Life on the Edge: Ecological Genetics of a High Arctic Insect Species and Its Circumpolar Counterpart
Abstract
1. Introduction
2. Materials and Methods
2.1. Aphid Sampling
2.2. Microsatellite Isolation and Aphid Genotyping
2.3. Genetic Analysis
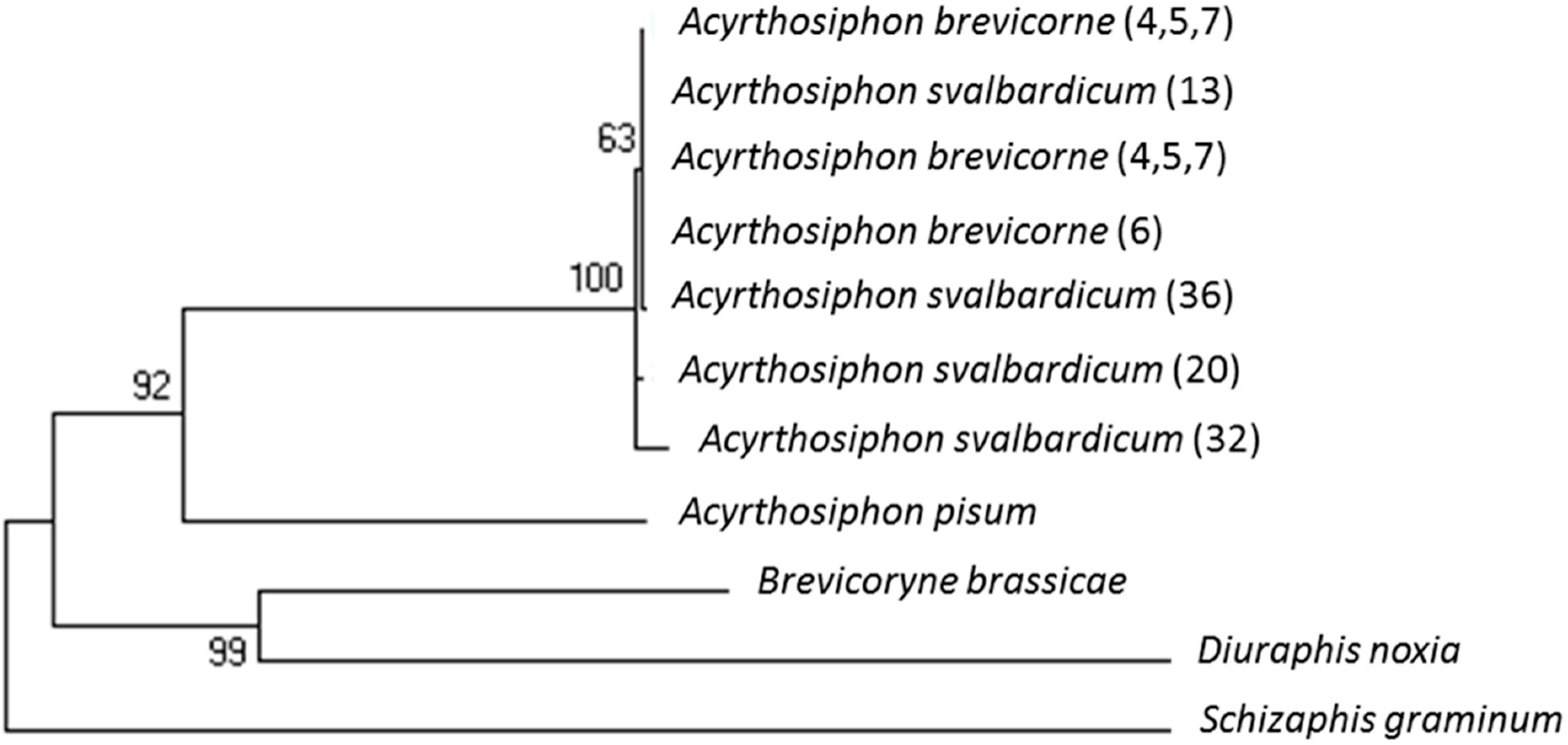
2.4. Molecular Divergence between A. brevicorne and A. svalbardicum
3. Results
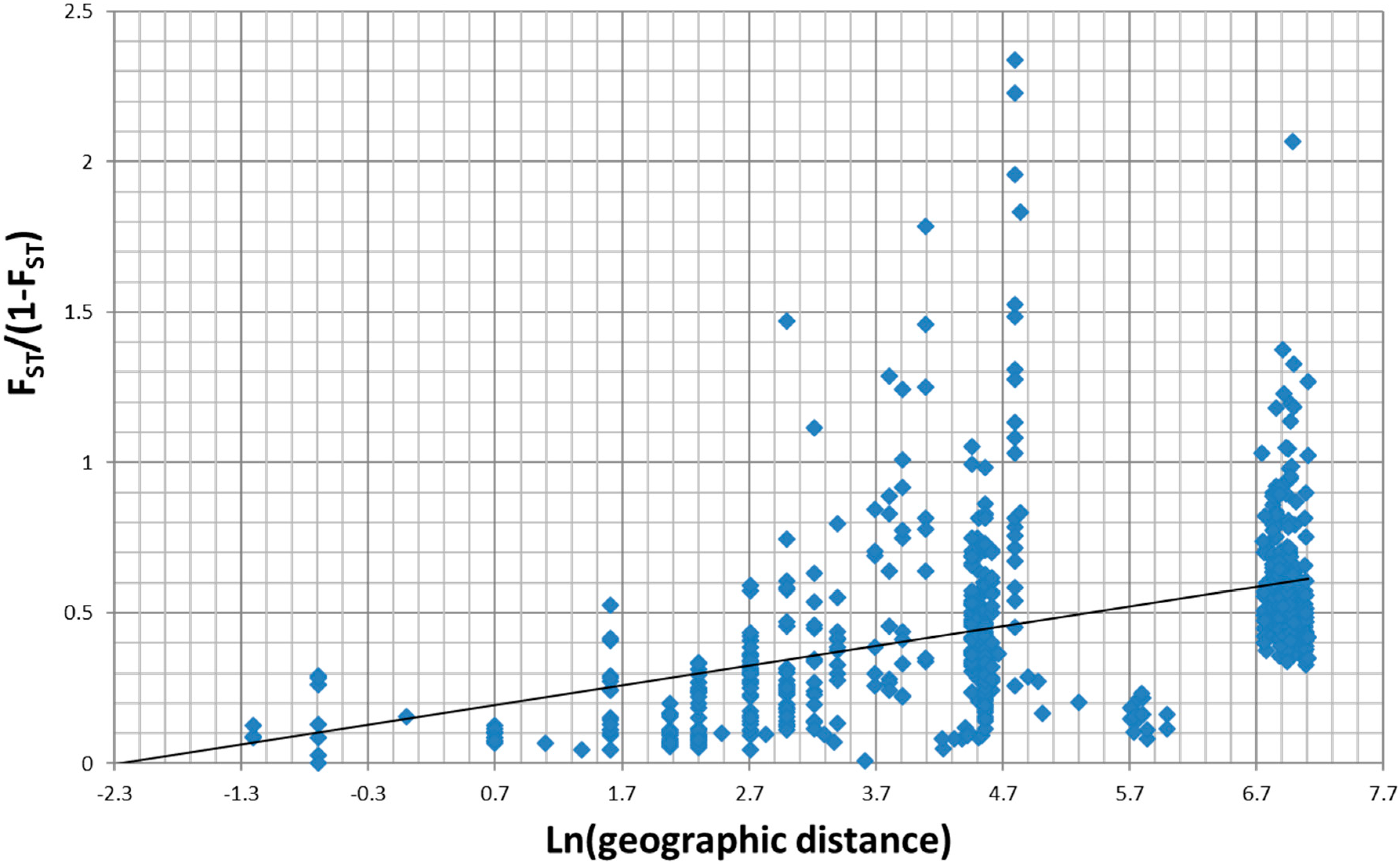
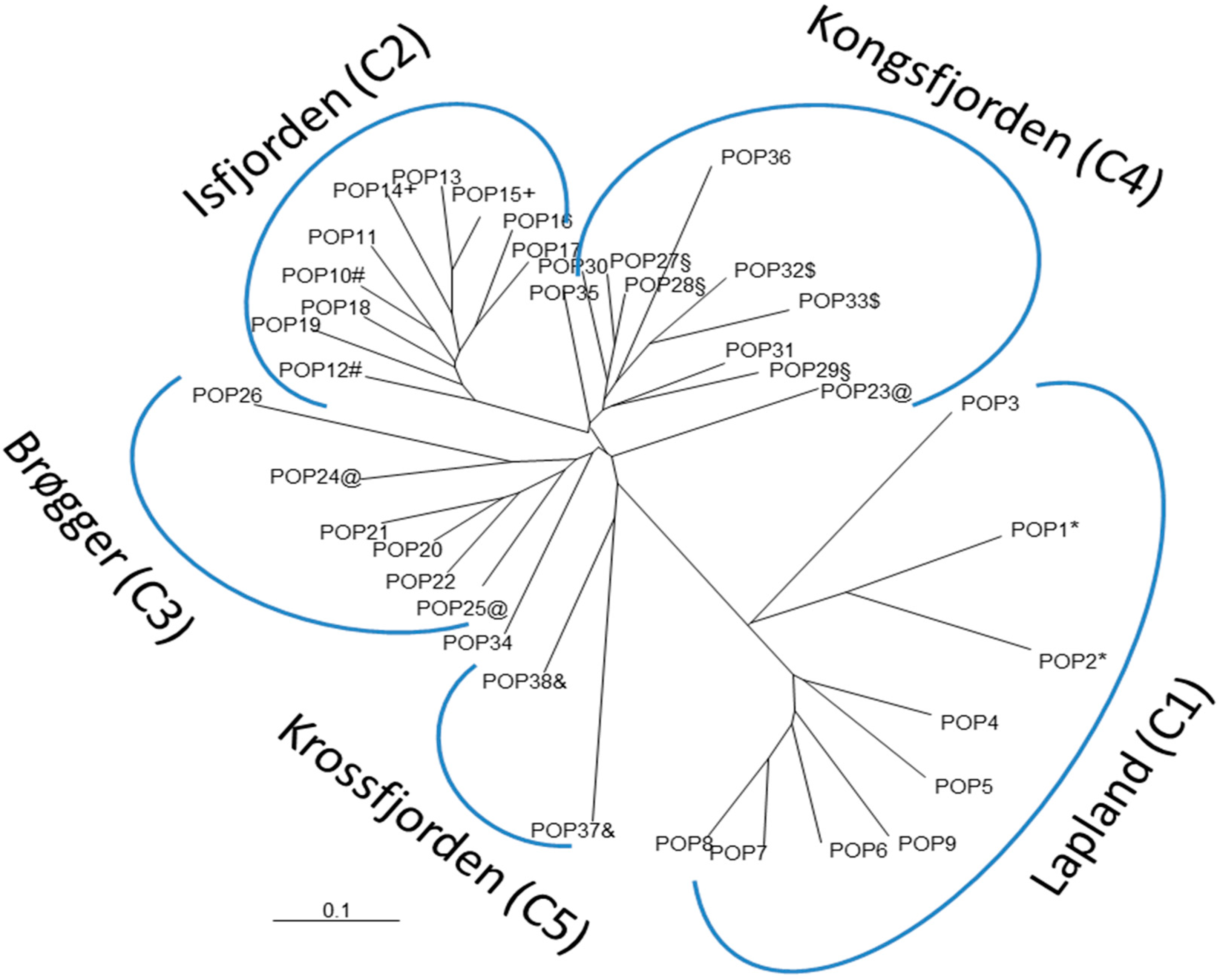
3.1. Overall Patterns of Geographic Distribution and Population Differentiation
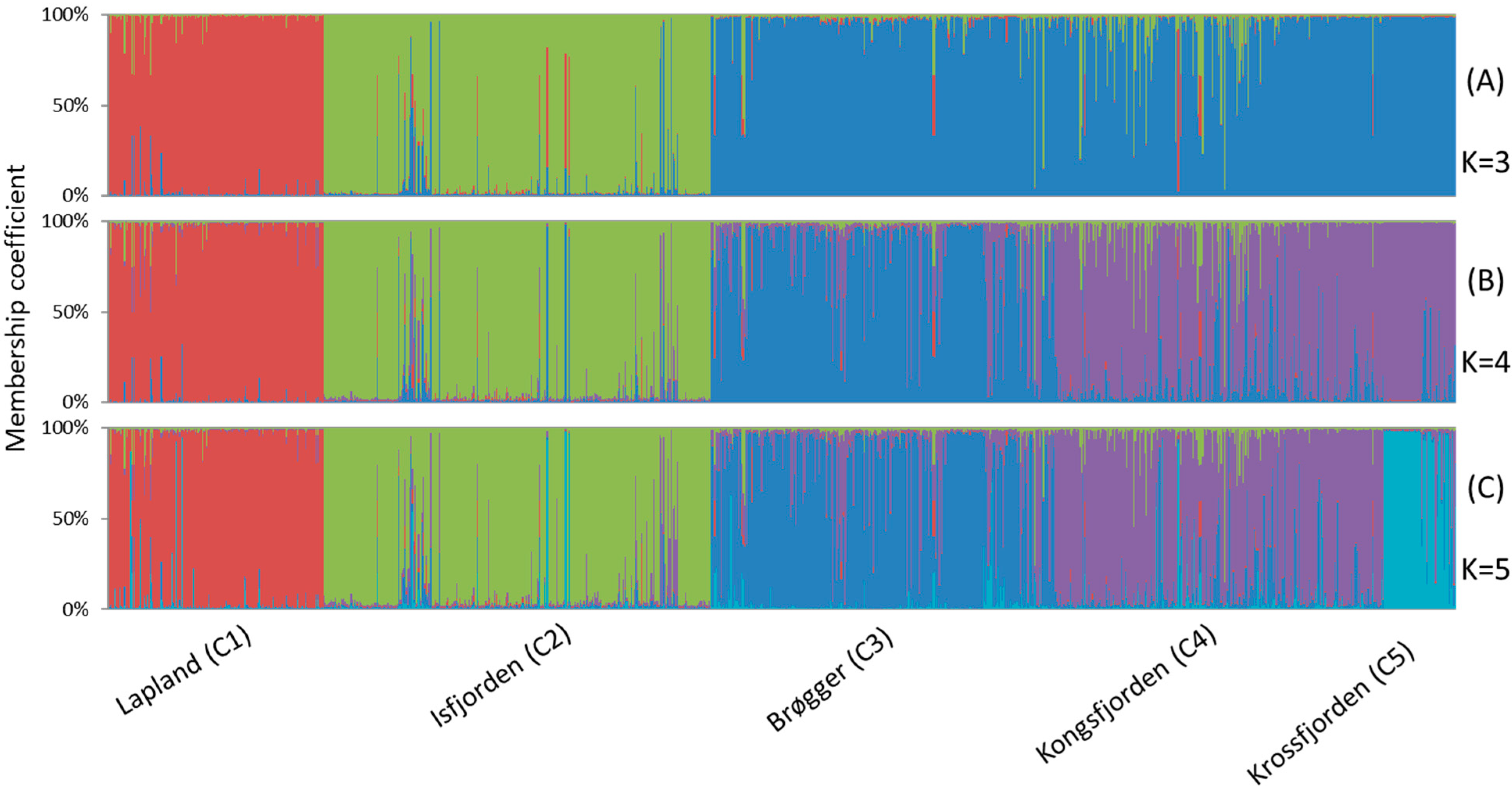
3.2. Bayesian Analysis of Population Structure
3.3. Within-Population Structure
3.4. Within-Population Structure
4. Discussion
4.1. Distribution and Range Limit of A. svalbardicum in Spitsbergen
4.2. Divergence between A. svalbardicum and A. brevicorne
4.3. Signatures of Inbreeding in Populations of A. brevicorne and A. svalbardicum?
4.4. Strong Spatial Differences in Population Structure of A. svalbardicum
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gaston, K.J. Geographic range limits of species. Proc. Roy. Soc. B-Biol. Sci. 2009, 276, 1391–1393. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Blows, M.W. Species borders—ecological and evolutionary perspectives. Trends Ecol. Evol. 1994, 9, 223–227. [Google Scholar] [CrossRef]
- Facon, B.; Genton, B.J.; Shykoff, J.; Jarne, P.; Estoup, A.; David, P. A general eco-evolutionary framework for understanding bioinvasions. Trends Ecol. Evol. 2006, 21, 130–135. [Google Scholar] [CrossRef]
- Strathdee, A.T.; Bale, J.S. Life on the edge: Insect ecology in arctic environments. Annu. Rev. Entomol. 1998, 43, 85–106. [Google Scholar] [CrossRef]
- Coulson, S.J.; Convey, P.; Aakra, K.; Aarvik, L.; Avila-Jimenez, M.L.; Babenko, A.; Biersma, E.M.; Bostrom, S.; Brittain, J.E.; Carlsson, A.M.; et al. The terrestrial and freshwater invertebrate biodiversity of the archipelagoes of the Barents sea, Svalbard, Franz Josef Land and Novaya Zemlya. Soil Biol. Bioch. 2014, 68, 440–470. [Google Scholar] [CrossRef]
- Alsos, I.G.; Eidesen, P.B.; Ehrich, D.; Skrede, I.; Westergaard, K.; Jacobsen, G.H.; Landvik, J.Y.; Taberlet, P.; Brochmann, C. Frequent long-distance plant colonization in the changing arctic. Science 2007, 316, 1606–1609. [Google Scholar] [CrossRef]
- Coulson, S.; Refseth, D. The terrestrial and freshwater invertebrate fauna of Svalbard (and Jan Mayen). A Cat. Terr. Mar. Anim. Svalbard 2004, 57–122. [Google Scholar]
- Strathdee, A.T.; Bale, J.S.; Block, W.C.; Webb, N.R.; Hodkinson, I.D.; Coulson, S.J. Extreme adaptive life-cycle in a high arctic aphid, Acyrthosiphon svalbardicum. Ecol. Entomol. 1993, 18, 254–258. [Google Scholar] [CrossRef]
- Hullé, M.; Bonhomme, J.; Maurice, D.; Simon, J.-C. Is the life cycle of high arctic aphids adapted to climate change? Polar Biol. 2008, 31, 1037–1042. [Google Scholar] [CrossRef]
- Strathdee, A.T.; Bale, J.S. Factors limiting the distribution of Acyrthosiphon svalbardicum (Hemiptera, Aphididae) on Spitsbergen. Polar Biol. 1995, 15, 375–380. [Google Scholar] [CrossRef]
- Hodkinson, I.; Coulson, S.; Bird, J.; Webb, N.R. Discovery of the rare alate morph of Acyrthosiphon svalbardicum Heikinheimo (Homoptera; Aphididae): Description and implications for species ecology. Norwegian J. Entomol. 2002, 49, 77–88. [Google Scholar]
- Simon, J.C.; Bonhomme, J.; Blackman, R.L.; Hullé, M. Winged morph of the high arctic aphid Acyrthosiphon svalbardicum (Hemiptera: Aphididae): Abundance, reproductive status, and ecological significance. Can. Entomol. 2008, 140, 385–387. [Google Scholar] [CrossRef]
- Eastop, V.F. Key for the identification of Acyrthosiphon (hemiptera: Aphididae). Bull. Brit. Mus. (Nat. Hist.) Entomol 1971, 26. [Google Scholar]
- Hille Ris Lambers, D. Additions to the aphid fauna of Greenland. Meddelelser om Grønland 1960, 159, 1–18. [Google Scholar]
- Heikinheimo, O. Notes on the arthropod fauna of Spitsbergen. II: 10. The aphid fauna of Spitsbergen. Ann. Entomol. Fenn. 1968, 34, 82–93. [Google Scholar]
- Strathdee, A.T.; Bale, J.S. Life cycle and morph production in the arctic aphid Acyrthosiphon brevicorne. Polar Biol. 1996, 16, 293–300. [Google Scholar] [CrossRef]
- Malausa, T.; Gilles, A.; Meglecz, E.; Blanquart, H.; Duthoy, S.; Costedoat, C.; Dubut, V.; Pech, N.; Castagnone-sereno, P.; Delye, C.; et al. High-throughput microsatellite isolation through 454 GS-FLX titanium pyrosequencing of enriched DNA libraries. Mol. Ecol. Res. 2011, 11, 638–644. [Google Scholar] [CrossRef]
- Meglécz, E.; Costedoat, C.; Dubut, V.; Gilles, A.; Malausa, T.; Pech, N.; Martin, J.-F. Qdd: A user-friendly program to select microsatellite markers and design primers from large sequencing projects. Bioinformatics 2009, 26, 403–404. [Google Scholar] [CrossRef]
- Boutin-Ganache, I.; Raposo, M.; Raymond, M.; Deschepper, C.F. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. BioTechniques 2001, 31, 24–26. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. Genepop (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. Genetix 4.05, Logiciel Sous Windows TM Pour la Génétique des Populations. Available online: http://kimura.univ-montp2.fr/genetix/ (accessed on 14 June 2014).
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Soft. 2015, 67, 48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Goudet, J. Fstat, A Program to Estimate and Test Gene Diversities and Fixation Indices. Available online: http://www2.unil.ch/popgen/softwares/fstat.htm (accessed on 14 June 2014).
- Jin, L.; Chakraborty, R. Estimation of genetic distance and coefficient of gene diversity from single-probe multilocus DNA fingerprinting data. Mol. Biol. Evol. 1994, 11, 120–127. [Google Scholar]
- Rousset, F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 1997, 145, 1219–1228. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Chen, R.; Jiang, L.-Y.; Liu, L.; Liu, Q.-H.; Wen, J.; Zhang, R.-L.; Li, X.-Y.; Wang, Y.; Lei, F.-M.; Qiao, G.-X. The gnd gene of Buchnera as a new, effective DNA barcode for aphid identification. Syst. Entomol. 2013, 38, 615–625. [Google Scholar] [CrossRef]
- Hall, T.A. Bioedit: A user-friendly biological sequence alignment editor and analysis program for windows 95⁄98⁄NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal w: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. Mega4: Molecular evolutionary genetics analysis (Mega) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Ávila-Jiménez, M.L.; Coulson, S.J. Can snow depth be used to predict the distribution of the high arctic aphid Acyrthosiphon svalbardicum (Hemiptera: Aphididae) on Spitsbergen? BMC Ecol. 2011, 11, 25. [Google Scholar] [CrossRef]
- Landvik, J.Y.; Brook, E.J.; Gualtieri, L.; Raisbeck, G.; Salvigsen, O.; Yiou, F. Northwest Svalbard during the last glaciation: Ice-free areas existed. Geology 2003, 31, 905–908. [Google Scholar] [CrossRef]
- Brochmann, C.; Gabrielsen, T.M.; Nordal, I.; Landvik, J.Y.; Elven, R. Glacial survival or tabula rasa? The history of north Atlantic biota revisited. Taxon 2003, 52, 417–450. [Google Scholar]
- Westergaard, K.B.; Alsos, I.G.; Popp, M.; Engelskjon, T.; Flatberg, K.I.; Brochmann, C. Glacial survival may matter after all: Nunatak signatures in the rare european populations of two west-arctic species. Mol. Ecol. 2011, 20, 376–393. [Google Scholar] [CrossRef]
- Skrede, I.; Eidesen, P.B.; Portela, R.P.; Brochmann, C. Refugia, differentiation and postglacial migration in arctic-alpine Eurasia, exemplified by the mountain avens (Dryas octopetala L.). Mol. Ecol. 2006, 15, 1827–1840. [Google Scholar] [CrossRef]
- Coeur d’acier, A.; Cruaud, A.; Artige, E.; Genson, G.; Clamens, A.-L.; Pierre, E.; Hudaverdian, S.; Simon, J.-C.; Jousselin, E.; Rasplus, J.-Y. DNA barcoding and the associated phylaphidb@se website for the identification of European aphids (Insecta: Hemiptera: Aphididae). PLoS ONE 2014, 9, e97620. [Google Scholar]
- Ding, M.H.; Wang, S.J.; Sun, W.J. Decadal climate change in Ny-Alesund, Svalbard, a representative area of the arctic. Condens. Matter 2018, 3. [Google Scholar] [CrossRef]
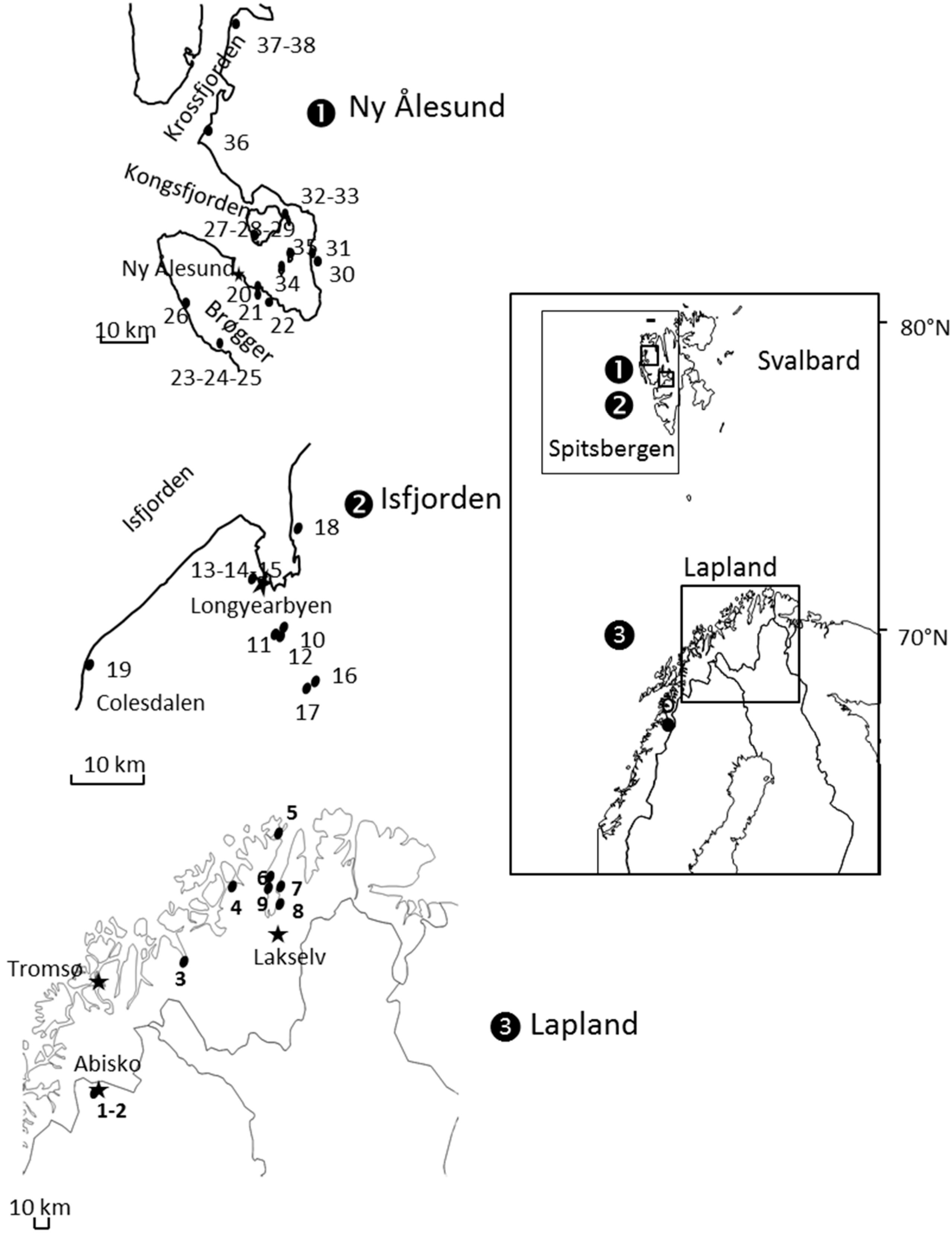
| Species (Zone) | Sample ID | Population | Area | Date | Latitude | Longitude |
|---|---|---|---|---|---|---|
| A. brevicorne (Lapland) | POP1 | Abisko1 | Abisko | 29/06/2006 | 68°21′127 | 18°49′324 |
| POP2 | Abisko2 | Abisko | 18/06/2010 | 68°21′128 | 18°49′325 | |
| POP3 | Gild_01 | Gildetun | 29/06/2012 | 69°54′174 | 21°35′969 | |
| POP4 | Ham_Kva_01 | Kvalsund | 01/07/2012 | 70°29′811 | 23°58′695 | |
| POP5 | Honn_01 | Honninsvag | 02/07/2012 | 70°59′793 | 25°58′246 | |
| POP6 | Lak_01 | Trollholmsund | 01/07/2012 | 70°16′914 | 25°06′776 | |
| POP7 | Lak_Bor_01 | Borselv | 03/07/2012 | 70°19′482 | 25°24′383 | |
| POP8 | Lak_NE_01 | Lakselv-NE | 03/07/2012 | 70°14′815 | 25°25′197 | |
| POP9 | Lak_Sarv_01 | Sarvvesvuotna | 01/07/2012 | 70°17′251 | 25°06′411 | |
| A. svalbardicum (Isfjorden) | POP10 | Endalen1-04 | Isfjorden | 30/06/2004 | 78°11′269 | 15°45′938 |
| POP11 | Endalen2-04 | Isfjorden | 30/06/2004 | 78°11′069 | 15°44′288 | |
| POP12 | Endalen09 | Isfjorden | 14/07/2009 | 78°11′131 | 15°45′939 | |
| POP13 | LYB1-04 | Isfjorden | 01/07/2004 | 78°12′995 | 15°36′631 | |
| POP14 | LYB2-04 | Isfjorden | 01/07/2004 | 78°12′995 | 15°36′631 | |
| POP15 | LYB09 | Isfjorden | 22/06/2009 | 78°12′995 | 15°36′631 | |
| POP16 | AdventS1 | Isfjorden | 03/07/2004 | 78°10′146 | 16°06′522 | |
| POP17 | AdventS2 | Isfjorden | 03/07/2004 | 78°10′026 | 16°06′412 | |
| POP18 | AdventN | Isfjorden | 20/07/2005 | 78°15′702 | 15°40′153 | |
| POP19 | Colesdalen | Isfjorden | 19/07/2005 | 78°04′602 | 15°15′092 | |
| A. svalbardicum (Ny Ålesund) | POP20 | Gasebu | Brøgger | 06/07/2009 | 78°54′590 | 12°04′338 |
| POP21 | Midre | Brøgger | 07/07/2004 | 78°54′573 | 12°04′339 | |
| POP22 | Corbel | Brøgger | 18/07/2004 | 78°53′698 | 12°10′215 | |
| POP23 | Daerten04 | Brøgger | 13/07/2004 | 78°51′179 | 11°49′826 | |
| POP24 | Daerten06 | Brøgger | 09/07/2006 | 78°51′179 | 11°49′826 | |
| POP25 | Daerten09 | Brøgger | 03/07/2009 | 78°51′179 | 11°49′826 | |
| POP26 | Traudalen | Brøgger | 15/07/2004 | 78°53′234 | 11°33′579 | |
| POP27 | Blomstrand04 | Kongsfjorden | 08/07/2004 | 78°57′801 | 12°02′768 | |
| POP28 | Blomstrand06 | Kongsfjorden | 11/07/2006 | 78°57′801 | 12°02′768 | |
| POP29 | Blomstrand09 | Kongsfjorden | 08/07/2009 | 78°57′801 | 12°02′768 | |
| POP30 | OssianE | Kongsfjorden | 09/07/2004 | 78°55′599 | 12°20′623 | |
| POP31 | OssianW | Kongsfjorden | 20/07/2004 | 78°55′599 | 12°27′413 | |
| POP32 | Gerdoya04 | Kongsfjorden | 24/07/2004 | 78°59′233 | 12°16′438 | |
| POP33 | Gerdoya09 | Kongsfjorden | 07/07/2009 | 78°59′233 | 12°16′438 | |
| POP34 | Storholmen | Kongsfjorden | 06/07/2009 | 78°55′424 | 12°12′324 | |
| POP35 | Observatoire | Kongsfjorden | 07/07/2009 | 78°55′526 | 12°16′387 | |
| POP36 | Kapp Guissez | Kongsfjorden | 25/07/2004 | 79°05′374 | 11°47′319 | |
| POP37 | CampZoe04 | Krossfjorden | 21/07/2004 | 79°11′841 | 11°56′485 | |
| POP38 | CampZoe09 | Krossfjorden | 29/06/2009 | 79°11′841 | 11°56′485 |
| Locus | Sequence Forward | Sequence Reverse | Size Range (bp) | Hobs | Hexp |
|---|---|---|---|---|---|
| AS23 | GGCACTGCTCTCATTACGGT | TTTTTCTTCGTCATCCCTCG | 125–190 | 0.648 | 0.715 |
| AS24 | GTCTGATGATGCGCTTGAAA | GAACCCAAACGAGGTGAAAA | 155–190 | 0.593 | 0.571 |
| AS26 | AGTCCGGAGGATAACAACGA | TCACGACCGAACACCATAAA | 155–205 | 0.415 | 0.458 |
| AS29 | CACCAAAAAGTCGGGGTAGA | GCCGTTGTTGAAGACTATTTCC | 160–180 | 0.529 | 0.528 |
| AS37 | CGACGGGCGAGTACCTATTA | TTTCAAGTAAACCGCTTCGG | 160–195 | 0.334 | 0.363 |
| AS41 | ATCTACCGCCACCACTTACG | TCGTCGAGATGCTATTGCTG | 225–145 | 0.307 | 0.324 |
| AS43 | GAAAAACGAGAAAACGCGAC | AGTCCCTGATGCAAACAACC | 250–305 | 0.162 | 0.244 |
| AS45 | TGAACCTGCTCAACAGCAAC | CCATGTCCTGACTCATCACG | 255–300 | 0.430 | 0.443 |
| Cluster | C1 | ||||
|---|---|---|---|---|---|
| C1 | 0 | C2 | |||
| C2 | 0.311 | 0 | C3 | ||
| C3 | 0.226 | 0.196 | 0 | C4 | |
| C4 | 0.281 | 0.185 | 0.097 | 0 | C5 |
| C5 | 0.277 | 0.398 | 0.216 | 0.232 | 0 |
| Species/Zone | Sample ID | Size | G/N | Hobs | Hexp | Allele Richness | Hetdef (p-val) | HetXs (p-val) | FIS | % LD |
|---|---|---|---|---|---|---|---|---|---|---|
| A. brevicorne (Lapland) | POP1 | 20 | 0.94 | 0.402 | 0.650 | 5.375 | 0.0000 | 1.0000 | 0.405 * | 25 |
| POP2 | 15 | 1.00 | 0.423 | 0.576 | 3.75 | 0.0000 | 1.0000 | 0.302 * | 7 | |
| POP3 | 9 | 1.00 | 0.453 | 0.409 | 2.75 | 0.3626 | 0.6374 | −0.048 | 0 | |
| POP4 | 19 | 1.00 | 0.449 | 0.486 | 4.125 | 0.0150 | 0.9850 | 0.108 * | 7 | |
| POP5 | 20 | 1.00 | 0.558 | 0.517 | 4.000 | 0.5875 | 0.4125 | −0.050 | 10 | |
| POP6 | 23 | 0.95 | 0.518 | 0.497 | 4.125 | 0.3210 | 0.6790 | −0.018 | 7 | |
| POP7 | 25 | 1.00 | 0.569 | 0.612 | 4.125 | 0.0152 | 0.9848 | 0.096 | 14 | |
| POP8 | 23 | 1.00 | 0.581 | 0.554 | 4.375 | 0.6860 | 0.3140 | −0.026 | 11 | |
| POP9 | 22 | 0.74 | 0.438 | 0.486 | 3.625 | 0.0006 | 0.9994 | 0.122 * | 57 | |
| A. svalbardicum (Isfjorden) | POP10 | 30 | 0.92 | 0.397 | 0.368 | 2.5 | 0.8958 | 0.1042 | −0.063 | 0 |
| POP11 | 29 | 1.00 | 0.300 | 0.286 | 2.375 | 0.7518 | 0.2482 | −0.027 | 0 | |
| POP12 | 35 | 0.85 | 0.433 | 0.458 | 3.125 | 0.0783 | 0.9217 | 0.068 | 43 | |
| POP13 | 28 | 0.82 | 0.432 | 0.394 | 2.75 | 0.5751 | 0.4249 | −0.050 | 20 | |
| POP14 | 30 | 1.00 | 0.427 | 0.407 | 3.000 | 0.4206 | 0.5794 | −0.007 | 0 | |
| POP15 | 23 | 0.96 | 0.399 | 0.407 | 3.375 | 0.0088 | 0.9912 | 0.028 | 29 | |
| POP16 | 30 | 1.00 | 0.372 | 0.482 | 3.375 | 0.0000 | 1.0000 | 0.246 * | 21 | |
| POP17 | 30 | 1.00 | 0.432 | 0.445 | 3.125 | 0.0409 | 0.9591 | 0.048 | 5 | |
| POP18 | 36 | 1.00 | 0.392 | 0.412 | 3.25 | 0.0023 | 0.9977 | 0.065 | 11 | |
| POP19 | 42 | 0.80 | 0.341 | 0.336 | 2.875 | 0.6112 | 0.3888 | −0.003 | 40 | |
| A. svalbardicum (Ny Ålesund) | POP20 | 30 | 1.00 | 0.430 | 0.470 | 3.000 | 0.0054 | 0.9946 | 0.102 * | 18 |
| POP21 | 30 | 1.00 | 0.531 | 0.475 | 3.125 | 0.9796 | 0.0204 | −0.102 | 14 | |
| POP22 | 24 | 0.96 | 0.490 | 0.447 | 2.875 | 0.6763 | 0.3237 | −0.073 | 4 | |
| POP23 | 30 | 0.90 | 0.409 | 0.429 | 2.25 | 0.2237 | 0.7763 | 0.062 | 14 | |
| POP24 | 33 | 0.97 | 0.440 | 0.435 | 3.125 | 0.6611 | 0.3389 | 0.004 | 11 | |
| POP25 | 38 | 1.00 | 0.505 | 0.513 | 3.625 | 0.4422 | 0.5578 | 0.029 | 36 | |
| POP26 | 30 | 0.92 | 0.395 | 0.342 | 2.000 | 0.5778 | 0.4222 | −0.137 | 30 | |
| POP27 | 47 | 1.00 | 0.512 | 0.487 | 3.5 | 0.1514 | 0.8486 | −0.040 | 18 | |
| POP28 | 48 | 1.00 | 0.494 | 0.507 | 3.875 | 0.0164 | 0.9836 | 0.035 | 11 | |
| POP29 | 27 | 0.96 | 0.586 | 0.530 | 3.125 | 0.9412 | 0.0588 | −0.087 | 19 | |
| POP30 | 30 | 1.00 | 0.532 | 0.516 | 3.625 | 0.8227 | 0.1773 | −0.015 | 11 | |
| POP31 | 30 | 1.00 | 0.557 | 0.511 | 3.75 | 0.9257 | 0.0743 | −0.073 | 7 | |
| POP32 | 30 | 1.00 | 0.503 | 0.490 | 3.375 | 0.6773 | 0.3227 | −0.009 | 7 | |
| POP33 | 20 | 1.00 | 0.356 | 0.343 | 2.625 | 0.6057 | 0.3943 | −0.012 | 10 | |
| POP34 | 30 | 1.00 | 0.448 | 0.441 | 3.625 | 0.7376 | 0.2624 | 0.000 | 5 | |
| POP35 | 29 | 1.00 | 0.478 | 0.559 | 3.5 | 0.0008 | 0.9992 | 0.162 * | 11 | |
| POP36 | 29 | 0.96 | 0.363 | 0.332 | 2.25 | 0.8856 | 0.1144 | −0.078 | 20 | |
| POP37 | 30 | 0.30 | 0.213 | 0.164 | 1.75 | 0.5554 | 0.4446 | −0.281 * | 20 | |
| POP38 | 29 | 1.00 | 0.438 | 0.429 | 2.875 | 0.5461 | 0.4539 | −0.004 | 29 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Simon, J.-C.; Mahéo, F.; Mieuzet, L.; Buchard, C.; Gauthier, J.-P.; Maurice, D.; Bonhomme, J.; Outreman, Y.; Hullé, M. Life on the Edge: Ecological Genetics of a High Arctic Insect Species and Its Circumpolar Counterpart. Insects 2019, 10, 427. https://doi.org/10.3390/insects10120427
Simon J-C, Mahéo F, Mieuzet L, Buchard C, Gauthier J-P, Maurice D, Bonhomme J, Outreman Y, Hullé M. Life on the Edge: Ecological Genetics of a High Arctic Insect Species and Its Circumpolar Counterpart. Insects. 2019; 10(12):427. https://doi.org/10.3390/insects10120427
Chicago/Turabian StyleSimon, Jean-Christophe, Frédérique Mahéo, Lucie Mieuzet, Christelle Buchard, Jean-Pierre Gauthier, Damien Maurice, Joël Bonhomme, Yannick Outreman, and Maurice Hullé. 2019. "Life on the Edge: Ecological Genetics of a High Arctic Insect Species and Its Circumpolar Counterpart" Insects 10, no. 12: 427. https://doi.org/10.3390/insects10120427
APA StyleSimon, J.-C., Mahéo, F., Mieuzet, L., Buchard, C., Gauthier, J.-P., Maurice, D., Bonhomme, J., Outreman, Y., & Hullé, M. (2019). Life on the Edge: Ecological Genetics of a High Arctic Insect Species and Its Circumpolar Counterpart. Insects, 10(12), 427. https://doi.org/10.3390/insects10120427





