Serotonin 2A Receptor SNP rs7330461 Association with Treatment Response to Pomaglumetad Methionil in Patients with Schizophrenia
Abstract
:1. Introduction
| Study | Author, Year | Duration (Weeks) | Study Design | Treatments |
|---|---|---|---|---|
| Study 2 | Adams et al. 2013 [7] | 24 weeks + 28-week extension | Phase 2, randomized, open-label | Pomaglumetad methionil (target: 40 mg BID) SOC (olanzapine (10, 15, or 20 mg, once daily; target = 15 mg/day), aripiprazole (10, 20 or 30 mg, once daily; target = 20 mg/day) or risperidone (2, 4 or 6 mg, once daily or BID; target = 4 mg/day)) |
| Study 3 | Downing et al. 2014 [8] | 6 weeks | Phase 2, randomized, double-blind | Pomaglumetad methionil (40 mg BID or 80 mg BID) Risperidone (2 mg BID) Placebo |
| Study 4 | Adams et al. 2014 [9] | 24 weeks | Phase 3, randomized, double-blind | Pomaglumetad methionil (flexible dosing: 20 mg to 80 mg BID) Aripiprazole (flexible dosing: 10, 15 or 30 mg) |
| Study 5 | Published on ClinicalTrials.gov., NCT01307800 | 6 weeks | Phase 3, randomized, double-blind | Pomaglumetad methionil (10 mg BID, 40 mg BID or 80 mg BID) Placebo |
2. Results and Discussion
2.1. Patient Baseline Demographics and Illness Characteristics in Studies 2 through 5
| Study | A/A | A/T | T/T | Overall p-Value | |
|---|---|---|---|---|---|
| Study 2 | Caucasian patients | N = 51 | N = 48 | N = 17 | |
| Age, mean (SD) years | 38.2 (11.8) | 41.5 (12.7) | 38.1 (10.6) | 0.344 1 | |
| Sex, male, n (%) | 32 (62.7) | 26 (54.2) | 13 (76.5) | 0.274 2 | |
| PANSS total score, mean (SD) | 85.3 (11.5) | 81.9 (12.2) | 79.5 (11.3) | 0.154 1 | |
| African American patients | N = 20 | N = 34 | N = 25 | ||
| Age, mean (SD) years | 44.9 (10.0) | 41.8 (11.9) | 41.0 (11.7) | 0.490 1 | |
| Sex, male, n (%) | 17 (85.0) | 28 (82.4) | 19 (76.0) | 0.814 2 | |
| PANSS total score, mean (SD) | 88.2 (11.4) | 83.5 (11.8) | 81.5 (14.5) | 0.206 1 | |
| Study 3 | Caucasian patients | N = 265 | N = 215 | N = 69 | |
| Age, mean (SD), years | 39.0 (11.9) | 41.3 (12.3) | 36.9 (11.2) | 0.016 1 | |
| Sex, male, n (%) | 152 (57.4) | 116 (54.0) | 50 (72.5) | 0.023 2 | |
| PANSS total score, mean (SD) | 84.3 (14.7) | 84.1 (13.9) | 88.0 (20.5) | 0.148 1 | |
| African American patients | N = 64 | N = 146 | N = 96 | ||
| Age, mean (SD) years | 41.9 (11.5) | 40.3 (11.1) | 40.8 (10.5) | 0.606 1 | |
| Sex, male, n (%) | 48 (75.0) | 105 (71.9) | 67 (69.8) | 0.782 2 | |
| PANSS total score, mean (SD) | 84.7 (13.6) | 82.1 (13.2) | 81.7 (13.5) | 0.328 1 | |
| Study 4 | Caucasian patients | N = 112 | N = 126 | N = 30 | |
| Age, mean (SD), years | 42.2 (12.1) | 43.0 (11.7) | 42.3 (12.1) | 0.881 1 | |
| Sex, male, n (%) | 63 (56.3) | 83 (65.9) | 19 (63.3) | 0.303 2 | |
| PANSS total score, mean (SD) | 83.1 (29.0) | 83.5 (32.9) | 84.0 (28.5) | 0.990 1 | |
| African American patients | N = 53 | N = 153 | N = 96 | ||
| Age, mean (SD), years | 44.1 (9.1) | 42.5 (10.0) | 43.4 (10.4) | 0.537 1 | |
| Sex, male, n (%) | 33 (62.3) | 100 (65.4) | 63 (65.6) | 0.909 2 | |
| PANSS total score, mean (SD) | 69.3 (16.5) | 74.5 (15.9) | 75.5 (15.3) | 0.060 1 | |
| Study 5 | Caucasian patients | N = 85 | N = 77 | N = 20 | |
| Age, mean (SD), years | 40.5 (10.9) | 41.5 (11.5) | 42.4 (10.1) | 0.751 1 | |
| Sex, male, n (%) | 59 (69.4) | 53 (68.8) | 12 (60.0) | 0.705 2 | |
| PANSS total score, mean (SD) | 80.8 (14.7) | 82.0 (13.6) | 80.6 (9.1) | 0.824 1 | |
| African American patients | N = 57 | N = 118 | N = 89 | ||
| Age, mean (SD), years | 39.9 (11.1) | 42.7 (10.5) | 41.4 (11.3) | 0.275 1 | |
| Sex, male, n (%) | 42 (73.7) | 82 (69.5) | 68 (76.4) | 0.557 2 | |
| PANSS total score, mean (SD) | 81.4 (15.3) | 82.1 (12.7) | 81.7 (13.4) | 0.938 1 | |
2.2. Effect of SNP rs7330461 Genotype in Studies 1, 2, 3 and 4
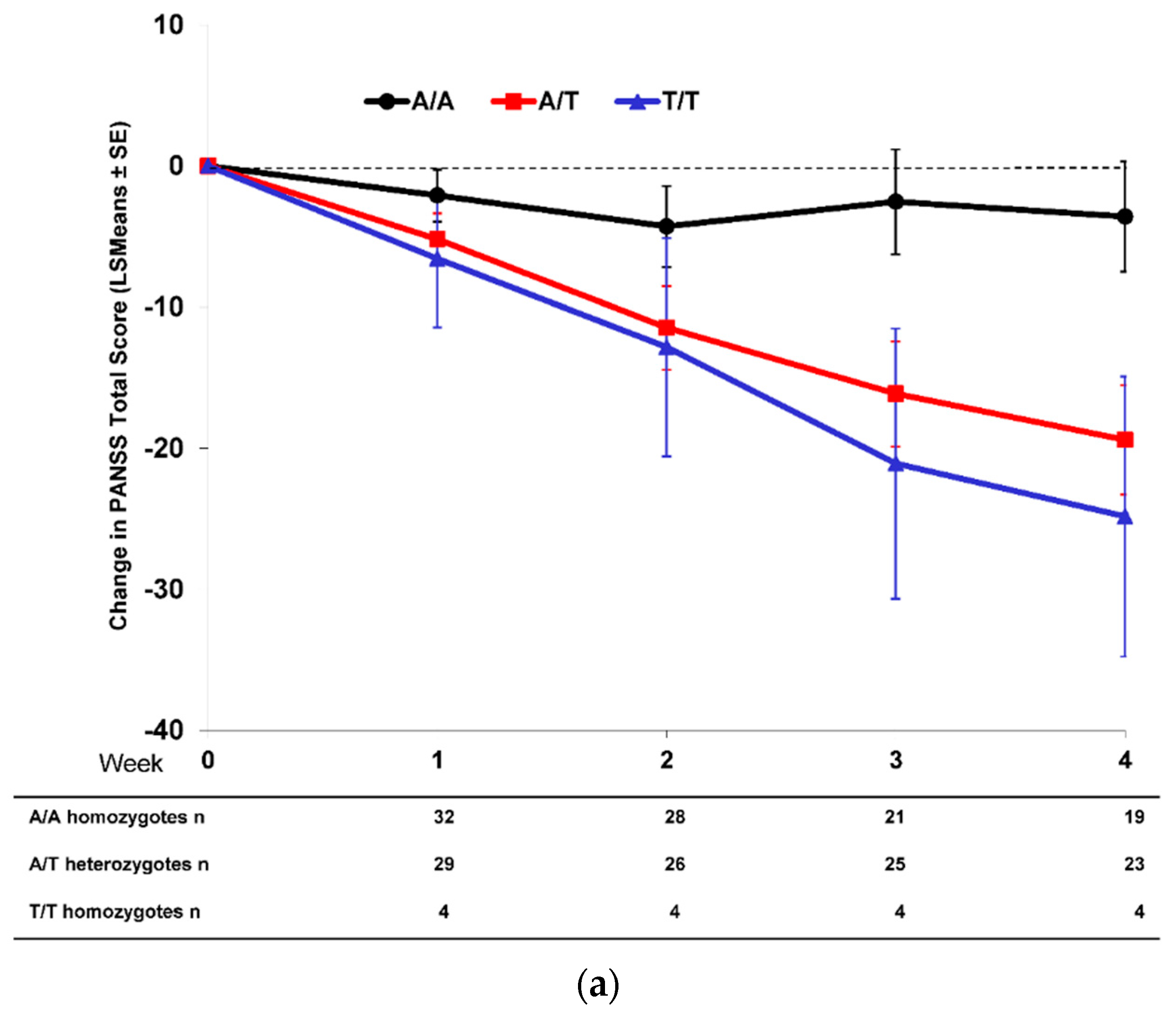
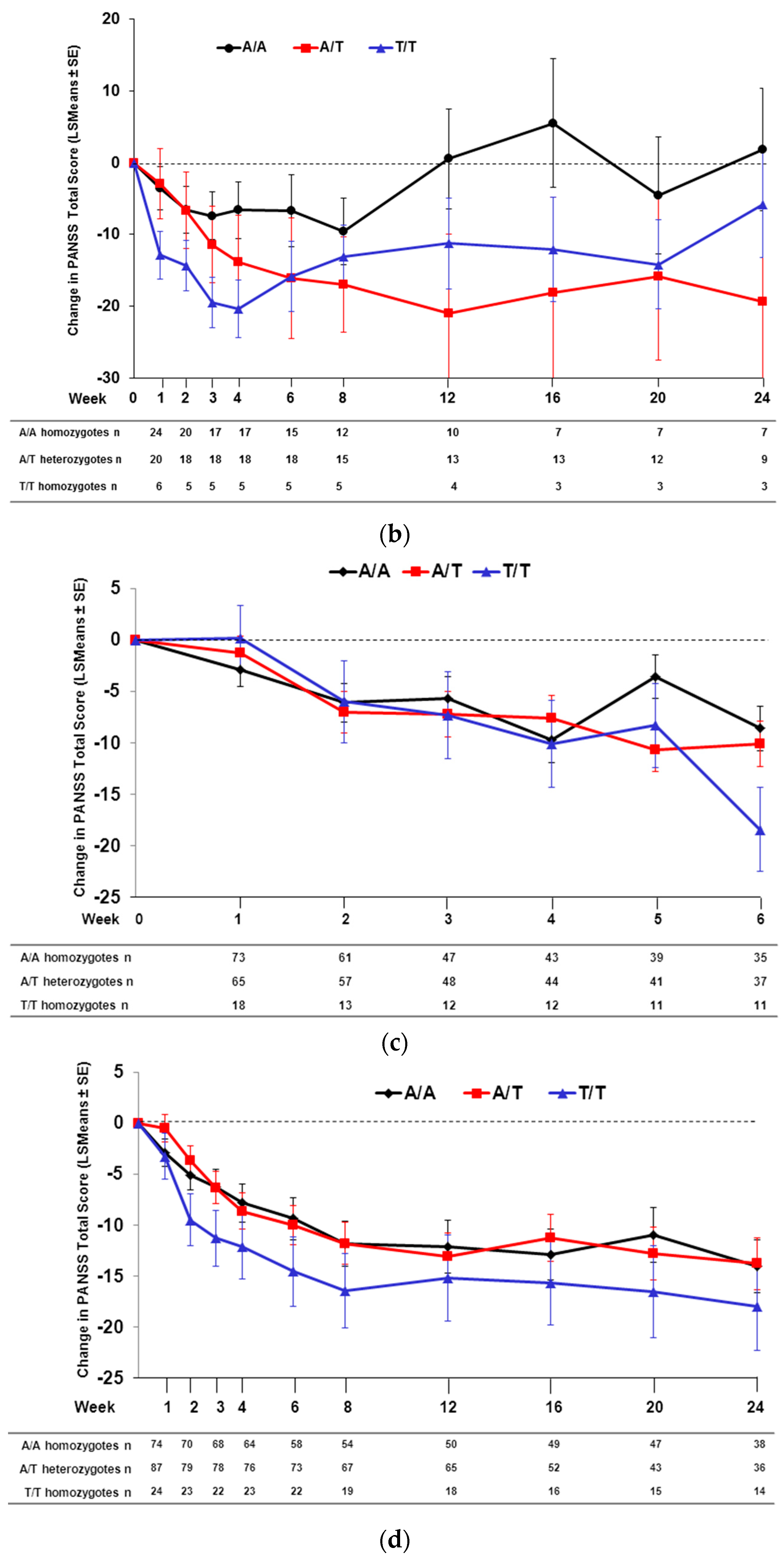
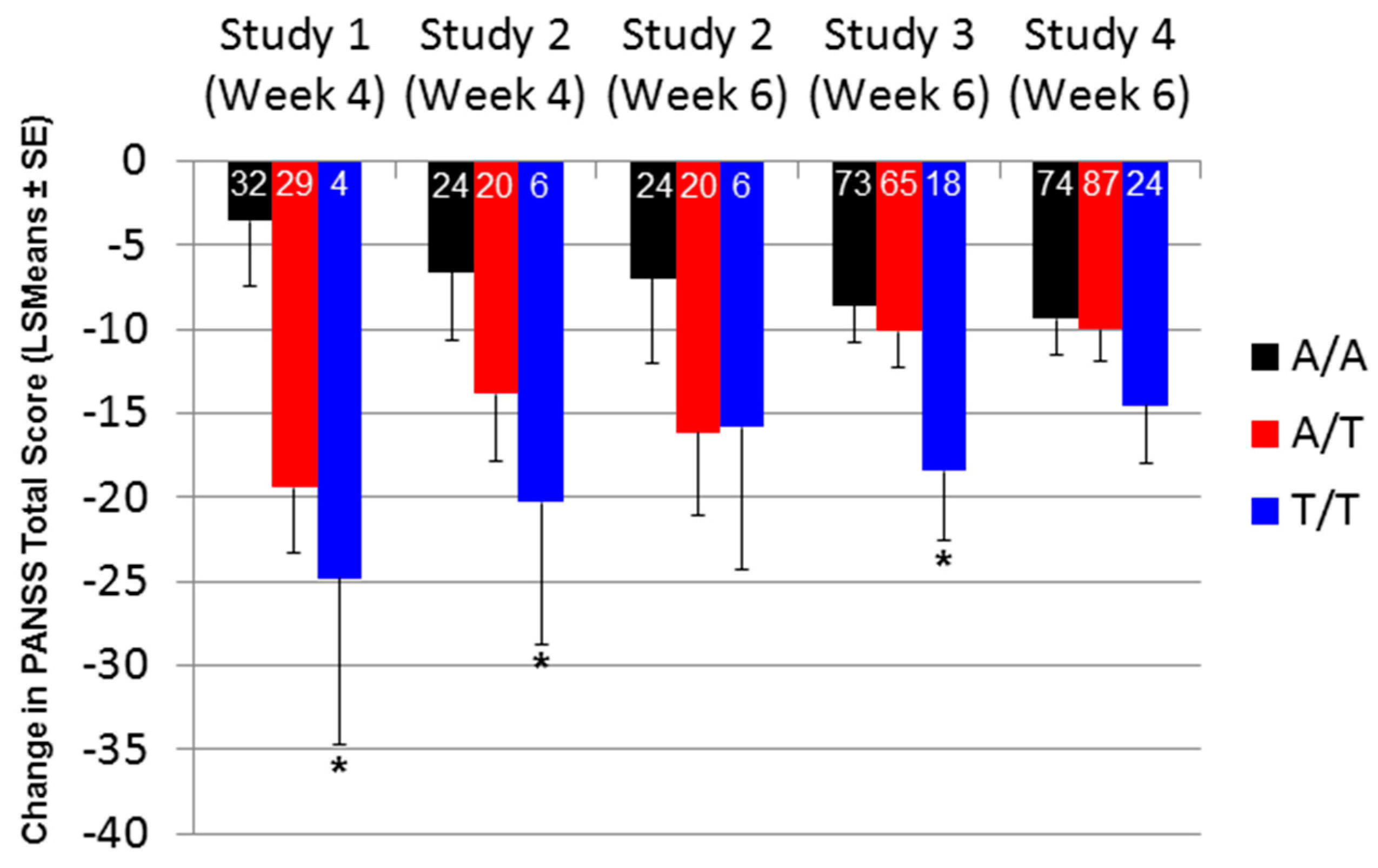
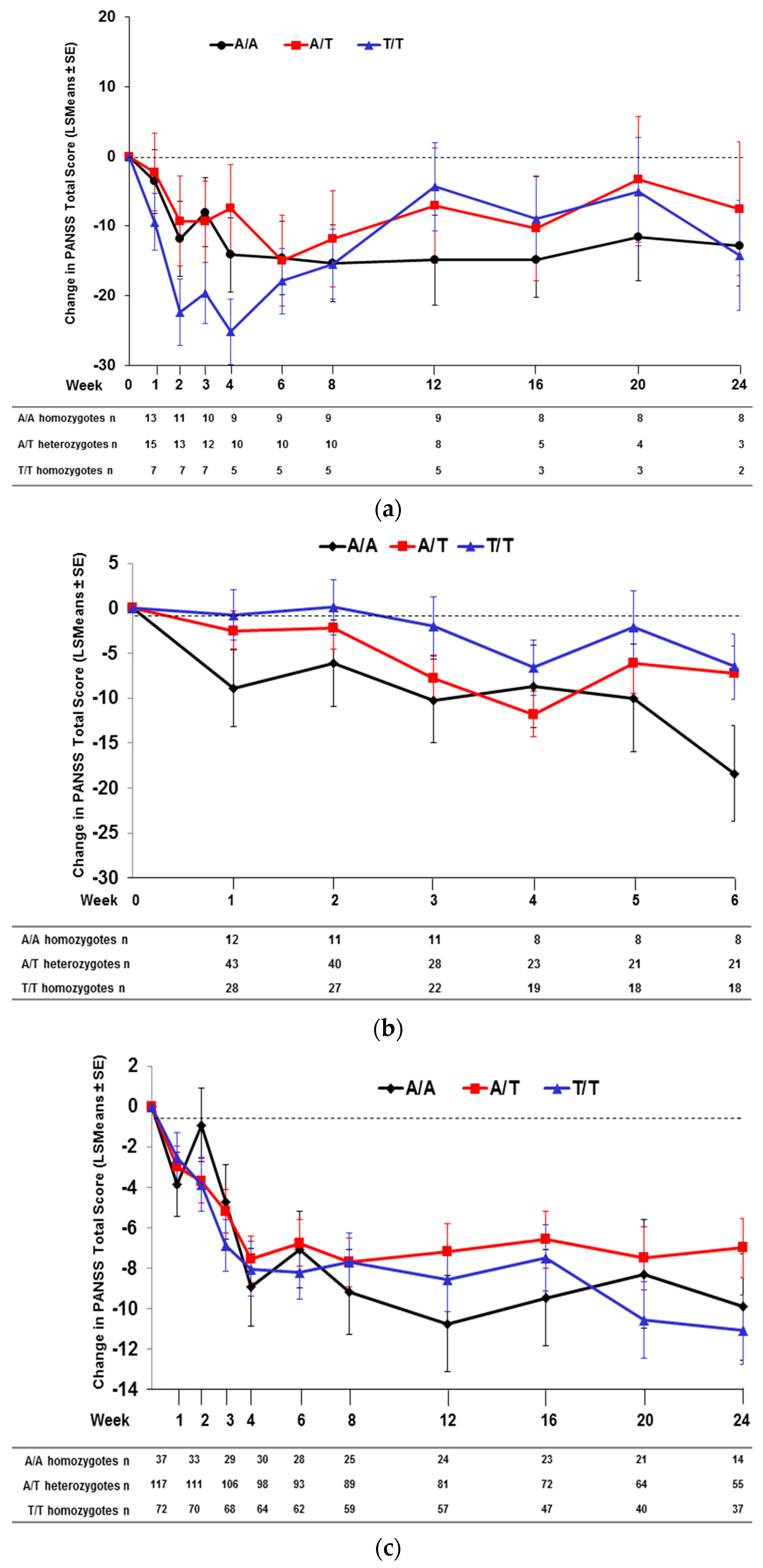
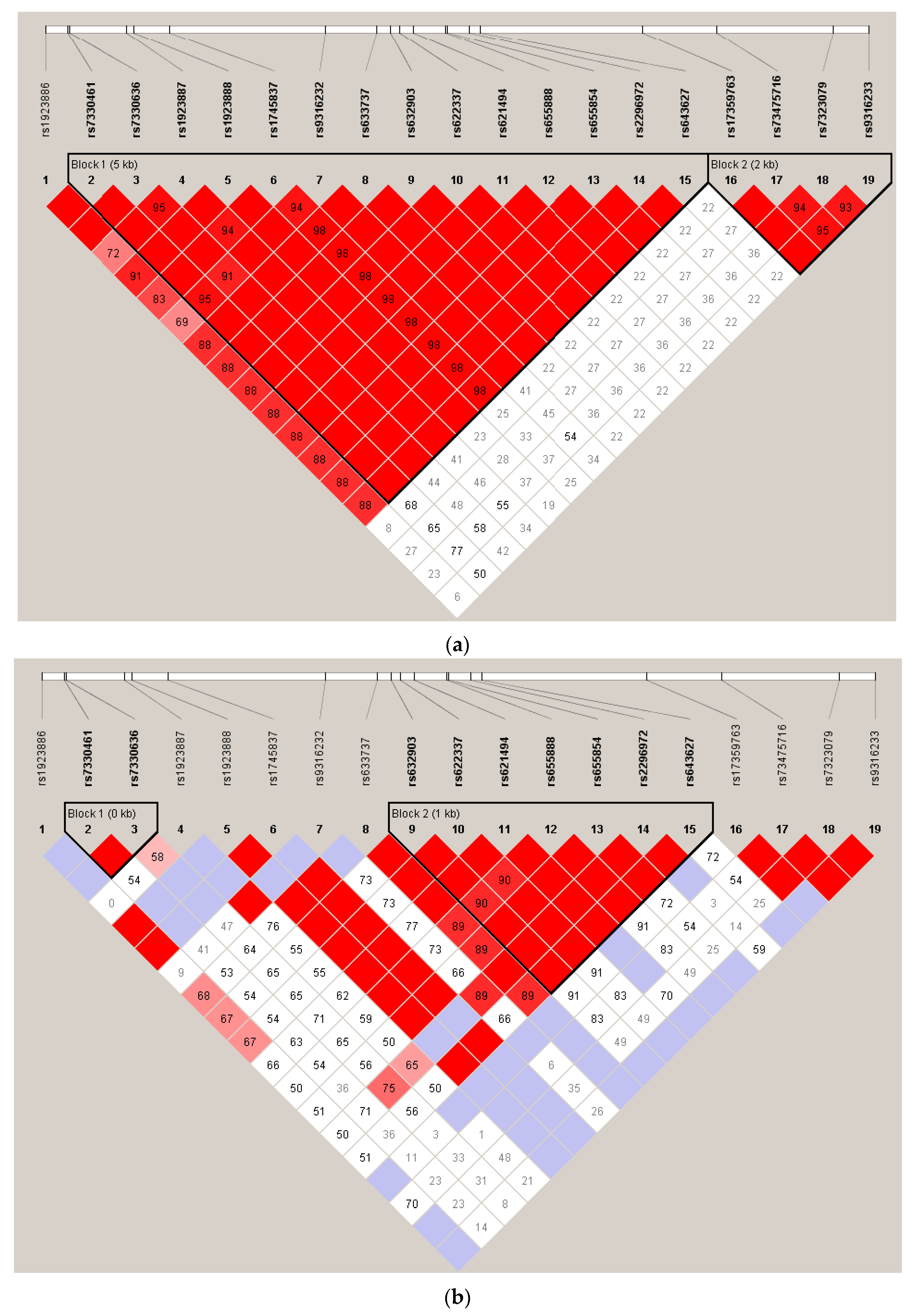
2.3. Treatment Response in Caucasian Patients Homozygous for the T/T Genotype in Study 3
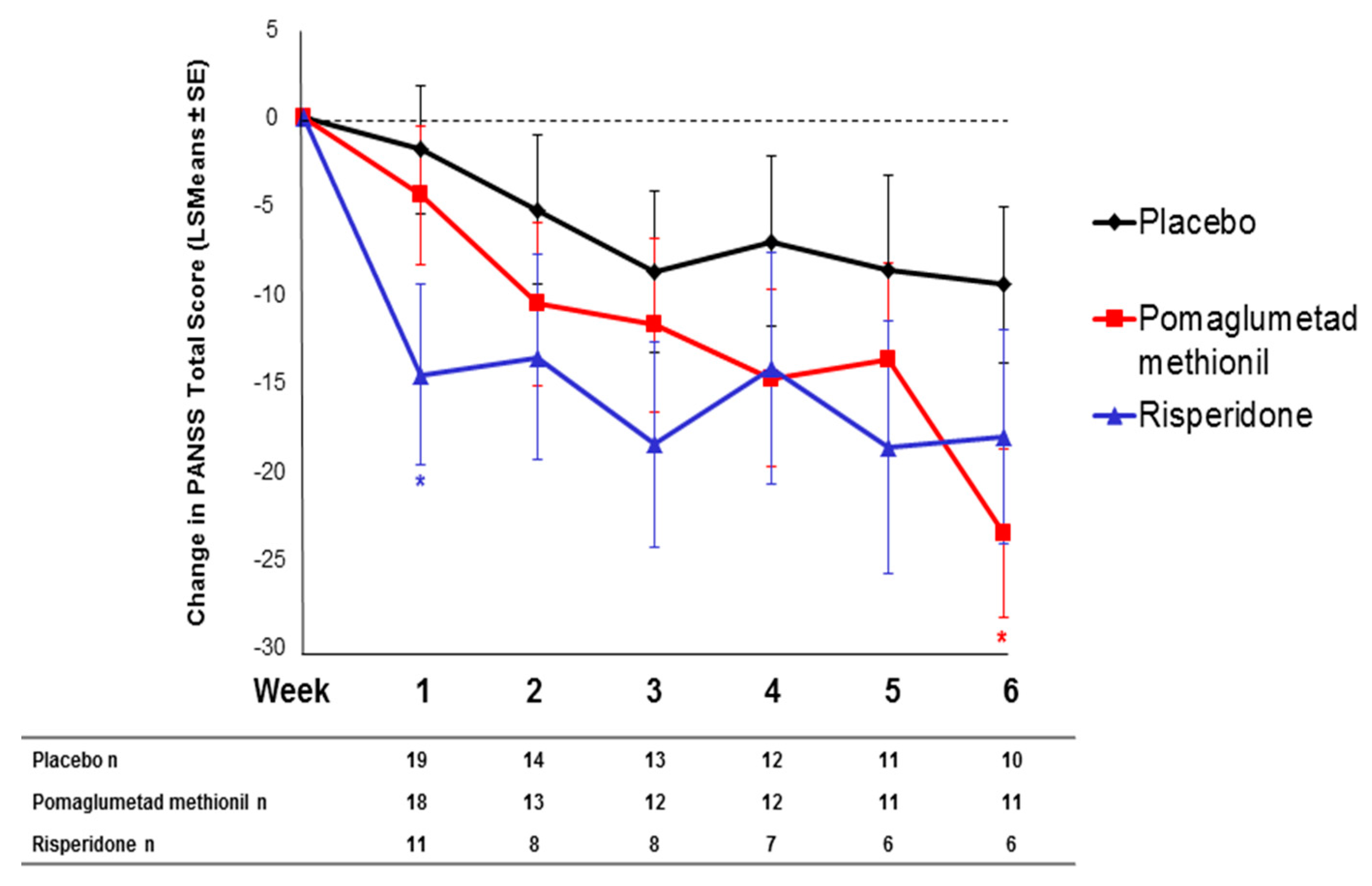
2.4. Effect of SNP rs7330461 Genotype in the Integrated Analysis
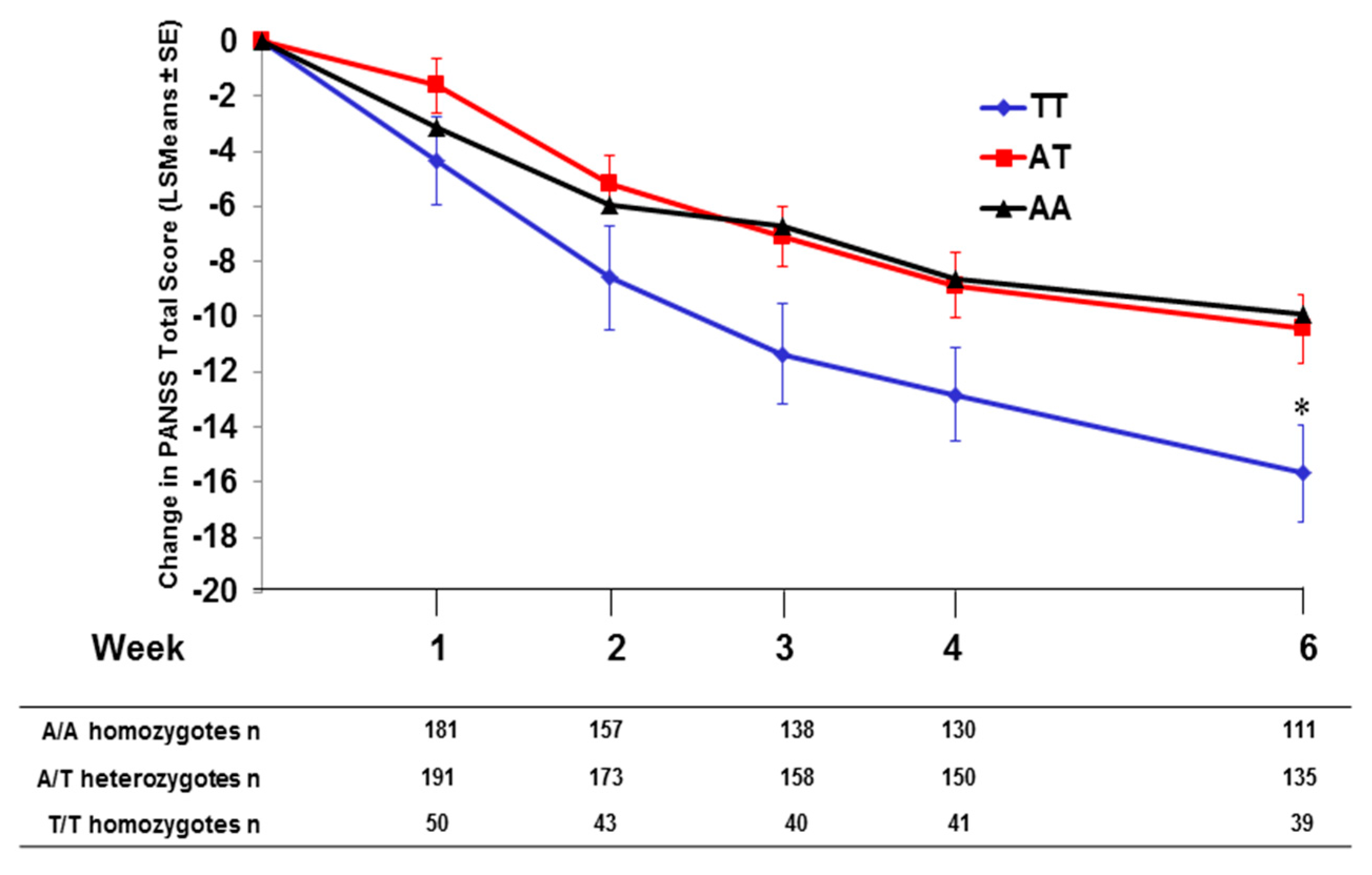
2.5. Pomaglumetad Methionil versus Placebo in the Integrated Analysis
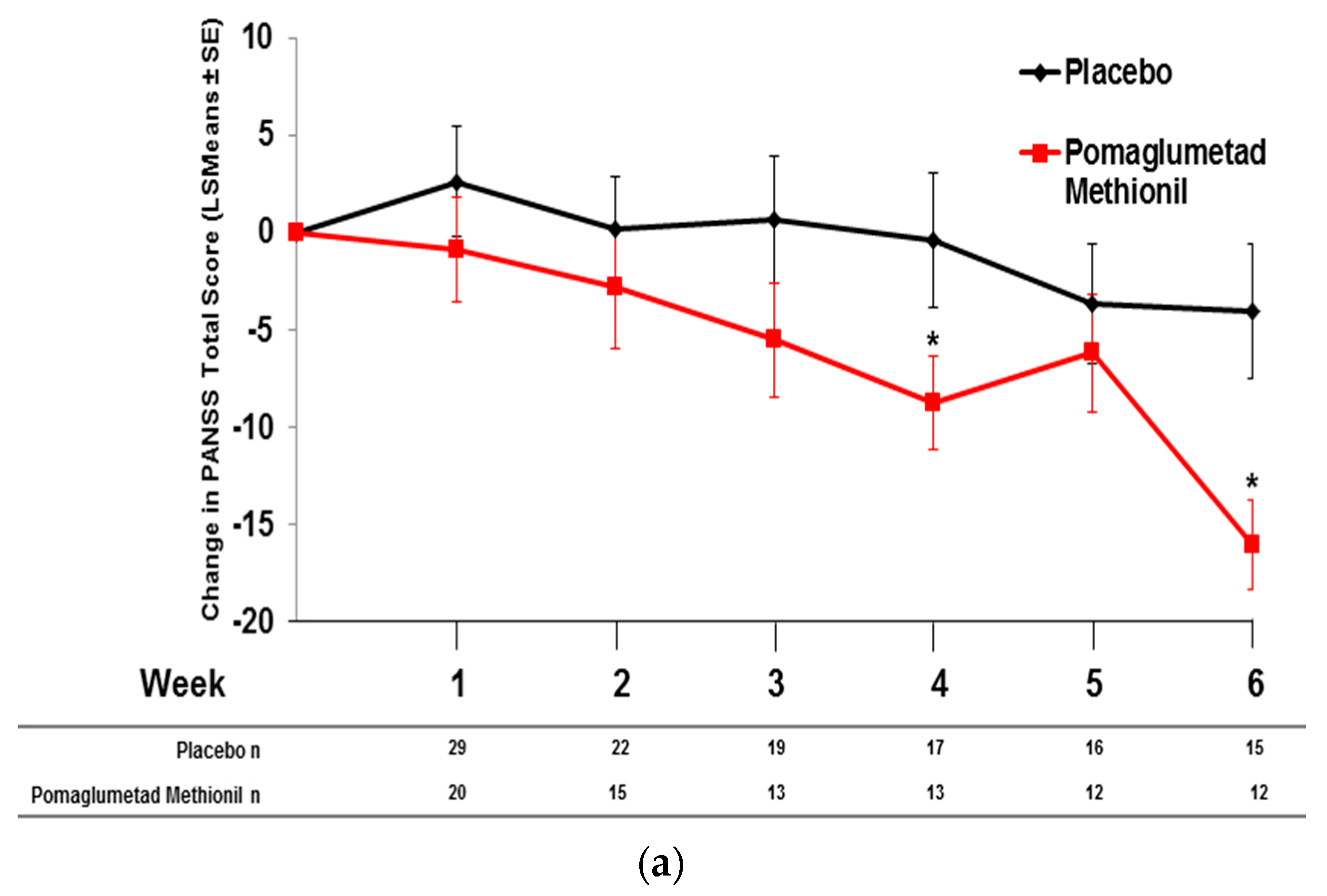
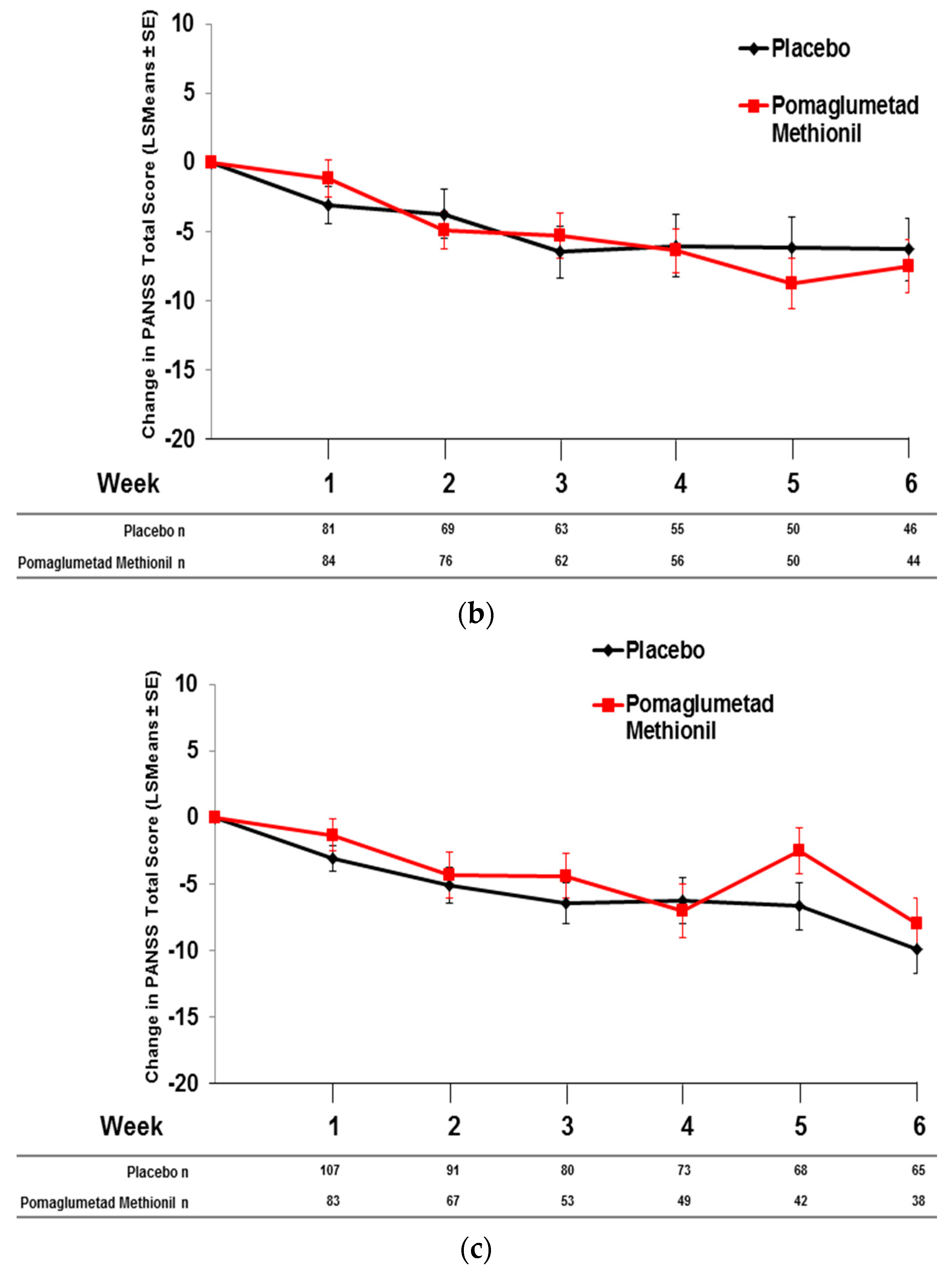
2.6. Pomaglumetad Methionil versus Standard-Of-Care in the Integrated Analysis

2.7. Potential Functional Effect of HTR2A SNP 7330461
2.8. Integration of Pharmacogenetic Analyses during Clinical Drug Development
3. Experimental Section
3.1. Clinical Trial Designs and Patients
3.2. Genotyping
3.3. Genotype Data Quality Control
| Study | Patients with rs7330461 Genotype, n/N (%) | HWE for rs7330461 |
|---|---|---|
| 2 | 253/261 (96.9) | 0.303 |
| 3 | 1011/1013 (99.8) | 0.105 |
| 4 | 676/678 (99.7) | 0.591 |
| 5 | 559/567 (98.6) | 0.513 |
3.4. Statistical Analyses
Integrated Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Rossler, W.; Salize, H.J.; van Os, J.; Riecher-Rössler, A. Size of burden of schizophrenia and psychotic disorders. Eur. Neuropsychopharmacol. 2005, 15, 399–409. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Faucett, J.; Morrison, S.; Brown, W.A. Comparative mortality risk in adult patients with schizophrenia, depression, bipolar disorder, anxiety disorders, and attention-deficit/hyperactivity disorder participating in psychopharmacology clinical trials. JAMA Psychiatry 2013, 70, 1091–1099. [Google Scholar] [CrossRef] [PubMed]
- Haro, J.M.; Novick, D.; Bertsch, J.; Karagianis, J.; Dossenbach, M.; Jones, P.B. Cross-national clinical and functional remission rates: Worldwide Schizophrenia Outpatient Health Outcomes (W-SOHO) study. Br. J. Psychiatry 2011, 199, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Patil, S.T.; Zhang, L.; Martenyi, F.; Lowe, S.L.; Jackson, K.A.; Andreev, B.V.; Avedisova, A.S.; Bardenstein, L.M.; Gurovich, I.Y.; Morozova, M.A.; et al. Activation of mGlu2/3 receptors as a new approach to treat schizophrenia: A randomized phase 2 clinical trial. Nat. Med. 2007, 13, 1102–1107. [Google Scholar] [CrossRef] [PubMed]
- Kinon, B.J.; Zhang, L.; Millen, B.A.; Osuntokun, O.O.; Williams, J.E.; Kollack-Walker, S.; Jackson, K.; Kryzhanovskaya, L.; Jarkova, N.; HBBI Study Group. A multicenter, inpatient, phase 2, double-blind, placebo-controlled dose-ranging study of LY2140023 monohydrate in patients with DSM-IV schizophrenia. J. Clin. Psychopharmacol. 2011, 31, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Mosolov, S.N.; Smulevich, A.B.; Neznanov, N.G.; Tochilov, V.A.; Andreev, B.V.; Avedisova, A.S.; Bardenshteĭn, L.M.; Gurovich, I.; Reznik, A.M.; Zharkova, N.B.; et al. The use of mGlu2/3 receptors as a new approach to treat schizophrenia: Results of a randomized double-blind trial. Zh. Nevrol. Psikhiatr. Im. S S. Korsakova. 2010, 110, 16–23. [Google Scholar] [PubMed]
- Adams, D.H.; Kinon, B.J.; Baygani, S.; Millen, B.A.; Velona, I.; Kollack-Walker, S.; Walling, D.P. A long-term, phase 2, multicenter, randomized, open-label, comparative safety study of LY2140023 versus atypical antipsychotic standard of care in patients with schizophrenia. BMC Psychiatry 2013, 13, 143. [Google Scholar] [CrossRef] [PubMed]
- Downing, A.M.; Kinon, B.J.; Millen, B.A.; Zhang, L.; Liu, L.; Morozova, M.A.; Brenner, R.; Rayle, T.J.; Nisenbaum, L.; Zhao, F.; et al. A double-blind, placebo-controlled comparator study of LY2140023 monohydrate in patients with schizophrenia. BMC Psychiatry 2014, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.H.; Zhang, L.; Millen, B.A.; Kinon, B.J.; Gomez, J.-C. Pomaglumetad methionil (LY2140023) and aripiprazole in patients with schizophrenia: A phase 3, multicenter, double-blind comparison. Schizophr. Res. Treatment 2014, 2014, 758212. [Google Scholar] [CrossRef] [PubMed]
- Eli Lilly and Company. ClinicalTrials.gov Identifier: NCT01307800—A Study of LY2140023 in Patients with Schizophrenia. 2013. Available online: https://clinicaltrials.gov/ct2/show/NCT01307800? (accessed on 15 December 2015). [Google Scholar]
- Liu, W.; Downing, A.M.; Munsie, L.M.; Chen, P.; Reed, M.R.; Ruble, C.L.; Landschulz, K.T.; Kinon, B.J.; Nisenbaum, L.K. Pharmacogenetic analysis of the mGlu2/3 agonist LY2140023 monohydrate in the treatment of schizophrenia. Pharmacogenomics J. 2012, 12, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Wischhof, L.; Koch, M. 5-HT2A and mGlu2/3 receptor interactions: On their relevance to cognitive function and psychosis. Behav. Pharmacol. 2015, 27, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Kinon, B.J.; Millen, B.A.; Zhang, L.; McKinzie, D.L. Exploratory analysis for a targeted patient population responsive to the metabotropic glutamate 2/3 receptor agonist pomaglumetad methionil in schizophrenia. Biol. Psychiatry 2015, 78, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar]
- The Broad Institute. Welcome to Ricopili. Available online: https://www.broadinstitute.org/mpg/ricopili/ (accessed on 15 December 2015).
- Rosenbloom, K.R.; Sloan, C.A.; Malladi, V.S.; Dreszer, T.R.; Learned, K.; Kirkup, V.M.; Wong, M.C.; Maddren, M.; Fang, R.; Heitner, S.G.; et al. ENCODE data in the UCSC genome browser: Year 5 update. Nucleic Acids Res. 2013, 41, D56–D63. [Google Scholar] [CrossRef] [PubMed]
- Fribourg, M.; Moreno, J.L.; Holloway, T.; Provasi, D.; Baki, L.; Mahajan, R.; Mahajan, R.; Park, G.; Adney, S.K.; Hatcher, C.; et al. Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell 2011, 147, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Kurita, M.; Holloway, T.; Garcia-Bea, A.; Kozlenkov, A.; Friedman, A.K.; Moreno, J.L.; Heshmati, M.; Golden, S.A.; Kennedy, P.J.; Takahashi, N.; et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat. Neurosci. 2012, 15, 1245–1254. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.L.; Holloway, T.; Albizu, L.; Sealfon, S.C.; González-Maeso, J. Metabotropic glutamate mGlu2 receptor is necessary for the pharmacological and behavioral effects induced by hallucinogenic 5-HT2A receptor agonists. Neurosci. Lett. 2011, 493, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Maeso, J.; Ang, R.L.; Yuen, T.; Chan, P.; Weisstaub, N.V.; López-Giménez, J.F.; Zhou, M.; Okawa, Y.; Callado, L.F.; Milligan, G.; et al. Identification of a serotonin/glutamate receptor complex implicated in psychosis. Nature 2008, 452, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Office of the Assistant Secretary for Planning and Evaluation. U.S. department of health and human services implementation guidance on data collection standards for race, ethnicity, sex, primary language, and disability status, 2011. U.S. Department of Health and Human Services. Available online: http://aspe.hhs.gov/datacncl/standards/ACA/4302/index.pdf (accessed 15 December 2015).
- Edwards, D.G.; Hsu, J.C. Multiple comparisons with the best treatment. J. Amer. Statist. Assoc. 1983, 78, 965–971. [Google Scholar] [CrossRef]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nisenbaum, L.K.; Downing, A.M.; Zhao, F.; Millen, B.A.; Munsie, L.; Kinon, B.J.; Adams, D.H.; Gomez, J.C.; Penny, M.A. Serotonin 2A Receptor SNP rs7330461 Association with Treatment Response to Pomaglumetad Methionil in Patients with Schizophrenia. J. Pers. Med. 2016, 6, 9. https://doi.org/10.3390/jpm6010009
Nisenbaum LK, Downing AM, Zhao F, Millen BA, Munsie L, Kinon BJ, Adams DH, Gomez JC, Penny MA. Serotonin 2A Receptor SNP rs7330461 Association with Treatment Response to Pomaglumetad Methionil in Patients with Schizophrenia. Journal of Personalized Medicine. 2016; 6(1):9. https://doi.org/10.3390/jpm6010009
Chicago/Turabian StyleNisenbaum, Laura K., AnnCatherine M. Downing, Fangyi Zhao, Brian A. Millen, Leanne Munsie, Bruce J. Kinon, David H. Adams, Juan Carlos Gomez, and Michelle Ann Penny. 2016. "Serotonin 2A Receptor SNP rs7330461 Association with Treatment Response to Pomaglumetad Methionil in Patients with Schizophrenia" Journal of Personalized Medicine 6, no. 1: 9. https://doi.org/10.3390/jpm6010009
APA StyleNisenbaum, L. K., Downing, A. M., Zhao, F., Millen, B. A., Munsie, L., Kinon, B. J., Adams, D. H., Gomez, J. C., & Penny, M. A. (2016). Serotonin 2A Receptor SNP rs7330461 Association with Treatment Response to Pomaglumetad Methionil in Patients with Schizophrenia. Journal of Personalized Medicine, 6(1), 9. https://doi.org/10.3390/jpm6010009




