Use of DNA Markers for Grape Phylloxera Population and Evolutionary Genetics: From RAPDs to SSRs and Beyond
Abstract
:1. Daktulosphaira vitifoliae: A Major Pest of Cultivated Grapevines
2. Relevance of Studying Population Structure in Grape Phylloxera
3. Use of DNA Markers for Grape Phylloxera Genetics
3.1. RAPDs and AFLPs
3.2. Cytochrome C Oxidase Subunit I (COI) Gene Sequencing
3.3. Microsatellites
4. What Is Next?
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Forneck, A.; Huber, L. (A)sexual reproduction—A review of life cycles of grape phylloxera, Daktulosphaira vitifoliae. Entomol. Exp. Appl. 2009, 131, 1–10. [Google Scholar] [CrossRef]
- Powell, K.S.; Cooper, P.D.; Forneck, A. The biology, physiology and host-plant interactions of grape phylloxera Daktulosphaira vitifoliae. In Advances in Insect Physiology; Johnson, S.N., Hiltpold, I., Turlings, T.C.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 159–218. [Google Scholar]
- Kellow, A.V.; Sedgley, M.; Van Heeswijck, R. Interaction between Vitis vinifera and grape phylloxera: Changes in root tissue during nodosity formation. Ann. Bot. 2004, 93, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Forneck, A.; Kleinmann, S.; Blaich, R.; Anvari, S.F. Histochemistry and anatomy of phylloxera (Daktulosphaira vitifoliae) nodosities on young roots of grapevine (Vitis spp.). Vitis 2002, 41, 93–97. [Google Scholar]
- Nabity, P.D.; Haus, M.J.; Bernbaum, M.R.; De Lucia, E.H. Leaf-galling phylloxera on grapes reprograms host metabolism and morphology. Proc. Natl. Acad. Sci. USA 2013, 110, 16663–16668. [Google Scholar] [CrossRef] [PubMed]
- Granett, J.; Walker, M.A.; Kocsis, L.; Omer, A.D. Biology and management of grape phylloxera. Annu. Rev. Entomol. 2001, 46, 387–412. [Google Scholar] [CrossRef] [PubMed]
- Eitle, M.W.; Loacker, J.; Meng-Reiterer, J.; Schuhmacher, R.; Griesser, M.; Forneck, A. Polyphenolic profiling of roots (Vitis spp.) under grape phylloxera (D. vitifoliae Fitch) attack. Plant Physiol. Biochem. 2019, 135, 174–181. [Google Scholar] [CrossRef] [PubMed]
- Griesser, M.; Lawo, N.C.; Crespo-Martinez, S.; Schoedl-Hummel, K.; Wieczorek, K.; Gorecka, M.; Liebner, F.; Zweckmair, T.; Pavese, N.S.; Kreil, D.; et al. Phylloxera (Daktulosphaira vitifoliae Fitch) alters the carbohydrate metabolism in root galls to allowing the compatible interaction with grapevine (Vitis ssp.) roots. Plant Sci. 2015, 234, 38–49. [Google Scholar] [CrossRef]
- Wapshere, A.J.; Helm, K.F. Phylloxera and Vitis: An experimentally testable coevolutionary hypothesis. Am. J. Enol. Vitic. 1987, 38, 216–222. [Google Scholar]
- Viala, P.; Ravaz, L. American Vines (Resistant Stock): Their Adaptation, Culture, Grafting and Propagation; Press of Freygang-Leary Co.: San Francisco, CA, USA, 1903. [Google Scholar]
- Bournier, A. Grape insects. Annu. Rev. Entomol. 1976, 22, 355–376. [Google Scholar] [CrossRef]
- Downie, D.A. Locating the sources of an invasive pest, grape phylloxera, using a mitochondrial DNA gene genealogy. Mol. Ecol. 2002, 11, 2013–2026. [Google Scholar] [CrossRef]
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Ollat, N.; Peccoux, A.; Papura, D.; Esmenjaud, D.; Marguerit, E.; Tandonnet, J.P.; Bordenave, L.; Cookson, S.J.; Barrieu, F.; Rossdeutsch, L.; et al. Rootstocks as a component of adaptation to environment. In Grapevine in a Changing Environment: A Molecular and Ecophysiological Perspective; Geros, H., Chaves, M.M., Medrano, H., Delrot, S., Eds.; Wiley Blackwell: Chichester, UK, 2016; pp. 68–108. [Google Scholar]
- Riaz, S.; Pap, D.; Uretsky, J.; Laucou, V.; Boursiquot, J.M.; Kocsis, L.; Walker, M.A. Genetic diversity and parentage analysis of grape rootstocks. Theor. Appl. Genet. 2019, 132, 1847–1860. [Google Scholar] [CrossRef] [PubMed]
- Migliaro, D.; De Lorenzis, G.; Di Lorenzo, G.S.; De Nardi, B.; Gardiman, M.; Failla, O.; Brancadoro, L.; Crespan, M. Grapevine non-vinifera genetic diversity assessed by SSR markers as a starting-point for new rootstock breeding programs. Am. J. Enol. Vitic. 2019, 10. [Google Scholar] [CrossRef]
- Arancibia, C.; Riaz, S.; Aguero, C.; Ramirez-Corona, B.; Alonso, R.; Buscema, F.; Martinez, L.; Walker, M.A. Grape phylloxera (Daktulosphaira vitifoliae Fitch) in Argentina: Ecological associations to diversity, population structure and reproductive mode. Aust. J. Grape Wine Res. 2018, 24, 284–291. [Google Scholar] [CrossRef]
- Lund, K.T.; Riaz, S.; Walker, M.A. Population structure, diversity and reproductive mode of the grape phylloxera (Daktulosphaira vitifoliae) across its native range. PLoS ONE 2017, 12, e0170678. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Lund, K.T.; Granett, J.; Walker, M.A. Population diversity of grape phylloxera in California and evidence for sexual reproduction. Am. J. Enol. Vitic. 2017, 68, 218–227. [Google Scholar] [CrossRef]
- Corrie, A.M.; Buchanan, G.A.; Van Heeswijck, R. DNA typing of populations of phylloxera (Daktulosphaira vitifoliae (Fitch)) from Australian vineyards. Aust. J. Grape Wine Res. 1997, 3, 50–56. [Google Scholar] [CrossRef]
- Downie, D.A. Evidence for multiple origins of grape phylloxera (Daktulosphaira vitifoliae Fitch) (Hemiptera: Phylloxeridae) in South African vineyards. Afr. Entomol. 2005, 13, 359–362. [Google Scholar]
- Yvon, M.; Peros, J.P. Variation in aggressiveness and genetic diversity of grape phylloxera in Southern France. J. Int. Sci. Vigne Vin. 2003, 37, 77–84. [Google Scholar] [CrossRef]
- Eitle, M.W.; Forneck, A. Comparison of bioassays to biotype grape phylloxera (Daktulosphaira vitifoliae Fitch) on Vitis ssp. Vitis 2017, 56, 141–146. [Google Scholar]
- Downie, D.A.; Fisher, J.R.; Granett, J. Grapes, galls, and geography: The distribution of nuclear and mithocondrial DNA variation across host-plant species and regions in a specialist herbivore. Evolution 2001, 55, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Corrie, A.M.; van Heeswijck, R.; Hoffmann, A.A. Evidence for host-associated clones of grape phylloxera Daktulosphaira vitifoliae (Hemiptera: Phylloxeridae) in Australia. Bull. Entomol. Res. 2003, 93, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Forneck, A.; Walker, M.A.; Blaich, R. Ecological and genetic aspects of grape phylloxera Daktulosphaira vitifoliae (Hemiptera: Phylloxeridae) performance on rootstock hosts. Bull. Entomol. Res. 2001, 91, 445–451. [Google Scholar] [PubMed]
- Forneck, A.; Powell, K.S.; Walker, M.A. Scientific opinion: Improving the definition of grape Phylloxera biotypes and standardizing biotype screening protocols. Am. J. Enol. Vitic. 2016, 67, 371–376. [Google Scholar] [CrossRef]
- Saxena, R.C.; Barrion, A.A. Biotypes of insect pests of agricultural crops. Int. J. Trop. Insect Sci. 1987, 8, 453–458. [Google Scholar] [CrossRef]
- Sunnucks, P.; de Barro, P.J.; Lushai, G.; Maclean, N.; Hales, D. Genetic structure of an aphid studied using microsatellites: Cyclic parthenogenesis, differentiated lineages and host specialization. Mol. Ecol. 1997, 6, 1057–1073. [Google Scholar] [CrossRef]
- Vorweck, S.; Forneck, A. Reproductive mode of grape phylloxera (Daktulosphaira vitifoliae, Homoptera: Phylloxeridae) in Europe: Molecular evidence for predominantly asexual populations and a lack of gene flow between them. Genome 2006, 49, 678–688. [Google Scholar] [CrossRef]
- Forneck, A.; Mammerler, R.; Tello, J.; Breuer, M.; Muller, J.; Fahrentrapp, J. First European leaf-feeding grape phylloxera (Daktulosphaira vitifoliae Fitch) survey in Swiss and German commercial vineyards. Eur. J. Plant Pathol. 2019. [Google Scholar] [CrossRef]
- Lushai, G.; De Barro, P.J.; Sherratt, D.T.N.; Maclean, N. Genetic variation within a parthenogenetic lineage. Insect Mol. Biol. 1998, 7, 337–344. [Google Scholar] [CrossRef]
- Corrie, A.M.; Crozier, R.H.; Heeswijck, R.V.; Hoffmann, A.A. Clonal reproduction and population genetic structure of grape phylloxera, Daktulosphaira vitifoliae, in Australia. Heredity 2002, 88, 203–211. [Google Scholar] [CrossRef]
- Rollins, L.A.; Woolnough, A.P.; Sherwin, W.B. Population genetic tools for pest management: A review. Wildl. Res. 2006, 33, 251–261. [Google Scholar] [CrossRef]
- MacDonald, C.; Loxdale, H. Molecular markers to study population structure and dynamics in beneficial insects (predators and parasitoids). Int. J. Pest Manag. 2004, 50, 215–224. [Google Scholar] [CrossRef]
- Hoffmann, M.; Ruehl, E.H.; Eisenbeis, G.; Huber, L. Indications for rootstock related ecological preferences of grape phylloxera (Daktulosphaira vitifoliae Fitch). Vitis 2015, 54, 137–142. [Google Scholar]
- Turley, M.; Granett, J.; Omer, A.D.; De Benedictis, J.A. Grape phylloxera (Homoptera: Phylloxeridae) temperature threshold for establishment of feeding sites and degree-day calculations. Environ. Entomol. 1996, 25, 842–847. [Google Scholar] [CrossRef]
- Korosi, G.A.; Mee, P.T.; Powell, K.S. Influence of temperature and humidity on mortality of grapevine phylloxera Daktulosphaira vitifoliae clonal lineages: A scientific validation of a disinfestation procedure for viticultural machinery. Aust. J. Grape Wine Res. 2012, 18, 43–47. [Google Scholar] [CrossRef]
- Corrie, A.M.; Hoffmann, A.A. Fine-scale genetic structure of grape phylloxera from the roots and leaves of Vitis. Heredity 2004, 92, 113–127. [Google Scholar] [CrossRef]
- Vorweck, S.; Forneck, A. Analysis of genetic variation within clonal lineages of grape phylloxera (Daktulosphaira vitifoliae Fitch) using AFLP fingerprinting and DNA sequencing. Genome 2007, 50, 660–667. [Google Scholar] [CrossRef]
- Wilson, A.C.C.; Sunnucks, P.; Hales, D.F. Heritable genetic variation and potential for adaptive evolution in asexual aphids (Aphidoidea). Biol. J. Linn. Soc. 2003, 79, 115–135. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Walker, M.A.; Hu, R.; Granett, J. New simple sequence repeat loci for the study of grape phylloxera (Daktulosphaira vitifoliae) genetics and host adaptation. Am. J. Enol. Vitic. 2006, 57, 33–40. [Google Scholar]
- Benheim, D.; Rochfort, S.; Robertson, E.; Potter, I.D.; Powell, K.S. Grape phylloxera (Daktulosphaira vitifoliae) —A review of potential detection and alternative management options. Ann. Appl. Biol. 2012, 161, 91–115. [Google Scholar] [CrossRef]
- Zhu, H.; Sun, Q.; Du, Y.; Gao, Z.; Zhai, H. Detection of grape phylloxera on grapevine roots with diagnostic polymerase chain reaction methods targeted to the internal transcribed space region 2 nuclear gene. Aust. J. Grape Wine Res. 2015, 21, 143–146. [Google Scholar] [CrossRef]
- Giblot-Ducray, D.; Correll, R.; Collins, C.; Nankivell, A.; Downs, A.; Pearce, I.; McKay, A.C.; Ophel-Keller, K.M. Detection of grape phylloxera (Daktulosphaira vitifoliae Fitch) by real-time quantitative PCR: Development of a soil sampling protocol. Aust. J. Grape Wine Res. 2016, 22, 469–477. [Google Scholar] [CrossRef]
- Herbert, K.; Powell, K.; McKay, A.; Di Hartley, H.; Ophel-Keller, K.M.; Schiffer, M.; Hoffmann, A. Developing and testing a diagnostic probe for grape phylloxera applicable to soil samples. J. Econ. Entomol. 2008, 101, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Triska, M.D.; Powell, K.S.; Collins, C.; Pearce, I.; Renton, M. Accounting for spatially heterogeneous conditions in local-scale surveillance strategies: Case study of the biosecurity insect pest, grape phylloxera (Daktulosphaira vitifoliae (Fitch)). Pest Manag. Sci. 2018, 74, 2724–2737. [Google Scholar] [CrossRef] [PubMed]
- Behura, S.K. Molecular marker systems in insects: Current trends and future avenues. Mol. Ecol. 2006, 15, 3087–3113. [Google Scholar] [CrossRef] [PubMed]
- Sunnucks, P. Efficient genetic markers for population biology. Trends Ecol. Evol. 2000, 15, 199–203. [Google Scholar] [CrossRef]
- Caterino, M.S.; Cho, S.; Sperling, F.A.H. The current state of insect molecular systematics: A thriving Tower of Babel. Annu. Rev. Entomol. 2000, 45, 1–54. [Google Scholar] [CrossRef] [PubMed]
- Loxdale, H.D.; Lushai, G. Molecular markers in entomology. Bull. Entomol. Res. 1998, 88, 577–600. [Google Scholar] [CrossRef]
- Lunt, D.H.; Zhang, D.-X.; Szymura, J.M.; Hewitt, G.M. The insect cytochrome oxidase I gene: Evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol. Biol. 2006, 5, 153–165. [Google Scholar] [CrossRef]
- Lin, H.; Downie, D.A.; Walker, M.A.; Granett, J.; English-Loeb, G. Genetic structure in native populations of grape phylloxera (Homoptera: Phylloxeridae). Ann. Entomol. Soc. Am. 1999, 92, 376–381. [Google Scholar] [CrossRef]
- Fong, G.; Walker, M.A.; Granett, J. RAPD assessment of California phylloxera diversity. Mol. Ecol. 1995, 4, 459–464. [Google Scholar] [CrossRef]
- Forneck, A.; Walker, M.A.; Blaich, R. Genetic structure of an introduced pest, grape phylloxera (Daktulosphaira vitifoliae Fitch), in Europe. Genome 2000, 43, 669–678. [Google Scholar] [CrossRef] [PubMed]
- Downie, D.A. Phylogeography in a galling insect, grape phylloxera, Daktulosphaira vitifoliae (Phylloxeridae) in the fragmented habitat of the Southwest USA. J. Biogeogr. 2004, 31, 1759–1768. [Google Scholar] [CrossRef]
- Bao, L.V.; Scatoni, I.B.; Gaggero, C.; Gutierrez, L.; Monza, J.; Walker, M.A. Genetic diversity of grape phylloxera leaf-galling populations on Vitis species in Uruguay. Am. J. Enol. Vitic. 2015, 66, 46–53. [Google Scholar] [CrossRef]
- Forneck, A.; Anhalt, U.C.M.; Mammerler, R.; Griesser, M. No evidence of superclones in leaf-feeding forms of austrian grape phylloxera (Daktulosphaira vitifoliae). Eur. J. Plant. Pathol. 2015, 142, 441–448. [Google Scholar] [CrossRef]
- Toth, H.L.; Taller, J.; Cernak, I.; Feher, E.; Kocsis, L. Diversity of Hungarian grape phylloxera (Daktulosphaira vitifoliae Fitch) populations. Acta Hortic. 2007, 733, 97–100. [Google Scholar] [CrossRef]
- Lynch, M.; Milligan, B.G. Analysis of population genetic structure with RAPD markers. Mol. Ecol. 1994, 3, 91–99. [Google Scholar] [CrossRef]
- Downie, D.A. Patterns of genetic variation in native grape phylloxera on two sympatric host species. Mol. Ecol. 2000, 9, 505–514. [Google Scholar] [CrossRef]
- Mueller, U.G.; Wolfenwarger, L.L. AFLP genotyping and fingerprinting. Trends Ecol. Evol. 1999, 14, 389–394. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; van de Lee, T.; Hornes, M.; Frijters, A.; Pot, J.; Peleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucl. Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef]
- Van de Zande, L.; Bijlsma, R. Limitations of the RAPD technique in phylogeny reconstruction in Drosophila. J. Evol. Biol. 1995, 8, 645–656. [Google Scholar] [CrossRef]
- Galtier, N.; Nabholz, B.; Glemin, S.; Hurst, G.D.D. Mitochondrial DNA as a marker of molecular diversity: A reappraisal. Mol. Ecol. 2009, 18, 4541–4550. [Google Scholar] [CrossRef] [PubMed]
- Hebert, P.D.N.; Ratnasingham, S.; De Waard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Y.; Yang, X.-M.; Lu, B.R.; Zhou, L.-H.; Wu, K.-M. Genetic variation and phylogeographic structure of the cotton aphid, Aphis gossypii, based on mitochondrial DNA and microsatellite markers. Sci. Rep. 2017, 7, 1920. [Google Scholar] [CrossRef] [PubMed]
- Miranda, E.A.; Batalha-Filho, H.; Congrains, C.; Carvalho, A.F.; Ferreira, K.M.; del Lama, M.A. Phylogeography of Partamona rustica (Hymenoptera, Apidae), an endemic stingless bee from the neotropical dry forest diagonal. PLoS ONE 2016, 11, e016441. [Google Scholar] [CrossRef] [PubMed]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Hebert, P.D.N.; Cywinska, A.; Ball, S.L.; deWaard, J.R. Biological identifications through DNA barcodes. Proc. R Soc. Lond. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cummings, M.A.; Krafsur, E.S. Spatial diversity in mitochondrial cytochrome c oxidase in house flies. Med. Vet. Entomol. 2005, 19, 53–59. [Google Scholar] [CrossRef]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. 2004, 5, 435–445. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Botta, R.; Costantini, L.; Crespan, M.; Dangl, G.S.; Eisenheld, C.; Ferreira-Monteiro, F.; et al. Development of a standard set of microsatellite reference alleles for identification of grape cultivars. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Schlötterer, C. Evolutionary dynamics of microsatellite DNA. Chromosoma 2000, 109, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Riaz, S.; Lund, K.; Lin, H.; Walker, M.A. Development and characterization of a large set of microsatellite markers for grape phylloxera (Daktulosphaira vitifoliae Fitch). Vitis 2014, 53, 95–101. [Google Scholar]
- Umina, P.A.; Corrie, A.M.; Herbert, K.S.; White, V.L.; Powell, K.S.; Hoffmann, A.A. The Use of DNA Markers for Pest Management—Clonal Linages and Population Biology of Grape phylloxera. In Proceedings of the III International Grapevine Phylloxera Symposium, Fremantle, Australia, 1 February 2007; pp. 133–195. [Google Scholar]
- Islam, M.S.; Roush, T.L.; Walker, M.A.; Granett, J.; Lin, H. Reproductive mode and fine-scale population genetic structure of grape phylloxera (Daktulosphaira vitifoliae) in a viticultural area in California. BMC Genet. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Forneck, A.; Dockner, V.; Mammerler, R.; Powell, K.S.; Kocsis, L.; Papura, D.; Fahrentrapp, J.; Riaz, S.; Walker, M.A. PHYLLI—An international database for grape phylloxera (Daktulosphaira vitifoliae Fitch). IOBC-WPRS Bull. 2017, 128, 45–51. [Google Scholar]
- Sun, Q.-H.; Chen, Y.-C.; Du, Y.-P.; Zhai, H. Genetic structure of grape phylloxera in China. Acta Hortic. 2011, 904, 25–32. [Google Scholar] [CrossRef]
- Temnykh, S.; DeClerck, G.; Lukashova, A.; Lipovich, L.; Cartinhour, S.; McCouch, S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): Frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001, 11, 1441–1452. [Google Scholar] [CrossRef]
- Bonin, A.; Bellemain, E.; Bronken Eidesen, P.; Pompanon, F.; Brochmann, C.; Taberlet, P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004, 13, 3261. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Estoup, A.; Jarne, P.; Cornuet, J.-M. Homoplasy and mutation model at microsatellite loci and their consequences for population genetics analysis. Mol. Ecol. 2002, 11, 1591–1601. [Google Scholar] [CrossRef]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Robertson, H.M.; Feder, J.L.; Varala, K.; Hudson, M.E.; Ragland, G.J.; Hahn, D.A.; Berlocher, S.H. Sympatric ecological speciation meets pyrosequencing: Sampling the transcriptome of the apple maggot Rhagoletis pomonella. BMC Genom. 2009, 10, 633. [Google Scholar] [CrossRef] [PubMed]
- Perry, K.D.; Pederson, S.M.; Baxter, S.W. Genome-wide SNP discovery in field and laboratory colonies of Australian Plutella species. Bioarxiv 2017. [Google Scholar] [CrossRef]
- Silva-Brandao, K.L.; Neto e Silva, O.A.B.; Brandao, M.M.; Omoto, C.; Sperling, F.A.H. Genotyping-by-sequencing approach indicates geographic distance as the main factor affecting genetic structure and gene flow in Brazilian populations of Grapholita molesta (Lepidoptera, Tortricidae). Evol. Appl. 2015, 8, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Black, W.C., IV; Vontas, J.G. Affordable assays for genotyping single nucleotide polymorphisms in insects. Insect Mol. Biol. 2007, 16, 377–387. [Google Scholar]
- Delmotte, F.; Papura, D.; Rispe, C.; Legeai, F.; Jaquiery, J.; Bretaudeau, A.; Tagu, D.; Powell, K.S.; Forneck, A. The grape phylloxera genome sequencing project. Acta Hortic. 2014, 1045, 15–20. [Google Scholar] [CrossRef]
- Stucki, D.; Malla, B.; Hostettler, S.; Huna, T.; Feldmann, J.; Yeboah-Manu, D.; Borrell, S.; Fenner, L.; Comas, I.; Coscolla, M.; et al. Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS ONE 2012, 7, e41253. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.A.; Martien, K.K.; Taylor, B.L. Assessing statistical power of SNPs for population structure and conservation studies. Mol. Ecol. Resour. 2009, 9, 66–73. [Google Scholar] [CrossRef]
- Van Inghelandt, D.; Melchinger, A.; Lebreton, C.; Stich, B. Population structure and genetic diversity in a commercial maize breeding program assessed with SSR and SNP markers. Theor. Appl. Genet. 2010, 120, 1289–1299. [Google Scholar] [CrossRef] [Green Version]
- Tello, J.; Roux, C.; Chouiki, H.; Laucou, V.; Sarah, G.; Weber, A.; Santoni, S.; Flutre, T.; Pons, T.; This, P.; et al. A novel high-density grapevine (Vitis vinifera L.) integrated linkage map using GBS in a half-diallel population. Theor. Appl. Genet. 2019, 132, 2237–2252. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.J.; Tay, W.T.; McGaughran, A.; Gordon, K.; Walsh, T.K. Population structure and gene flow in the global pest, Helicoverpa armigera. Mol. Ecol. 2016, 25, 5296–5311. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.M.; Battlay, P.; Gledhill-Smith, R.S.; Good, R.T.; Lumb, C.; Fournier-Level, A.; Robin, C. Insights into DDT resistance from the Drosophila melanogaster genetic reference panel. Genetics 2017, 207, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Magwire, M.M.; Fabian, D.K.; Schweyen, H.; Cao, C.; Longdon, B.; Bayer, F.; Jiggins, F.M. Genome-wide association studies reveal a simple genetic basis of resistance to naturally coevolving viruses in Drosophila melanogaster. PLoS Genet. 2012, 8, e1003057. [Google Scholar] [CrossRef] [PubMed]
- Rispe, C.; Legeai, F.; Papura, D.; Bretaudeau, A.; Hudaverdian, S.; Le Trionnaire, G.; Tagu, D.; Jaquiery, J.; Delmotte, F. De novo transcriptome assembly of the grapevine phylloxera allows identification of genes differentially expressed between leaf- and root-feeding forms. BMC Genom. 2016, 17, 219. [Google Scholar] [CrossRef] [PubMed]
- Delmotte, F.; Forneck, A.; Powell, K.; Rispe, C.; Tagu, D. Proposal to sequence the genome of the grape Phylloxera (Daktulosphaira vitifoliae Fitch). Available online: http://bipaa.genouest.org/is/wp-content/uploads/2015/10/White-Paper-Phylloxera_25may2011-1.pdf (accessed on 24 September 2019).
- Forneck, A.; Jin, Y.; Walker, A.; Blaich, R. Karyotype studies on grape phylloxera (Daktulosphaira vitifoliae Fitch). Vitis 1999, 38, 123–125. [Google Scholar]

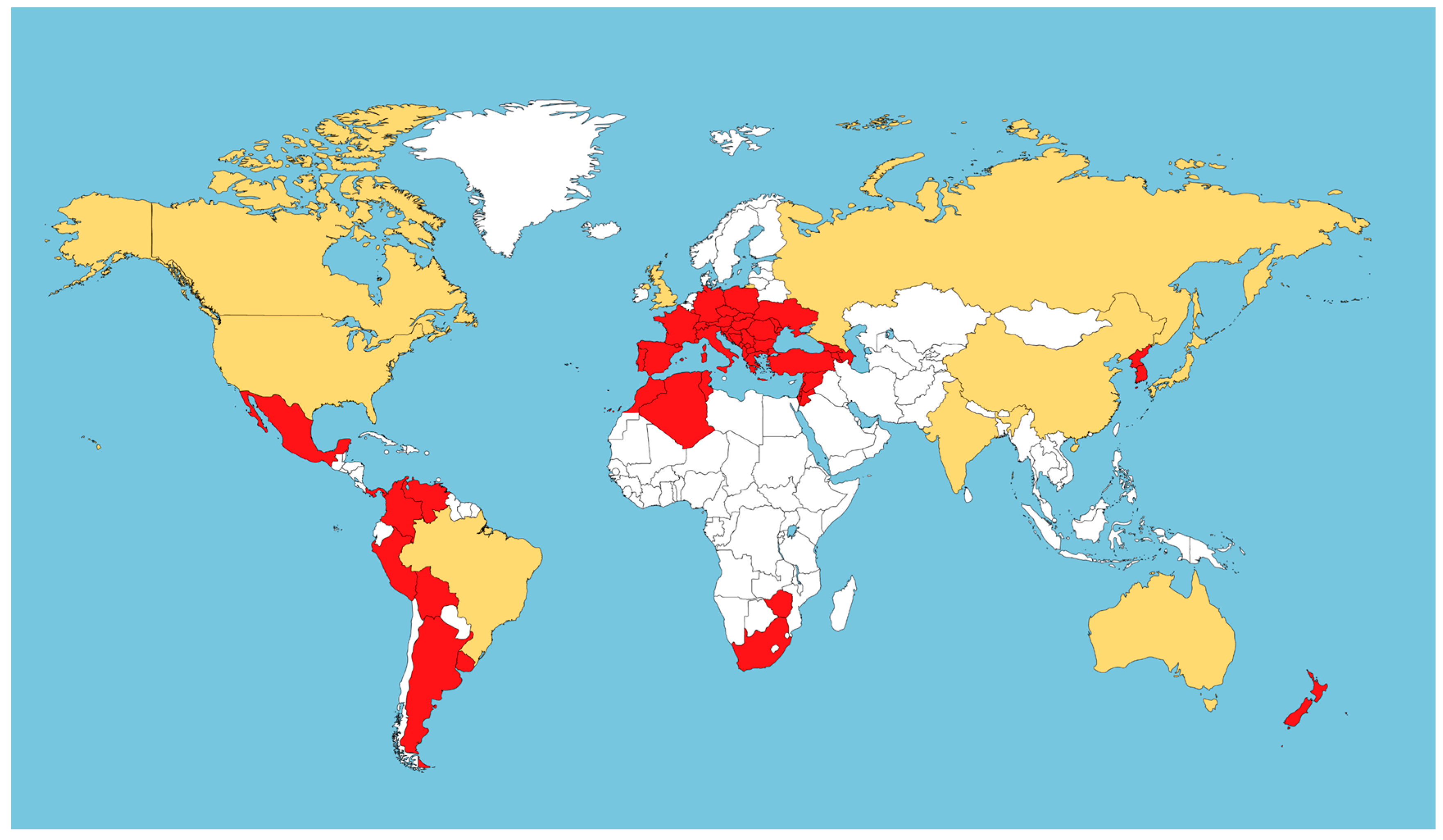
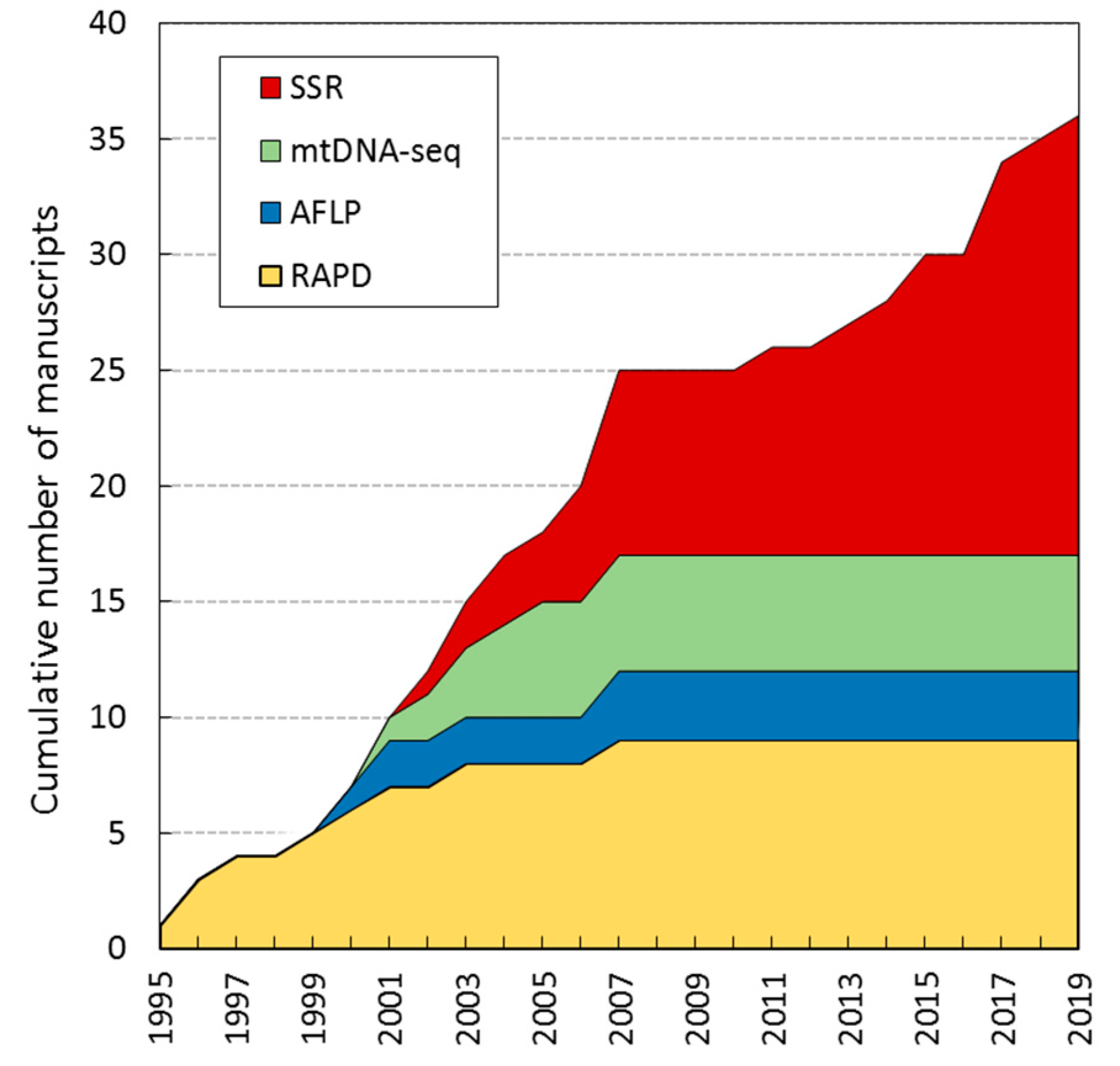
| Biotype/ Feeding Tissue | Vitis vinifera | Rootstocks and Hybrids (V. vin. x American Vitis species) | Rootstocks (American Vitis species) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| A | |||||||||
| Root | T | N | - | t | n | - | - | n | - |
| Leaves | - | G | G | ||||||
| B | |||||||||
| Root | t | n | T | N | - | n | - | ||
| Leaves | - | - | - | ||||||
| C | |||||||||
| Root | - | n | - | T | N | P | - | N | P |
| Leaves | - | G | G | ||||||
| D | |||||||||
| Root | t | n | - | - | N | - | - | N | - |
| Leaves | - | ? | G | ||||||
| E | |||||||||
| Root | T | N | - | T | N | ? | T | N | P |
| Leaves | - | - | - | ||||||
| F | |||||||||
| Root | T | N | - | ? | ? | ? | ? | n | ? |
| Leaves | G | ? | G | ||||||
| G | |||||||||
| Root | ? | ? | ? | ? | N | ? | - | N | ? |
| Leaves | G | G | G | ||||||
| SSR 1 | Motif | Flanking Primer Sequences (5′→3′) | Source | Reference |
|---|---|---|---|---|
| Dvit1 | (CA)n (CG)n | F: CGTTCGTTCTGGTATCGTTATT R: TAACGACCCGACTGAAATGTAG | [33] | [17,25,30,33,39,40,57,58,77,78] |
| Dvit2 | (CT)n (AT)n | F: GCTTAATTTTGTGTCTCAAGTTA R: TAATGCTTCGTTTTCTAAGTGC | [33] | [25,30,33,39,40,57,58,77,78] |
| Dvit3 | (AT)n (GT)n | F: CCAAAACAACCAAGATTTTCTCC R: GATCCAAACTATGACAAACACCC | [33] | [19,25,30,33,39,40,58,77,78,79] |
| Dvit4 | (AAT)n | F: TCTTCAAAAATGTTACATGAT R: TATACAATGAATGGTATCAATTC | [33] | [19,25,30,33,39,40,58,77,80] |
| Dvit5 | (A)n | F: GAAATCCGTTCGGTGAGAGC R: TATGGTCAATGGTCAATCCGTC | [30] | [30,40,58,77] |
| Dvit6 | (AAT)n | F: TGGACGATGGTTTTCATAGC R: TTGATTGTCATTGGTTTTGC | [30] | [19,30,31,40,58,77,79] |
| DVSSR1 | (CA)n | F CGGCGACGAGTTAAACTATC R TCGTTGTATAGATCTGTGTTGC | [42] | [42,80] |
| DVSSR2 | (CA)n | F TCGCTACTACCAGCCGATCAG R TGAACAATGAAAGCCCTGGTGG | [42] | [42,80] |
| DVSSR3 | (CA)n | F: AGCATGTGAGGTGCAAGGC R: CCTCGGGCGGAACAATCG | [42] | [19,42,57] |
| DVSSR4 | (CT)n | F: TGGTATTCACCTTGGAGCCTAG R: GCTACTGAAACCCCTTCAACAC | [42] | [19,31,42,57,78,79,80] |
| DVSSR6 | (CTT)n | F: GTTTACTGAAATAAGGGCTGG R: AGTTGTGATTATAAGCCGAGG | [42] | [19,42,78] |
| DVSSR7 | (GCA)n | F: GTGAGTTGACTGTTGATTCG R: CGCAATTCATTCGGTTACC | [42] | [19,42,78] |
| DVSSR9 | (GCA)n | F: CGCAATTCATTCGGTTACC R: GTGAGTTGACTGTTGATTCG | [42] | [42,80] |
| DVSSR16 | (A)n | F: AGACCAGACGCGAGCAATG R: ACCATCAATGAAAGCCTTGTCG | [42] | [42,78,80] |
| DVSSR17 | (CGTTTCTG)n | F: CTCTGTGTAGCCAAGTCAAC R: TATCCTACGCCAGTAAGAAG | [42] | [19,42,78,80] |
| DV4 | (GTT)n | unpublished | - | [31,79] |
| DV8 | (TG)n | unpublished | - | [31,79] |
| DV11 | (CT)n | unpublished | - | [79] |
| PhyII_6 2 | (TA)n | F: TTATTGTCAGTTAGGTCTGAGATACC R: ATTGTTTTCGACCCGTCATTAT | [76] | [17,18,76] |
| PhyII_10 2 | (AT)n | F: CCTTCTCACTTCACATCAAAGC R: TCCAAAAGCTATATGATCCCCTA | [76] | [17,18,76] |
| PhyII_13 2 | (AC)n | F: GCGTATAAACGATGGCGTTAAA R: TCTTCTTCACGTTTGCTCAGAA | [76] | [17,18,76] |
| PhyII_16 2 | (AT)n | F: CTGGTGGCTTTGGTGGTAAG R: CTCGATCTTGCCTGCTACCTAT | [76] | [17,18,19,76] |
| PhyII_23 2 | (AT)n | F: CGTATGCCCTTCTAACACGATT R: CGGGATATTCGATTAAATGCTG | [76] | [17,18,19,76] |
| PhyII_26 2 | (AT)n | F: TTACTATTTGGCCGTCAAGTCA R: GCTGAAAGAGCAACAAATTCAA | [76] | [76] |
| PhyII_28 2 | (AT)n | F: CCGAGAGCAAGAGAAAACTGAG R: TCGTACATTCAAGTTACTTTTACACA | [76] | [76] |
| PhyII_29 2 | (AT)n | F: CCAATCATTTTACTAGGCTCGTG R: GAGGCGATAGCAGAGTATGGAG | [76] | [76] |
| PhyII_31 2 | (TG)n | F: CGTCGCCCTTATATCAAATTCT R: GCGGTGATGGACTGTAGAAAAT | [76] | [17,18,76] |
| PhyII_32 2 | (GT)n | F: ACGTATTAATGGGCGTCGTTAT R: TTAAAATATTGCCGCAAGTTCA | [76] | [19,76] |
| PhyII_34 2 | (AC)n | F: AAGCCGGTCTGCAATATTATGT R: TTTCGTTTACACAAGAATGGTATG | [76] | [17,18,76] |
| PhyII_36 2 | (AC)n | F: CGTACCCCACACAGAGTATTCA R: CCCTCATACACTCACACTCGAA | [76] | [17,18,76] |
| PhyIII_15 2 | (TGT)n | F: TTCCAGTAGTTGCTGTTATTCCTG R: AACCACAGAATTTTCCTTTTGTTC | [76] | [17,18,76] |
| PhyIII_19 2 | (ATT)n | F: CGCCGATTTATGTATCAACTCA R: GACTGTTTCGTACCGCACATAA | [76] | [18,76] |
| PhyIII_302 | (TCT)n | F: ACCGTTATGAACAAAAGCAGGA R: GGTTTTGCCTTCAGACTCCTT | [76] | [17,18,31,76,79] |
| PhyIII_362 | (TAA)n | F: CGTCCTTCTTGCGTGATATTTT R: GGCGGAATAAATGAGAAAAGTG | [76] | [18,19,31,76,79] |
| PhyIII_38 2 | (GAA)n (GAC)n | F: TTGATGAAAATGCTCCTTGTTTT R: CTGGTGGTTCAGTATTCTCGTC | [76] | [76] |
| PhyIII_42 2 | (TA)n (CGG)n | F: GTATATACGGTGGCGGTAGGAC R: CGTACTCAAGTCGCTATACCCTA | [76] | [17,18,76] |
| PhyIII_46 2 | (CCA)n | F: TCTCGCACGGCTATTGTAGTTA R: TCTGTTGCAATGCCTAAAAGAA | [76] | [17,18,76] |
| PhyIII_49 2 | (TAA)n | F: CCATCTTAAATCTTTGGCTCGT R: ACGGAACTACACACGCACATAC | [76] | [17,18,76] |
| PhyIII_53 2 | (ATA)n | F: CACTCATGATTGCAATTTTTCC R: TTGCACATAGTGTGATACATTTCC | [76] | [17,18,76] |
| PhyIII_552 | (ATT)n | F: CGTATGATCGTCACAGAGGAAA R: CGATTCCGCTTTAAACAATACC | [76] | [17,18,31,76,79] |
| PhyIII_61 2 | (ATA)n | F: GTACCGGCCGAAAATTGTATT R: ACCTCCACCTAAGCGAGAAAC | [76] | [17,18,19,76] |
| PhyIII_63 2 | (AGC)n | F: GTGTGGTAATTTATGGGCGTTT R: CAAAGTGGGCACGATAAAATTG | [76] | [17,18,76] |
| PhyIII_65 2 | (ATT)n | F: TTTACTATCATAGCTTTCCACTTGAAC R: GGGTATTTTTGGGTTTAATTCTACTG | [76] | [17,18,76] |
| PhyIII_69 2 | (TAA)n (ATT)n | F: CTTTCTCTCCCGATTGTCCTT R: GGCCTTTAACGCGTAGGTAGAC | [76] | [18,19,76] |
| P hyIII_86 2 | (TAT)n | F: AACAAAGTCCACTTTCGCTGTT R: CACGGTCTGCATAAATCACTGT | [76] | [76] |
| PhyIII_87 2 | (ATT)n | F: TTCAGAATCGACGTCAGCTAAT R: CATTCGACTCTAGCAATACCAAA | [76] | [17,18,76] |
| PhyIV_4 2 | (AATA)n | F: CAGGCATCTCAAATGGATTAGC R: TGCGTCATTTCATTAACTTACACTT | [76] | [17,18,19,76,79] |
| Region | SSRs (n) | Samples (n) | Multilocus Genotypes (n) | Main Genetic Groups (n) | Ref. |
|---|---|---|---|---|---|
| Native range | 32 | 549 | 466 | 5 | [18] |
| California | 15 | 296 | 145 | 4 | [19] |
| Europe | 6 | 360 | 195 | - | [30] |
| Switzerland-Germany | 7 | 335 | 203 | 1 | [31] |
| Austria | 6 | 315 | 223 | - | [58] |
| Australia | 4 | 361 | 45 | - | [33] |
| Uruguay | 4 | 69 | - | - | [57] |
| Argentina | 21 | 129 | 17 | 2 | [17] |
| China | 7 | 31 | 15 | 2 | [80] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tello, J.; Forneck, A. Use of DNA Markers for Grape Phylloxera Population and Evolutionary Genetics: From RAPDs to SSRs and Beyond. Insects 2019, 10, 317. https://doi.org/10.3390/insects10100317
Tello J, Forneck A. Use of DNA Markers for Grape Phylloxera Population and Evolutionary Genetics: From RAPDs to SSRs and Beyond. Insects. 2019; 10(10):317. https://doi.org/10.3390/insects10100317
Chicago/Turabian StyleTello, Javier, and Astrid Forneck. 2019. "Use of DNA Markers for Grape Phylloxera Population and Evolutionary Genetics: From RAPDs to SSRs and Beyond" Insects 10, no. 10: 317. https://doi.org/10.3390/insects10100317
APA StyleTello, J., & Forneck, A. (2019). Use of DNA Markers for Grape Phylloxera Population and Evolutionary Genetics: From RAPDs to SSRs and Beyond. Insects, 10(10), 317. https://doi.org/10.3390/insects10100317





