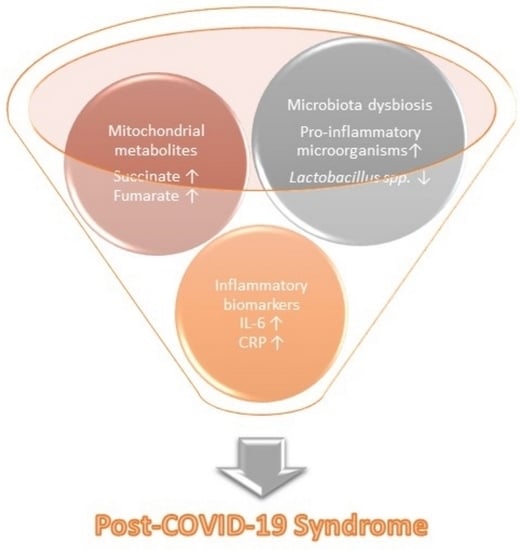
Promising Markers of Inflammatory and Gut Dysbiosis in Patients with Post-COVID-19 Syndrome
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Post-COVID-19 Rehabilitation Complex
2.3. Sample Collection
2.4. Sample Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinical Condition of Patients
3.2. Laboratory Parameters
3.3. Low-Molecular-Weight Metabolites in Serum
3.4. Gut Microbiota Taxonomy
3.5. COVID-19 Vaccination
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anaya, J.-M.; Rojas, M.; Salinas, M.L.; Rodríguez, Y.; Roa, G.; Lozano, M.; Rodríguez-Jiménez, M.; Montoya, N.; Zapata, E.; Monsalve, D.M.; et al. Post-COVID syndrome. A case series and comprehensive review. Autoimmun. Rev. 2021, 20, 102947. [Google Scholar] [CrossRef] [PubMed]
- Pasini, E.; Corsetti, G.; Romano, C.; Scarabelli, T.M.; Chen-Scarabelli, C.; Saravolatz, L.; Dioguardi, F.S. Serum Metabolic Profile in Patients with Long-COVID (PASC) Syndrome: Clinical Implications. Front. Med. 2021, 8, 714426. [Google Scholar] [CrossRef] [PubMed]
- Colarusso, C.; Maglio, A.; Terlizzi, M.; Vitale, C.; Molino, A.; Pinto, A.; Vatrella, A.; Sorrentino, R. Post-COVID-19 Patients Who Develop Lung Fibrotic-like Changes Have Lower Circulating Levels of IFN-β but Higher Levels of IL-1α and TGF-β. Biomedicines 2021, 9, 1931. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Tang, N.; Peluso, M.J.; Iyer, N.S.; Torres, L.; Donatelli, J.L.; Munter, S.E.; Nixon, C.C.; Rutishauser, R.L.; Rodriguez-Barraquer, I.; et al. Characterization and Biomarker Analyses of Post-COVID-19 Complications and Neurological Manifestations. Cells 2021, 10, 386. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, M.A.F.; das Neves, P.F.M.; Lima, S.S.; Lopes, J.D.C.; Torres, M.K.D.S.; Vallinoto, I.M.V.C.; Bichara, C.D.A.; dos Santos, E.F.; de Brito, M.T.F.M.; da Silva, A.L.S.; et al. Cytokine Profiles Associated with Acute COVID-19 and Long COVID-19 Syndrome. Front. Cell. Infect. Microbiol. 2022, 12, 931. [Google Scholar] [CrossRef] [PubMed]
- Yong, S.J.; Halim, A.; Halim, M.; Liu, S.; Aljeldah, M.; Al Shammari, B.R.; Alwarthan, S.; Alhajri, M.; Alawfi, A.; Alshengeti, A.; et al. Inflammatory and vascular biomarkers in post-COVID-19 syndrome: A systematic review and meta-analysis of over 20 biomarkers. Rev. Med. Virol. 2023, 33, e2424. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-J.; Liu, S.-H.; Manachevakul, S.; Lee, T.-A.; Kuo, C.-T.; Bello, D. Biomarkers in long COVID-19: A systematic review. Front. Med. 2023, 10, 7. [Google Scholar] [CrossRef]
- Lorkiewicz, P.; Waszkiewicz, N. Biomarkers of Post-COVID Depression. J. Clin. Med. 2021, 10, 4142. [Google Scholar] [CrossRef]
- Ghini, V.; Meoni, G.; Pelagatti, L.; Celli, T.; Veneziani, F.; Petrucci, F.; Vannucchi, V.; Bertini, L.; Luchinat, C.; Landini, G.; et al. Profiling metabolites and lipoproteins in COMETA, an Italian cohort of COVID-19 patients. PLoS Pathog. 2022, 18, e1010443. [Google Scholar] [CrossRef]
- Holmes, E.; Wist, J.; Masuda, R.; Lodge, S.; Nitschke, P.; Kimhofer, T.; Loo, R.L.; Begum, S.; Boughton, B.; Yang, R.; et al. Incomplete Systemic Recovery and Metabolic Phenoreversion in Post-Acute-Phase Nonhospitalized COVID-19 Patients: Implications for Assessment of Post-Acute COVID-19 Syndrome. J. Proteome Res. 2021, 20, 3315–3329. [Google Scholar] [CrossRef]
- Alharbi, K.S.; Singh, Y.; Hassan Almalki, W.; Rawat, S.; Afzal, O.; Alfawaz Altamimi, A.S.; Kazmi, I.; Al-Abbasi, F.A.; Alzarea, S.I.; Singh, S.K.; et al. Gut Microbiota Disruption in COVID-19 or Post-COVID Illness Association with severity biomarkers: A Possible Role of Pre/Pro-biotics in manipulating microflora. Chem. Biol. Interact. 2022, 358, 109898. [Google Scholar] [CrossRef] [PubMed]
- Jena, R. Effect of Strelnikova Exercise on Respiratory Parameters among Children with LRTI In Selected Hospital Bhubaneswar. Eur. J. Mol. Clin. Med. 2020, 7, 1058–1065. [Google Scholar]
- Kokhan, S.; Vlasava, S.; Kolokoltsev, M.; Bayankin, O.; Kispayev, T.; Trofimova, N.; Suslina, I.; Romanova, E. Postcovid physical rehabilitation at the sanatorium. J. Phys. Educ. Sport 2022, 22, 607–613. [Google Scholar] [CrossRef]
- Pautova, A.K.; Bedova, A.Y.; Sarshor, Y.N.; Beloborodova, N.V. Determination of Aromatic Microbial Metabolites in Blood Serum by Gas Chromatography–Mass Spectrometry. J. Anal. Chem. 2018, 73, 160–166. [Google Scholar] [CrossRef]
- Zurabov, F.M.; Chernevskaya, E.A.; Beloborodova, N.V.; Zurabov, A.Y.; Petrova, M.V.; Yadgarov, M.Y.; Popova, V.M.; Fatuev, O.E.; Zakharchenko, V.E.; Gurkova, M.M.; et al. Bacteriophage Cocktails in the Post-COVID Rehabilitation. Viruses 2022, 14, 2614. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Erta, M.; Quintana, A.; Hidalgo, J. Interleukin-6, a Major Cytokine in the Central Nervous System. Int. J. Biol. Sci. 2012, 8, 1254–1266. [Google Scholar] [CrossRef]
- Povar-Echeverría, M.; Auquilla-Clavijo, P.E.; Andrès, E.; Martin-Sánchez, F.J.; Laguna-Calle, M.V.; Calvo-Elías, A.E.; Lorenzo-Villalba, N.; Méndez-Bailón, M. Interleukin-6 Could Be a Potential Prognostic Factor in Ambulatory Elderly Patients with Stable Heart Failure: Results from a Pilot Study. J. Clin. Med. 2021, 10, 504. [Google Scholar] [CrossRef]
- Smolen, J.S.; Maini, R.N. Interleukin-6: A new therapeutic target. Arthritis Res. Ther. 2006, 8, S5. [Google Scholar] [CrossRef]
- Wei, Q.; Lin, H.; Wei, R.-G.; Chen, N.; He, F.; Zou, D.-H.; Wei, J.-R. Tocilizumab treatment for COVID-19 patients: A systematic review and meta-analysis. Infect. Dis. Poverty 2021, 10, 71. [Google Scholar] [CrossRef]
- Alvarez, M.; Trent, E.; Goncalves, B.D.S.; Pereira, D.G.; Puri, R.; Frazier, N.A.; Sodhi, K.; Pillai, S.S. Cognitive dysfunction associated with COVID-19: Prognostic role of circulating biomarkers and microRNAs. Front. Aging Neurosci. 2022, 14, 1164. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.; O’neill, L.A. Succinate: A metabolic signal in inflammation. Trends Cell Biol. 2014, 24, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Beloborodova, N.; Pautova, A.; Sergeev, A.; Fedotcheva, N. Serum Levels of Mitochondrial and Microbial Metabolites Reflect Mitochondrial Dysfunction in Different Stages of Sepsis. Metabolites 2019, 9, 196. [Google Scholar] [CrossRef] [PubMed]
- Pautova, A.K.; Samokhin, A.S.; Beloborodova, N.V.; Revelsky, A.I. Multivariate Prognostic Model for Predicting the Outcome of Critically Ill Patients Using the Aromatic Metabolites Detected by Gas Chromatography-Mass Spectrometry. Molecules 2022, 27, 4784. [Google Scholar] [CrossRef]
- Beloborodov, N.V.; Khodakova, A.S.; Bairamov, I.T.; Olenin, A.Y. Microbial origin of phenylcarboxylic acids in the human body. Biochemistry 2009, 74, 1350–1355. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef]
- Herebian, D.; Seibt, A.; Smits, S.H.J.; Rodenburg, R.J.; Mayatepek, E.; Distelmaier, F. 4-Hydroxybenzoic acid restores CoQ10 biosynthesis in human COQ2 deficiency. Ann. Clin. Transl. Neurol. 2017, 4, 902–908. [Google Scholar] [CrossRef]
- Nardini, M.; Natella, F.; Scaccini, C.; Ghiselli, A. Phenolic acids from beer are absorbed and extensively metabolized in humans. J. Nutr. Biochem. 2006, 17, 14–22. [Google Scholar] [CrossRef]
- Fukumoto, T.; Nishiumi, S.; Fujiwara, S.; Yoshida, M.; Nishigori, C. Novel serum metabolomics-based approach by gas chromatography/triple quadrupole mass spectrometry for detection of human skin cancers: Candidate biomarkers. J. Dermatol. 2017, 44, 1268–1275. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.Y.; Sim, G.Y.; Ahn, J.-H. Synthesis of 4-Hydroxybenzoic Acid Derivatives in Escherichia coli. J. Agric. Food Chem. 2020, 68, 9743–9749. [Google Scholar] [CrossRef]
- Chernevskaya, E.; Meglei, A.; Buyakova, I.; Kovaleva, N.; Gorshkov, K.; Zakharchenko, V.; Beloborodova, N. Taxonomic dysbiosis of gut microbiota and serum biomarkers reflect severity of central nervous system injury. Bull. Russ. State Med. Univ. 2020, 5, 54–61. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.-Y.; Zhang, F.; Liu, Q.; Li, A.Y.; Chung, A.C.; Cheung, C.P.; Tso, E.Y.; Fung, K.S.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients with COVID-19 during Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef] [PubMed]
- Petakh, P.; Kobyliak, N.; Kamyshnyi, A. Gut microbiota in patients with COVID-19 and type 2 diabetes: A culture-based method. Front. Cell. Infect. Microbiol. 2023, 13, 108. [Google Scholar] [CrossRef]
- Righi, E.; Lambertenghi, L.; Gorska, A.; Sciammarella, C.; Ivaldi, F.; Mirandola, M.; Sartor, A.; Tacconelli, E. Impact of COVID-19 and Antibiotic Treatments on Gut Microbiome: A Role for Enterococcus spp. Biomedicines 2022, 10, 2786. [Google Scholar] [CrossRef]
- Abed, J.; Maalouf, N.; Manson, A.L.; Earl, A.M.; Parhi, L.; Emgård, J.E.M.; Klutstein, M.; Tayeb, S.; Almogy, G.; Atlan, K.A.; et al. Colon Cancer-Associated Fusobacterium nucleatum May Originate from the Oral Cavity and Reach Colon Tumors via the Circulatory System. Front. Cell. Infect. Microbiol. 2020, 10, 400. [Google Scholar] [CrossRef]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; DeVinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Wolff, L.; Martiny, D.; Deyi, V.Y.M.; Maillart, E.; Clevenbergh, P.; Dauby, N. COVID-19–Associated Fusobacterium nucleatum Bacteremia, Belgium. Emerg. Infect. Dis. 2021, 27, 975–977. [Google Scholar] [CrossRef]
- Dhar, D.; Mohanty, A. Gut microbiota and COVID-19-possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef]
- Łoniewski, I.; Skonieczna-Żydecka, K.; Sołek-Pastuszka, J.; Marlicz, W. Probiotics in the Management of Mental and Gastrointestinal Post-COVID Symptomes. J. Clin. Med. 2022, 11, 5155. [Google Scholar] [CrossRef]
- Beloborodova, N.V.; Grechko, A.V.; Gurkova, M.M.; Zurabov, A.Y.; Zurabov, F.M.; Kuzovlev, A.N.; Megley, A.Y.; Petrova, M.V.; Popova, V.M.; Redkin, I.V.; et al. Adaptive Phage Therapy in the Treatment of Patients with Recurrent Pneumonia (Pilot Study). Gen. Reanimatol. 2021, 17, 4–14. [Google Scholar] [CrossRef]
- D’Ettorre, G.; Ceccarelli, G.; Marazzato, M.; Campagna, G.; Pinacchio, C.; Alessandri, F.; Ruberto, F.; Rossi, G.; Celani, L.; Scagnolari, C.; et al. Challenges in the Management of SARS-CoV2 Infection: The Role of Oral Bacteriotherapy as Complementary Therapeutic Strategy to Avoid the Progression of COVID-19. Front. Med. 2020, 7, 389. [Google Scholar] [CrossRef] [PubMed]
- Wisnivesky, J.P.; Govindarajulu, U.; Bagiella, E.; Goswami, R.; Kale, M.; Campbell, K.N.; Meliambro, K.; Chen, Z.; Aberg, J.A.; Lin, J.J. Association of Vaccination with the Persistence of Post-COVID Symptoms. J. Gen. Intern. Med. 2022, 37, 1748–1753. [Google Scholar] [CrossRef] [PubMed]
- Peghin, M.; De Martino, M.; Palese, A.; Gerussi, V.; Bontempo, G.; Graziano, E.; Visintini, E.; D’Elia, D.; Dellai, F.; Marrella, F.; et al. Post–COVID-19 syndrome and humoral response association after 1 year in vaccinated and unvaccinated patients. Clin. Microbiol. Infect. 2022, 28, 1140–1148. [Google Scholar] [CrossRef] [PubMed]
- Freund, O.; Eviatar, T.; Bornstein, G. Concurrent myopathy and inflammatory cardiac disease in COVID-19 patients: A case series and literature review. Rheumatol. Int. 2022, 42, 905–912. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Reference Values | Patients on Admission (n = 28) | Patients after 14 Days (n = 28) | p-Value |
|---|---|---|---|---|
| White Blood Cell Count (WBC), ×109/L | 3.8–11.8 | 5.6 (5.3–6.8), 3.0–10.9 n (c < RV) = 1 | 5.6 (4.9–6.1), 3.2–12.0 n (c > RV) = 1 n (c < RV) = 1 | 0.946 |
| Red Blood Cell Count (RBC), ×1012/L | 3.63–5.63 | 4.56 (4.18–5.15), 3.71–5.56 n (c > RV) = 6 | 4.74 (4.33–4.99), 3.99–5.50 n (c > RV) = 4 | 0.060 |
| Hemoglobin (Hb), g/L | 109–163 | 134 (125–150), 95–169 n (c > RV) = 7 n (c < RV) = 1 | 140 (129–147), 95–163 n (c > RV) = 6 n (c < RV) = 1 | 0.027 |
| Hematocrit (Hct), % | 31.2–47.1 | 40.6 (37.2–44.7), 31.1–49.6 n (c > RV) = 7 n (c < RV) = 1 | 42.6 (39.1–43.6), 30.7–48.1 n (c > RV) = 6 n (c < RV) = 1 | 0.027 |
| Mean Cell Volume (MCV), fL | 75.5–95.3 | 89.4 (86.2–92.4), 62.2–96.9 n (c > RV) = 3 n (c < RV) = 1 | 88.2 (86.5–92.6), 62.8–97.9 n (c > RV) = 2 n (c < RV) = 1 | 0.840 |
| Mean Cell Hemoglobin (MCH), pg/cell | 24.7–33.4 | 29.8 (28.5–30.9), 19.0–33.7 n (c > RV) = 1 n (c < RV) = 1 | 29.5 (28.8–30.7), 19.4–33.5 n (c > RV) = 1 n (c < RV) = 1 | 0.961 |
| Mean Cell Hemoglobin Concentration (MCHC), g/L | 323–356 | 333 (326–336), 305–348 n (c < RV) = 3 | 332 (330–336), 310–342 n (c < RV) = 3 | 0.807 |
| Red Blood Cell Distribution Width (RDW), % | 12.3–17.7 | 14.1 (13.0–15.0), 10.2–20.7 n (c > RV) = 1 n (c < RV) =2 | 14.25 (13.2–14.7), 12.4–21.2 n (c > RV) = 1 | 0.893 |
| Platelet Count (Plt), ×109/L | 179–408 | 232 (201–295), 72–436 n (c > RV) = 1 n (c < RV) = 3 | 249 (206–287), 140–437 n (c > RV) = 1 n (c < RV) = 3 | 0.162 |
| Mean Platelet Volume (MPV), fL | 7.9–10.8 | 9.3 (8.4–9.9), 7.5–10.8 n (c < RV) = 1 | 8.9 (8.4–9.7), 7.3–11.0 n (c < RV) = 1 | 0.807 |
| Neutrophil, % | 42.7–76.8 | 55.2 (48.1–59.5), 37.4–71.3 n (c < RV) = 2 | 54.3 (47.6–57.3), 34.9–74.6 n (c < RV) = 3 | 0.809 |
| Lymphocytes, % | 16.0–45.9 | 32.4 (29.1–40.2), 18.6–48.8 n (c > RV) = 3 | 33.4 (30.9–41.4), 12.1–52.5 n (c > RV) = 2 n (c < RV) = 1 | 0.809 |
| Monocytes, % | 4.3–10.9 | 8.3 (6.9–9.4), 5.9–12.7 n (c > RV) = 3 | 8.0 (6.9–10.1), 5.2–16.3 n (c > RV) = 2 | 0.764 |
| Eosinophils, % | 0.5–7.0 | 2.5 (1.5–3.6), 1.1–8.8 n (c > RV) = 1 | 2.3 (1.8–3.6), 0.9–13.3 n (c > RV) = 1 | 0.627 |
| Basophil, % | 0.2–1.3 | 0.7 (0.6–0.9), 0.1–1.7 n (c > RV) = 2 n (c< RV) = 1 | 0.9 (0.6–1.1), 0.4–5.3 n (c > RV) = 2 | 0.321 |
| Absolute Neutrophil, ×109/L | 1.9–8.2 | 3.3 (2.6–3.9), 1.7–6.2 n (c < RV) = 2 | 3.1 (2.4–3.6), 1.5–7.0 n (c < RV) = 2 | 0.493 |
| Absolute Lymphocytes, ×109/L | 1.1–3.1 | 1.9 (1.6–2.5), 0.8–3.5 n (c > RV) = 2 n (c < RV) = 2 | 1.9 (1.6–2.4), 0.7–3.7 n (c > RV) = 2 n (c < RV) = 3 | 0.605 |
| Absolute Monocytes, ×109/L | 0.2–0.9 | 0.5 (0.4–0.6), 0.3–0.8 | 0.5 (0.4–0.5), 0.3–1.0 n (c > RV) = 1 | 0.388 |
| Absolute Eosinophils, ×109/L | <0.5 | 0.2 (0.1–0.2), <0.1–0.6 n (c > RV) = 1 | 0.1 (0.1–0.2), <0.1–0.7 n (c > RV) = 1 | 0.110 |
| Absolute Basophil, ×109/L | <0.10 | 0.01 (<0.10–0.10), <0.10–0.10 | <0.10 (<0.10–0.10), <0.10–0.30 n (c > RV) = 2 | 0.110 |
| Erythrocyte Sedimentation Rate (ESR), mm/hr | <20 | 14 (8–23), 3–37 n (c > RV) = 7 | 12 (5–18), 2–40 n (c > RV) = 6 | 0.100 |
| Parameter | Reference Values | Patients on Admission (n = 28) | Patients after 14 Days (n = 28) | p-Value |
|---|---|---|---|---|
| Prothrombin time (PT), sec | 9.4–12.5 | 10.9 (10.4–11.8), 9.4–16.4 n (c > RV) = 4 | 11.0 (10.5–11.6), 9.2–15.5 n (c > RV) = 2 n (c < RV) = 1 | 0.039 |
| Prothrombin by Quik % | 80–140 | 118 (86–134), 66–167 n (c > RV) = 3 n (c < RV) = 2 | 115 (96–129), 75–173 n (c > RV) = 4 n (c < RV) = 2 | 0.224 |
| International Normalised Ratio (INR) | 0.90–1.20 | 1.03 (0.94–1.08), 0.86–1.50 n (c > RV) = 4 n (c < RV) = 1 | 1.01 (0.94–1.06), 0.85–1.42 n (c > RV) = 2 n (c < RV) = 1 | 0.073 |
| Fibrinogen Activity, g/L | 2.38–4.98 | 3.21 (2.55–3.44), 2.04–3.81 n (c < RV) = 3 | 3.10 (2.55–3.53), 2.22–4.61 n (c < RV) = 5 | 0.786 |
| Activated Partial Thromboplastin Time (PTT), sec | 25.0–36.5 | 29.0 (27.7–33.4), 23.3–41.4 n (c > RV) = 4 n (c < RV) = 2 | 28.7 (27.9–32.1), 23.8–39.4 n (c > RV) = 2 n (c < RV) = 1 | 0.306 |
| Thrombin time (TT), sec | 11.0–17.8 | 16.2 (13.4–17.6), 12.1–19.2 n (c > RV) = 5 | 16.9 (14.3–17.4), 13.1–18.7 n (c > RV) = 5 | 0.118 |
| D-dimer, μ/mL | <0.49 | 0.21 (0.14–0.32), 0.07–1.89 n (c > RV) = 3 | 0.24 (0.19–0.41), 0.10–1.35 n (c > RV) = 4 | 0.085 |
| Parameter | Reference Values | Patients on Admission (n = 28) | Patients after 14 Days (n = 28) | p-Value |
|---|---|---|---|---|
| Bilirubin, μmol/L | 5.0–21.0 | 14.3 (9.2–18.4), 5.4–38.1 n (c > RV) = 4 | 12.1 (9.0–14.5), 3.0–39.0 n (c > RV) = 3 n (c < RV) = 1 | 0.106 |
| Total Protein, g/L | 66.0–83.0 | 68.1 (66.8–72.4), 62.1–81.2 n (c < RV) = 1 | 71.3 (68.4–72.7), 61.3–75.6 n (c < RV) = 2 | 0.158 |
| Creatinine, μmol/L | 58.0–110.0 | 84.1 (79.6–92.4), 69.3–110.7 n (c > RV) = 1 | 87.2 (81.3–93.4), 62.8–120.4 n (c > RV) = 1 | 0.750 |
| Glucose, mmol/L | 4.1–5.9 | 5.3 (5.0–5.9), 4.1–9.1 n (c > RV) = 5 | 5.5 (5.0–6.0), 4.0–8.5 n (c > RV) = 7 n (c < RV) = 1 | 0.733 |
| Cholesterol, mmol/L | < 5.2 | 5.4 (4.4–6.5), 2.4–8.4 n (c > RV) = 16 | 5.3 (4.3–6.3), 2.3–7.6 n (c > RV) = 14 | 0.234 |
| Lactate Dehydrogenase (LDH), U/L | <247.0 | 194.0 (164.4–213.1), 131.4–306.3 n (c > RV) = 2 | 197.5 (166.3–226.8), 128.7–334.7 n (c > RV) = 3 | 0.046 |
| Alanine Transaminase (ALT), U/L | <50.0 | 19.7 (14.1–28.4), 9.2–44.5 n (c > RV) = 2 | 19.2 (14.4–28.8), 8.8–68.0 n (c > RV) = 3 | 0.232 |
| Aspartate Transaminase (AST), U/L | <50.0 | 20.3 (18.8–23.9),15.5–91.7 n (c > RV) = 3 | 22.6 (19.1–29.5), 14.0–121.4 n (c > RV) = 3 | 0.005 |
| C-Reactive Protein (CRP), mg/L | <5.0 | 0.6 (0.1–0.9), 0.1–14.0 n (c > RV) = 4 | 0.6 (0.1–0.8), 0.1–8.4 n (c > RV) = 2 | 0.925 |
| Uric acid, μmol/L | 154.7–428.0 | 321.6 (256.0–386.7), 196.5–516.0 n (c > RV) = 8 | 321.1 (243.1–385.9),157.3–567.7 n (c > RV) = 6 | 0.524 |
| Interleukin-6 (IL-6), pg/mL | <7.0 | 11.7 (8.1–15.5), 1.5–61.9 n (c > RV) = 23 | 12.2 (8.7–17.7), 2.7–58.2 n (c > RV) = 25 | 0.255 |
| Neuron-specific Enolase (NSE), ng/mL | <16.0 | 9.3 (4.9–11.8), 0.1–22.0 n (c > RV) = 2 | 9.6 (5.7–13.0), 0.1–48.0 n (c > RV) = 4 | 0.038 |
| Acid, µmol/L | Healthy Volunteers (n = 48) | Patients on Admission (n = 28) | Patients after 14 Days (n = 28) | p-Value |
|---|---|---|---|---|
| Benzoic | <0.5 (<0.5–<0.5), <0.5–0.6 n (c > 0.5) = 2 | <0.5 (<0.5–<0.5), <0.5–0.8 n (c > 0.5) = 6 | <0.5 (<0.5–0.5), <0.5–1.0 n (c > 0.5) = 7 | - |
| Phenylpropionic | <0.5 (<0.5–0.5), <0.5–3.0 n (c > 0.5) = 15 | <0.5 (<0.5–0.5), <0.5–2.4 n (c > 0.5) = 8 | 0.5 (<0.5–0.8), <0.5–2.0 n (c > 0.5) = 15 | - |
| Phenyllactic | <0.5 (<0.5–<0.5), <0.5–0.7 n (c > 0.5) = 2 | not detected | <0.5 (<0.5–<0.5), <0.5–0.5 n (c > 0.5) = 1 | - |
| 4-Hydroxybenzoic | not detected | 6.8 (5.2–8.6), 2.0–13.0 n (c > 0.5) = 28 | 3.3 (1.5–6.7), 1.1–13.6 n (c > 0.5) = 28 | 0.003 |
| 4-Hydroxyphenylacetic | <0.5 (<0.5–<0.5), <0.5–1.2 n (c > 0.5) = 5 | <0.5 (<0.5–0.5), <0.5–1.6 n (c > 0.5) = 8 | <0.5 (<0.5–<0.5), <0.5–0.8 n (c > 0.5) = 6 | - |
| 4-Hydroxyphenyllactic | 1.2 (0.9–1.5), 0.7–2.5 n (c > 0.5) = 48 | 1.1 (0.9–1.3), 0.7–2.7 n (c > 0.5) = 28 | 1.1 (0.9–1.4), 0.6–2.5 n (c > 0.5) = 28 | 0.908 |
| Succinic | 4.8 (4.4–6.0), 3.3–12.4 n (c > 0.5) = 48 | 14.0 (10.0–15.5), 8.0–25.0 n (c > 0.5) = 28 | 12.0 (9.2–16.0), 6.0–29.0 n (c > 0.5) = 28 | 0.181 |
| Fumaric | 1.3 (1.1–1.5), 0.8–2.3 n (c > 0.5) = 48 | 2.4 (1.9–3.0),1.5–5.5 n (c > 0.5) = 28 | 1.8 (1.6–2.2),1.3–11.6 n (c > 0.5) = 28 | 0.121 |
| Parameter, lg CFU/g | Reference Values | Patients on Admission (n = 28) | Patients after 14 Days (n = 28) | p-Value |
|---|---|---|---|---|
| Total bacterial mass | 1011–1013 | 2 × 1013 (1 × 1013–5 × 1013), 6 × 1011–4 × 1015 n (c > 104) = 28 n (c > RV) = 17 | 2 × 1013 (1 × 1013–3 × 1013), 2 × 1012–1 × 1014 n (c > 104) = 28 n (c > RV) =16 | 0.327 |
| Lactobacillus spp. | 107–108 | 4 × 107 (3 × 106–5 × 108), 1 × 105–4 × 1010 n (c > 105) = 28 n (c > RV) = 11 n (c < RV) = 10 | 2 × 107 (5 × 106–1 × 108), 1 × 105–4 × 109 n (c > 105) = 28 n (c > RV) = 7 n (c < RV) = 10 | 0.311 |
| Bifidobacterium spp. | 109–1010 | 3 × 1010 (9 × 109–2 × 1011), 2 × 108–3 × 1012 n (c > 105) = 28 n (c > RV) = 19 n (c < RV) = 1 | 2 × 1010 (3 × 109–7 × 1010), 8 × 107–3 × 1011 n (c > 105) = 28 n (c > RV) = 16 n (c < RV) = 4 | 0.019 |
| Escherichia coli | 106–108 | 3 × 108 (3 × 107–2 × 109), 3 × 106–5 × 1011 n (c > 105) = 28 n (c > RV) = 17 | 6 × 107 (3 × 107–1 × 108), 9 × 105–1 × 1010 n (c > 105) = 28 n (c > RV) = 6 n (c < RV) = 1 | 0.023 |
| Bacteroides spp. | 109–1012 | 2 × 1013 (1 × 1013–5 × 1013), 6 × 1011–4 × 1015 n (c > 104) = 28 n (c > RV) = 27 | 2 × 1013 (1 × 1013–3 × 1013), 2 × 1012–1 × 1014 n (c > 104) = 28 n (c > RV) = 28 | 0.327 |
| Faecalibacterium prausnitzii | 108–1011 | 4 × 1011 (6 × 1010–6 × 1011), 1 × 107–5 × 1013 n (c > 104) = 28 n (c > RV) = 16 n (c < RV) = 1 | 2 × 1011 (6 × 1010–6 × 1011), 1 × 1010–3 × 1013 n (c > 104) = 28 n (c > RV) = 15 | 0.524 |
| Bacteroides thetaiotaomicron | Any quantity is allowed | 9 × 108 (<105–9 × 109), <105–1 × 1011 n (c > 105) = 20 | 8 × 108 (7 × 106–4 × 109), <105–6 × 1010 n (c > 105) = 22 | 0.833 |
| Akkermansia muciniphila | <1011 | <105 (<105–<105), <105–2 × 107 n (c > 105) = 4 | <105 (<105–5 × 104), <105–6 × 108 n (c > 105) = 7 | - |
| Enterococcus spp. | <108 | <105 (<105–<105), <105–4 × 1012 n (c > 105) = 1 n (c > RV) = 1 | not detected | - |
| Escherichia coli enteropathogenic | <104 | <104 (<104–<104), <104–5 × 106 n (c > 104) = 2 n (c > RV) = 2 | not detected | - |
| Candida spp. | <104 | <104 (<104–<104), <104–3 × 107 n (c > 104) = 3 n (c > RV) = 3 | <104 (<104–<104), <104–3 × 106 n (c > 104) = 3 n (c > RV) = 3 | - |
| Klebsiella oxytoca | <104 | not detected | not detected | - |
| Staphylococcus aureus | <104 | <104 (<104–7 × 105), <104–5 × 107 n (c > 104) = 9 n (c > RV) = 9 | <104 (<104–9 × 105), <104–8 × 106 n (c > 104) = 10 n (c > RV) = 10 | - |
| Clostridium difficile | not detected | not detected | not detected | - |
| Clostridium perfringens | not detected | <105 (<105–<105), <105–1 × 107 n (c > 105) = 3 n (c > RV) = 3 | <105 (<105–<105), <105–8 × 106 n (c > 105) = 4 n (c > RV) = 4 | - |
| Proteus vulgaris/mirabilis | <104 | <105 (<105–<105), <105–2 × 109 n (c > 105) = 3 n (c > RV) = 3 | <105 (<105–1 × 105), <105–3 × 107 n (c > 105) = 7 n (c > RV) = 7 | - |
| Enterobacter spp. | <104 | 2 × 106 (<105–3 × 107), <105–7 × 1010 n (c > 105) = 17 n (c > RV) = 17 | 8 × 106 (1 × 106–5 × 107), <105–3 × 1010 n (c > 105) = 22 n (c > RV) = 22 | 0.403 |
| Citrobacter spp. | <104 | <105 (<105–<105), <105–5 × 1013 n (c > 105) = 3 n (c > RV) = 3 | <105 (<105–<105), <105–6 × 105 n (c > 105) = 2 n (c > RV) = 2 | - |
| Fusobacterium nucleatum | not detected | <105 (<105–<105), <105–1 × 107 n (c >105) = 6 n (c > RV) = 6 | <105 (<105–<105), <105–6 × 105 n (c >105) = 3 n (c > RV) = 3 | - |
| Parvimonas micra | not detected | <105 (<105–<105), <105–2 × 106 n (c > 105) = 4 n (c > RV) = 4 | <105 (<105–<105), <105–1 × 106 n (c > 105) = 4 n (c > RV) = 4 | - |
| Salmonella spp. | not detected | not detected | not detected | - |
| Shigella spp. | not detected | not detected | not detected | - |
| Bacteroides fragilis/Faecalibacterium prausnitzii Ratio | 0.01–100 | 106 (58–293) 13–40,000 n (c > RV) = 14 | 121 (63–250) 40–900 n (c > RV) = 17 | 0.387 |
| Parameter | Reference Values | Vaccinated Patients (n = 20) | Unvaccinated Patients (n = 19) | p-Value |
|---|---|---|---|---|
| Glucose, mmol/L | 4.1–5.9 | 5.2 (4.8–5.4), 4.1–7.4 n (c > RV) = 2 | 5.7 (5.3–6.4), 4.9–10.5 n (c > RV) = 7 | 0.004 |
| Alanine Transaminase (ALT), U/L | <50.0 | 16.2 (12.6–20.8), 9.2–43.4 | 21.6 (17.9–26.6), 10.0–44.5 | 0.030 |
| Bacteroides spp. | 109–1012 | 2 × 1013 (7 × 1012–4 × 1013), 6 × 1011–4 × 1015 n (c > RV) = 18 | 4 × 1013 (1 × 1013–2 × 1014), 2 × 1012–8 × 1014 n (c > RV) =19 | 0.030 |
| Bacteroides fragilis/Faecalibacterium prausnitzii Ratio | 0.01–100 | 88 (33–191) 1–4000 n (c > RV) = 8 | 750 (111–1667) 43–400,000 n (c > RV) = 15 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorokina, E.; Pautova, A.; Fatuev, O.; Zakharchenko, V.; Onufrievich, A.; Grechko, A.; Beloborodova, N.; Chernevskaya, E. Promising Markers of Inflammatory and Gut Dysbiosis in Patients with Post-COVID-19 Syndrome. J. Pers. Med. 2023, 13, 971. https://doi.org/10.3390/jpm13060971
Sorokina E, Pautova A, Fatuev O, Zakharchenko V, Onufrievich A, Grechko A, Beloborodova N, Chernevskaya E. Promising Markers of Inflammatory and Gut Dysbiosis in Patients with Post-COVID-19 Syndrome. Journal of Personalized Medicine. 2023; 13(6):971. https://doi.org/10.3390/jpm13060971
Chicago/Turabian StyleSorokina, Ekaterina, Alisa Pautova, Oleg Fatuev, Vladislav Zakharchenko, Alexander Onufrievich, Andrey Grechko, Natalia Beloborodova, and Ekaterina Chernevskaya. 2023. "Promising Markers of Inflammatory and Gut Dysbiosis in Patients with Post-COVID-19 Syndrome" Journal of Personalized Medicine 13, no. 6: 971. https://doi.org/10.3390/jpm13060971
APA StyleSorokina, E., Pautova, A., Fatuev, O., Zakharchenko, V., Onufrievich, A., Grechko, A., Beloborodova, N., & Chernevskaya, E. (2023). Promising Markers of Inflammatory and Gut Dysbiosis in Patients with Post-COVID-19 Syndrome. Journal of Personalized Medicine, 13(6), 971. https://doi.org/10.3390/jpm13060971