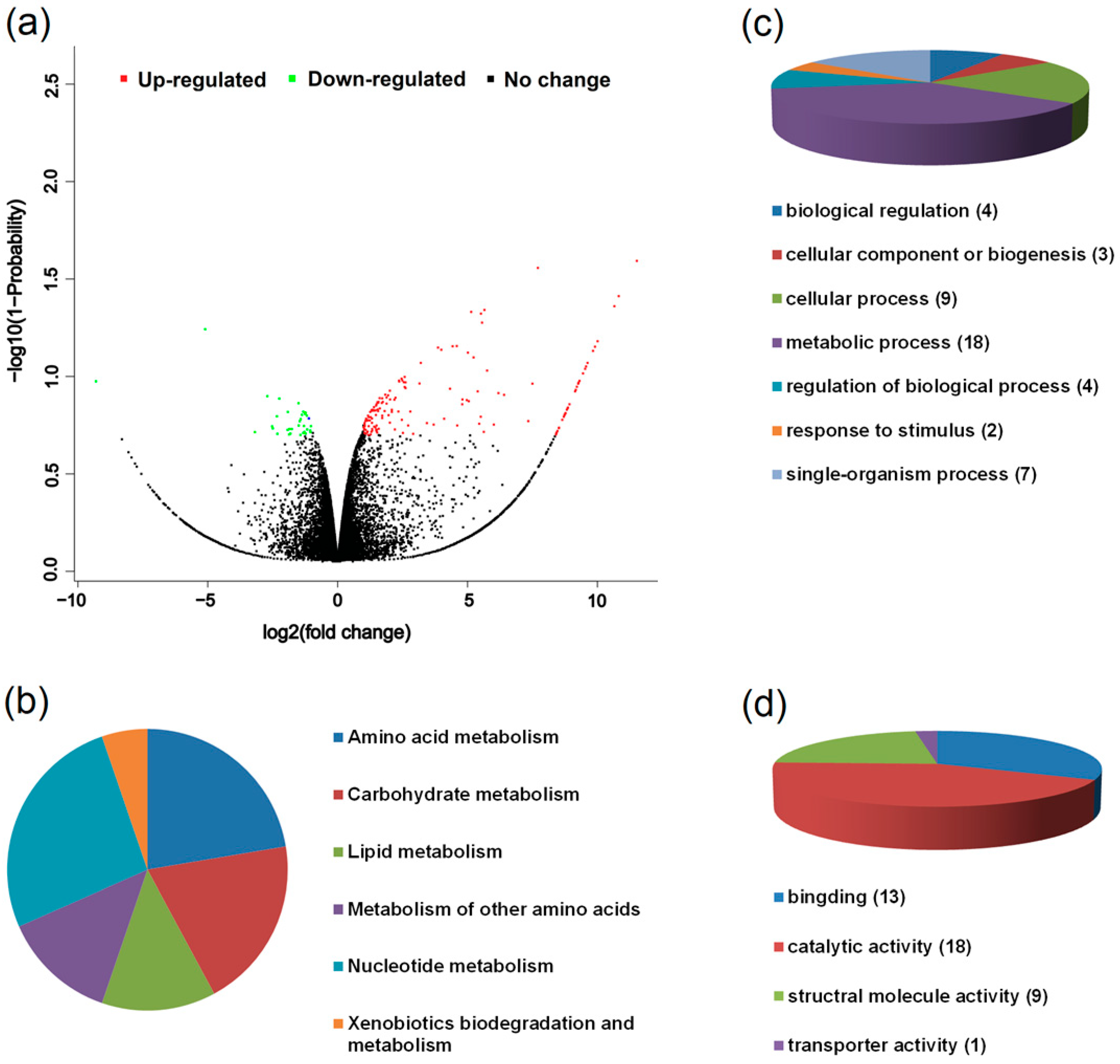
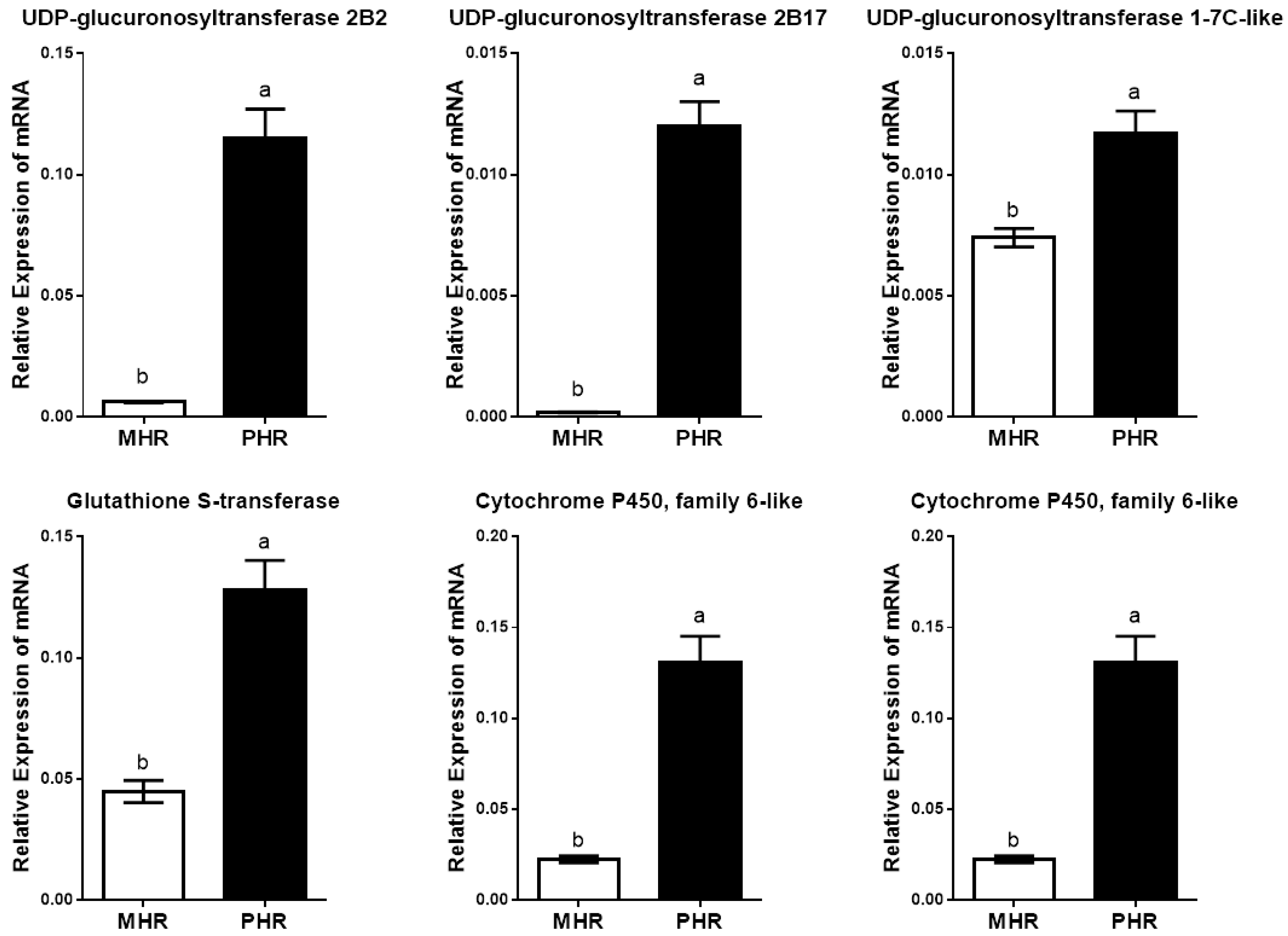
3.3. Transcriptomic Characteristics of the Two Host Races of A. pisum on M. sativa
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dres, M.; Mallet, J. Host races in plant-feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. Lond. B 2002, 357, 471–492. [Google Scholar] [CrossRef]
- Peccoud, J.; Simon, J.-C. The pea aphid complex as a model of ecological speciation. Ecol. Entomol. 2010, 35, 119–130. [Google Scholar] [CrossRef]
- Carletto, J.; Lombaert, E.; Chavigny, P.; Brevault, T.; Lapchin, L.; Vanlerberghe-Masutti, F. Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol. Ecol. 2009, 18, 2198–2212. [Google Scholar] [CrossRef] [PubMed]
- Peccoud, J.; Ollivier, A.; Plantegenest, M.; Simon, J.C. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc. Natl. Acad. Sci. USA 2009, 106, 7495–7500. [Google Scholar] [CrossRef] [PubMed]
- Peccoud, J.; Mahéo, F.; de la Huerta, M.; Laurence, C.; Simon, J.-C.; Leather, S.R. Genetic characterisation of new host-specialised biotypes and novel associations with bacterial symbionts in the pea aphid complex. Insect Conserv. Divers. 2015, 8, 484–492. [Google Scholar] [CrossRef]
- Ferrari, J.; Godfray, H.C.J.; Faulconbridge, A.S.; Prior, K.; Via, S. Population differentiation and genetic variation in host choice among pea aphids from eight host plant genera. Evolution 2006, 60, 1574–1584. [Google Scholar] [CrossRef]
- Eyres, I.; Jaquiery, J.; Sugio, A.; Duvaux, L.; Gharbi, K.; Zhou, J.-J.; Legeai, F.; Nelson, M.; Simon, J.-C.; Smadja, C.M.; et al. Differential gene expression according to race and host plant in the pea aphid. Mol. Ecol. 2016, 25, 4197–4215. [Google Scholar] [CrossRef]
- Via, S. Reproductive isolation between sympatric races of pea aphids. I. Gene flow restriction and habitat choice. Evolution 1999, 53, 1446–1457. [Google Scholar] [CrossRef]
- Blackman, R.L.; Eastop, V.F.; Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops: An Identification and Information Guide, 2nd ed.; Wiley: New York, NY, USA, 2000; pp. 1–466. [Google Scholar]
- Hogenhout, S.A.; Bos, J.I. Effector proteins that modulate plant—Insect interactions. Curr. Opin. Plant Biol. 2011, 14, 422–428. [Google Scholar] [CrossRef]
- Jaouannet, M.; Rodriguez, P.A.; Thorpe, P.; Lenoir, C.J.; MacLeod, R.; Escudero-Martinez, C.; Bos, J.I. Plant immunity in plant-aphid interactions. Front. Plant Sci. 2014, 5, 663. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Dai, H.; Zhang, Y.; Chandrasekar, R.; Luo, L.; Hiromasa, Y.; Sheng, C.; Peng, G.; Chen, S.; Tomich, J.M.; et al. Armet is an effector protein mediating aphid-plant interactions. FASEB J. 2015, 29, 2032–2045. [Google Scholar] [CrossRef] [PubMed]
- Elzinga, D.A.; De Vos, M.; Jander, G. Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol. Plant Microbe Interact. 2014, 27, 747–756. [Google Scholar] [CrossRef] [PubMed]
- Atamian, H.S.; Chaudhary, R.; Dal Cin, V.; Bao, E.; Girke, T.; Kaloshian, I. In Planta Expression or Delivery of Potato Aphid Macrosiphum euphorbiae Effectors Me10 and Me23 Enhances Aphid Fecundity. Mol. Plant Microbe Interact. 2013, 26, 67–74. [Google Scholar] [CrossRef]
- Sanchez-Arcos, C.; Reichelt, M.; Gershenzon, J.; Kunert, G. Modulation of Legume Defense Signaling Pathways by Native and Non-native Pea Aphid Clones. Front. Plant Sci. 2016, 7, 1872. [Google Scholar] [CrossRef]
- Egan, S.P.; Ott, J.R. Host plant quality and local adaptation determine the distribution of a gall-forming herbivore. Ecology 2007, 88, 2868–2879. [Google Scholar] [CrossRef]
- Schwarzkopf, A.; Rosenberger, D.; Niebergall, M.; Gershenzon, J.; Kunert, G. To feed or not to feed: Plant factors located in the epidermis, mesophyll, and sieve elements influence pea aphid’s ability to feed on legume species. PLoS ONE 2013, 8, e75298. [Google Scholar] [CrossRef]
- Züst, T.; Agrawal, A.A. Mechanisms and evolution of plant resistance to aphids. Nat. Plants 2016, 2, 15206. [Google Scholar] [CrossRef]
- Ahmad, S.; Veyrat, N.; Gordon-Weeks, R.; Zhang, Y.; Martin, J.; Smart, L.; Glauser, G.; Erb, M.; Flors, V.; Frey, M.; et al. Benzoxazinoid Metabolites Regulate Innate Immunity against Aphids and Fungi in Maize. Plant Physiol. 2011, 157, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Saheed, S.A.; Cierlik, I.; Larsson, K.A.; Delp, G.; Bradley, G.; Jonsson, L.M.; Botha, C.E. Stronger induction of callose deposition in barley by Russian wheat aphid than bird cherry-oat aphid is not associated with differences in callose synthase or beta-1,3-glucanase transcript abundance. Physiol. Plant. 2009, 135, 150–161. [Google Scholar] [CrossRef]
- Yan, H.Y.; Guo, H.G.; Sun, Y.C.; Ge, F. Plant phenolics mediated bottom-up effects of elevated CO2 on Acyrthosiphon pisum and its parasitoid Aphidius avenae. Insect Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Heidel-Fischer, H.M.; Vogel, H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect Sci. 2015, 8, 8–14. [Google Scholar] [CrossRef]
- Ramsey, J.S.; Elzinga, D.A.; Sarkar, P.; Xin, Y.-R.; Ghanim, M.; Jander, G. Adaptation to Nicotine Feeding in Myzus persicae. J. Chem. Ecol. 2014, 40, 869–877. [Google Scholar] [CrossRef]
- Tjallingii, W.F.; Esch, T.H. Fine-structure of aphid stylet routes in plant-tissues in correlation with epg signals. Physiol. Entomol. 1993, 18, 317–328. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Gillespie, K.M. Estimation of total phenolic content and other oxidation substrates in plant tissues using Folin-Ciocalteu reagent. Nat. Protoc. 2007, 2, 875–877. [Google Scholar] [CrossRef] [PubMed]
- Febvay, G.; Rahbe, Y.; Rynkiewicz, M.; Guillaud, J.; Bonnot, G. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, acyrthosiphon pisum, reared on different diets. J. Exp. Biol. 1999, 202, 2639–2652. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Tarazona, S.; Garcia-Alcalde, F.; Dopazo, J.; Ferrer, A.; Conesa, A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011, 21, 2213–2223. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ji, R.; Wang, Y.; Cheng, Y.; Zhang, M.; Zhang, H.B.; Zhu, L.; Fang, J.; Zhu-Salzman, K. Transcriptome Analysis of Green Peach Aphid (Myzus persicae): Insight into Developmental Regulation and Inter-Species Divergence. Front. Plant Sci. 2016, 7, 1562. [Google Scholar] [CrossRef]
- Wang, W.; Luo, L.; Lu, H.; Chen, S.; Kang, L.; Cui, F. Angiotensin-converting enzymes modulate aphid-plant interactions. Sci. Rep. 2015, 5, 8885. [Google Scholar] [CrossRef] [PubMed]
- Guerrieri, E.; Digilio, M.C. Aphid-plant interactions: A review. J. Plant Interact. 2008, 3, 223–232. [Google Scholar] [CrossRef]
- Oliver, K.M.; Degnan, P.H.; Burke, G.R.; Moran, N.A. Facultative Symbionts in Aphids and the Horizontal Transfer of Ecologically Important Traits. Annu. Rev. Entomol. 2010, 55, 247–266. [Google Scholar] [CrossRef]
- Kawecki, T.J.; Ebert, D. Conceptual issues in local adaptation. Ecol. Lett. 2004, 7, 1225–1241. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Stam, R.; Warbroek, T.; Bos, J.I.B. Mp10 and Mp42 from the Aphid Species Myzus persicae Trigger Plant Defenses in Nicotiana benthamiana Through Different Activities. Mol. Plant Microbe Interact. 2014, 27, 30–39. [Google Scholar] [CrossRef]
- Cui, N.; Lu, H.; Wang, T.; Zhang, W.; Kang, L.; Cui, F. Armet, an aphid effector protein, induces pathogen resistance in plants by promoting the accumulation of salicylic acid. Philos. Trans. R. Soc. B 2019, 374, 20180314. [Google Scholar] [CrossRef]
- Tsuchida, T.; Koga, R.; Shibao, H.; Matsumoto, T.; Fukatsu, T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol. Ecol. 2002, 11, 2123–2135. [Google Scholar] [CrossRef]
- Leonardo, T.E.; Muiru, G.T. Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc. R. Soc. Lond. B 2003, 270, S209–S212. [Google Scholar] [CrossRef]
- Simon, J.C.; Carre, S.; Boutin, M.; Prunier-Leterme, N.; Sabater-Munoz, B.; Latorre, A.; Bournoville, R. Host-based divergence in populations of the pea aphid: Insights from nuclear markers and the prevalence of facultative symbionts. Proc. R. Soc. Lond. B 2003, 270, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, J.; Darby, A.C.; Daniell, T.J.; Godfray, H.C.J.; Douglas, A.E. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol. Entomol. 2004, 29, 60–65. [Google Scholar] [CrossRef]
- Simon, J.-C.; Mieuzet, L.; Frantz, A.; Calcagno, V.; Plantegenest, M. Complex trait differentiation between host-populations of the pea aphid Acyrthosiphon pisum (Harris): Implications for the evolution of ecological specialisation. Biol. J. Linn. Soc. 2009, 97, 718–727. [Google Scholar] [CrossRef]
- Leonardo, T.E. Removal of a specialization-associated symbiont does not affect aphid fitness. Ecol. Lett. 2004, 7, 461–468. [Google Scholar] [CrossRef]
- Tsuchida, T.; Koga, R.; Fukatsu, T. Host plant specialization governed by facultative symbiont. Science 2004, 303, 1989. [Google Scholar] [CrossRef]
- Ferrari, J.; Scarborough, C.L.; Godfray, H.C.J. Genetic variation in the effect of a facultative symbiont on host-plant use by pea aphids. Oecologia 2007, 153, 323–329. [Google Scholar] [CrossRef]
- Leszczynski, B.; Tjallingii, F.; Dixon, A.; Swiderski, R. Effect of methoxyphenols on grain aphid feeding behaviour. Entomol. Exp. Appl. 1995, 76, 157–162. [Google Scholar] [CrossRef]
- Lattanzio, V.; Arpaia, S.; Cardinali, A.; Di Venere, D.; Linsalata, V. Role of endogenous flavonoids in resistance mechanism of Vigna to aphids. J. Agric. Food Chem. 2000, 48, 5316–5320. [Google Scholar] [CrossRef] [PubMed]
- Łukasik, I.; Goławska, S.; Wójcicka, A. Antioxidant defense mechanisms of cereal aphids based on ascorbate and ascorbate peroxidase. Biologia 2009, 64, 994–998. [Google Scholar] [CrossRef]
- Lukasik, I.; Golawska, S.; Wojcicka, A.; Golawski, A. Effect of host plants on antioxidant system of pea aphid Acyrthosiphon pisum. Bull. Insectol. 2011, 64, 153–158. [Google Scholar]
- Golawska, S.; Lukasik, I. Antifeedant activity of luteolin and genistein against the pea aphid, Acyrthosiphon pisum. J. Pest Sci. 2012, 85, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Han, Y.; Teng, W.; Li, Y.; Li, W. QTL underlying the resistance to soybean aphid (Aphis glycines Matsumura) through isoflavone-mediated antibiosis in soybean cultivar ‘Zhongdou 27’. Theor. Appl. Genet. 2011, 123, 1459–1465. [Google Scholar] [CrossRef]
- Yan, H.; Guo, H.; Yuan, E.; Sun, Y.; Ge, F. Elevated CO2 and O3 alter the feeding efficiency of Acyrthosiphon pisum and Aphis craccivora via changes in foliar secondary metabolites. Sci. Rep. 2018, 8, 9964. [Google Scholar] [CrossRef]
- Cabrera, H.M.; Muñoz, O.; Zúñiga, G.E.; Corcuera, L.J.; Argandoña, V.H. Changes in ferulic acid and lipid content in aphid-infested barley. Phytochemistry 1995, 39, 1023–1026. [Google Scholar] [CrossRef]
- Bentivenha, J.P.F.; Canassa, V.F.; Baldin, E.L.L.; Borguini, M.G.; Lima, G.P.P.; Lourencao, A.L. Role of the Rutin and Genistein Flavonoids in Soybean Resistance to Piezodorus guildinii (Hemiptera: Pentatomidae). Arthropod-Plant Interact. 2018, 12, 311–320. [Google Scholar] [CrossRef]
- Cui, L.; Yuan, H.; Wang, Q.; Wang, Q.; Rui, C. Sublethal effects of the novel cis-nitromethylene neonicotinoid cycloxaprid on the cotton aphid Aphis gossypii Glover (Hemiptera: Aphididae). Sci. Rep. 2018, 8, 8915. [Google Scholar] [CrossRef] [PubMed]
- Castaneda, L.E.; Figueroa, C.C.; Fuentes-Contreras, E.; Niemeyer, H.M.; Nespolo, R.F. Physiological approach to explain the ecological success of ‘superclones’ in aphids: Interplay between detoxification enzymes, metabolism and fitness. J. Insect Physiol. 2010, 56, 1058–1064. [Google Scholar] [CrossRef]
- Chrzanowski, G.; Leszczynski, B.; Czerniewicz, P.; Sytykiewicz, H.; Matok, H.; Krzyzanowski, R.; Sempruch, C. Effect of phenolic acids from black currant, sour cherry and walnut on grain aphid (Sitobion avenae F.) development. Crop Prot. 2012, 35, 71–77. [Google Scholar] [CrossRef]
- Francis, F.; Vanhaelen, N.; Haubruge, E. Glutathione S-transferases in the adaptation to plant secondary metabolites in the Myzus persicae aphid. Arch. Insect Biochem. Physiol. 2005, 58, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Mukanganyama, S.; Figueroa, C.C.; Hasler, J.A.; Niemeyer, H.M. Effects of DIMBOA on detoxification enzymes of the aphid Rhopalosiphum padi (Homoptera: Aphididae). J. Insect Physiol. 2003, 49, 223–229. [Google Scholar] [CrossRef]
| Medicago sativa | ||
|---|---|---|
| Parameters | Medicago Host Race | Pisum Host Race |
| Nonpenetration 1 | 47.00 ± 10.20 a | 86.29 ± 10.20 a |
| Pathway 2 | 148.75 ± 25.86 a | 175.79 ± 25.86 a |
| Total pd (potential drops) 3 | 14.12 ± 2.02 b | 26.66 ± 2.02 a |
| Time to first pd | 8.60 ± 3.79 a | 8.45 ± 4.83 a |
| Number of pds before first E1 | 65.09 ± 10.08 a | 96.43 ± 10.08 a |
| Total E1 (phloem salivation) 4 | 32.26 ± 7.98 a | 21.29 ± 7.98 a |
| Number of E1 | 4.85 ± 0.96 a | 4.57 ± 0.96 a |
| Time to first E1 | 100.64 ± 17.50 a | 125.90 ± 17.50 a |
| Total E2 (phloem ingestion) 5 | 231.70 ± 35.11 a | 123.25 ± 35.11 b |
| Number of E2 | 2.73 ± 0.51 a | 1.79a ± 0.35 a |
| Time to first E2 | 168.96 ± 30.98 b | 248.06 ± 30.98 a |
| Total G (xylem ingestion) 6 | 6.16 ± 3.43 a | 21.37 ± 8.85 a |
| Gene ID | Putative Function | log2 Fold Change 1 | Probability 2 |
|---|---|---|---|
| LOC100166729 | UDP-glucuronosyltransferase 2B2 | 4.42 × up | 0.93 |
| LOC100169601 | UDP-glucuronosyltransferase 2B17 | 6.40 × up | 0.88 |
| LOC100159691 | UDP-glucuronosyltransferase 1-7C-like | 2.58 × up | 0.89 |
| LOC100570856 | glutathione S-transferase | 1.74 × up | 0.87 |
| LOC100572007 | cytochrome P450, family 6-like | 2.61 × up | 0.89 |
| LOC100569567 | cytochrome P450, family 6 | 2.47 × up | 0.83 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, E.; Yan, H.; Gao, J.; Guo, H.; Ge, F.; Sun, Y. Increases in Genistein in Medicago sativa Confer Resistance against the Pisum Host Race of Acyrthosiphon pisum. Insects 2019, 10, 97. https://doi.org/10.3390/insects10040097
Yuan E, Yan H, Gao J, Guo H, Ge F, Sun Y. Increases in Genistein in Medicago sativa Confer Resistance against the Pisum Host Race of Acyrthosiphon pisum. Insects. 2019; 10(4):97. https://doi.org/10.3390/insects10040097
Chicago/Turabian StyleYuan, Erliang, Hongyu Yan, Jing Gao, Huijuan Guo, Feng Ge, and Yucheng Sun. 2019. "Increases in Genistein in Medicago sativa Confer Resistance against the Pisum Host Race of Acyrthosiphon pisum" Insects 10, no. 4: 97. https://doi.org/10.3390/insects10040097
APA StyleYuan, E., Yan, H., Gao, J., Guo, H., Ge, F., & Sun, Y. (2019). Increases in Genistein in Medicago sativa Confer Resistance against the Pisum Host Race of Acyrthosiphon pisum. Insects, 10(4), 97. https://doi.org/10.3390/insects10040097





