Abstract
Numerous reports have explored the roles of different genetic variants in miRNA biogenesis mechanisms and the progression of various types of carcinomas. The goal of this study is to explore the association between XPO5*rs34324334 and RAN*rs14035 gene variants and susceptibility to hepatocellular carcinoma (HCC). In a cohort of 234 participants (107 HCC patients and 127 unrelated cancer-free controls) from the same geographic region, we characterized allelic discrimination using PCR-RFLP and performed subgroup analysis and multivariate regression. We found that the frequency of the XPO5*rs34324334 (A) variant was correlated with elevated risk of HCC under allelic (OR = 10.09, p-value < 0.001), recessive (OR = 24.1, p-value < 0.001), and dominant (OR = 10.1, p-value < 0.001) models. A/A genotype was associated with hepatitis C cirrhosis (p-value = 0.012), ascites (p-value = 0.003), and higher levels of alpha-fetoproteins (p-value = 0.011). Carriers of the RAN*rs14035 (T) variant were more likely to develop HCC under allelic (OR = 1.76, p-value = 0.003) and recessive (OR = 3.27, p-value < 0.001) models. Our results suggest that XPO5*rs34324334 and RAN*rs14035 variants are independent risk factors for developing HCC.
1. Introduction
Hepatocellular carcinoma (HCC) is the second-highest cause of cancer-related mortality diagnosed worldwide [1]. It is the most challenging carcinoma to treat, and rates continue to grow rapidly: Global incidence is projected to reach over one million cases by 2025 [2,3]. The prevalence of HCC in Africa is highest among Egyptian individuals, which may be due to various genetic factors or to Egypt’s high hepatitis C virus (HCV) burden [4,5,6]. Although studies have sought to explain the involvement of hereditary and epigenetic modifications in the course of the disease, the molecular pathogenesis of HCC remains poorly understood [7,8].
MicroRNAs (miRNAs) are a subclass of small non-coding RNAs. Their ability to interact with mRNAs, which regulate cell proliferation, migration, and infiltration, has led to speculation that they are involved in the progression of HCC [9]. Genetic variations within the biogenesis of miRNA machinery may be associated with various carcinomas, including HCC [10]. miRNA biogenesis is a sophisticated process that begins intranuclearly with transcription of pri-miRNAs that submitted to modifications by microprocessors, forming pre-miRNAs. The export of miRNAs to the cytoplasm, with the aid of exportin 5 (XPO5) and its partner RanGTP, is believed to be correlated with limiting the rate of translocation of these miRNAs [11,12]. In the cytoplasm, miRNAs are shuttled via ribonuclease III (Dicer) and the transactivation response element RNA-binding protein (TRBP), which are incorporated into RNA-induced silencing complex (RISC) with Argonaute proteins [13,14,15].
The exportin 5 (XPO5) gene is situated at chromosome 6p21.1 with 32 exons, while RanGTP (RAN) is located at chromosome 12q24.33 with 7 exons and is thought to be one of the nucleocytoplasmic transporters belonging to the karyopherin β family [16,17]. The XPO5 gene encoded a specific protein known as exportin 5 protein that is responsible for the export of pri-miRNAs to the cytoplasm through the nuclear membrane that elucidates the critical function of this protein in the shuttling mechanism of different miRNAs. Thus, the genetic variations of the XPO5 gene could imply various epigenetic alterations and posttranscriptional modifications that altered the expression patterns of the miRNAs, leading to the progression of different cancer diseases [18]. Additionally, the transport of pre-miRNAs to cytosol triggers hydrolysis of RanGTP to RanGDP, mediated by RANBP1 and RANGAP1, respectively [19,20,21].
RAN (ras-related nuclear protein) is a small GTP-binding protein belonging to the RAS superfamily that is critical for protein transport through the nuclear pore complex [22]. RanGTP provokes microtubule assembly and is involved in normal mitotic spindle assembly and chromosome segregation [23]. It can also inhibit human vaccinia-related kinase (VRK1/2), which is involved in transcription regulation, nuclear membrane assembly, chromatin condensation, and the cell cycle [24]. RAN participates in DNA synthesis and, together with exportin 5, protects pre-miRNA from decay by nucleases, as it is surrounded by machinery on all sides [25,26]. Once pre-miRNA is translocated to the cytosol, RanGTP converts to RanGDP, causing conformational changes and releasing pre-miRNA [27].
During carcinogenesis, the dysregulation of the nuclear export of target pre-miRNAs could cause extensive expression of numerous abnormal miRNAs within tumor tissues, including miR-138 [28,29]. Additionally, the mutations in the XPO5 gene might disrupt the transportation process and inhibit miRNA maturation, causing more pre-miRNAs to be retained and trapped in the nucleus [30]. The mutated form of the XPO5 protein lacks the C-terminus region that has a crucial function in the joining of pre-miRNAs with XPO5 and RanGTP, causing the aggregation of the pre-miRNAs within the nucleus [31]. Single nucleotide variants (SNVs) within miRNA machinery genes, especially XPO5 and RAN, are widely reported to be associated with elevated risk of HCC, but the mechanism remains unclear [12,32]. Egypt has the highest incidence of HCC in the Middle East with nearly ~45.9/100,000 within males and ~22.7/100,000 within females. Additionally, HCC among Egyptians is considered the second-most prevalent carcinoma, after breast cancer [33,34]. The present work was designed to clarify the relationship of XPO5 (c.722G>A; rs34324334) and RAN (c.*770C>T; rs14035) variants with susceptibility to HCC among Egyptian subjects from the Delta Region of Egypt. Additionally, we performed various bioinformatic analyses regarding the XPO5 and RAN genes to focus light on the importance of these potential variants in the progression of carcinomas.
2. Materials and Methods
2.1. Study Population
Our case–control design study comprised 234 participants [107 HCC patients and 127 healthy controls]; of these, HCC patients (87 [81.3%] male and 20 [18.7%] female) had a median age (interquartile range [IQR]) of 53.0 (45.0–61.0) years. HCC patients were diagnosed using multiphase magnetic resonance imaging (MRI) and/or computerized tomography (CT), in accordance with the guidelines of the American Association for the Study of Liver Disease (AASLD) [35]. The Institutional Review Board of the Faculty of Medicine at Tanta University in Tanta, Egypt, authorized the project (approval number 34230/11/20). All procedures performed in the study were in accordance with Helsinki Declaration guidelines. Recruitment and management of HCC patients were performed at the Outpatient Clinic of the Oncology Department at Tanta University Hospital from February 2021 to January 2022. A group of 127 cancer-free controls, matched for age (median 54.0 years, interquartile range [IQR] 49.0–58.0) and sex (94 [74.0%] male and 33 [26.0%] female), was recruited as volunteer blood donors. HCC patients with a history of other carcinomas, autoimmune disorders, diabetes, or kidney disease were excluded from the study. Clinical and demographic characteristics, including age, sex, age of onset, smoking status, splenomegaly status, and ascites status, were acquired from electronic medical health archives.
Biochemical measurements, including hepatic aminotransferases, albumin, total bilirubin, INR (International Normalized Ratio), and creatinine, were determined using a fully automated chemical analyzer. Estimations of serological measurements, including hepatitis C virus autoantibodies (HCV Abs) and alpha-fetoprotein (AFP), were carried out using chemiluminescent techniques (Abbott PRISM Immunoassay Analyzer, Corston, Bath, UK). Hematological parameters were estimated with the aid of a multiparameter hematology counter system (CELL-DYN 3700 SL [Abbott Diagnostics]).
2.2. Genomic DNA Extraction and Amplification Analysis
Genomic DNA was extracted and purified with GeneJET Whole Blood Genomic DNA Purification Mini Kit (Thermo Fisher Scientific; Waltham, MA, USA). The concentration and absorbance of purified DNA was measured using a full spectrum NanoDropTM ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
The allelic discrimination for the XPO5 (rs34324334; c.722G>A) and RAN (rs14035; c.*770C>T) variants were determined using the polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) method [36,37]. The electronic NCBI primer BLAST tool was applied to design the primers used in this work. The forward and reverse primers for the XPO5 (rs34324334; c.722G>A) variant [F1: 5′-ATG AGA AGA CAC TCA GCG GCT C-3′, and R1: 5′-ACC TGT TAC CGA GGA CTT CAT G-3′] were processed to generate a PCR product of 218 bp. The two primers used for amplification of the RAN (rs14035; c.*770C>T) variant (F2: 5′-AAG CAG TGT TTG CTC CAC CTT C-3′, and R2: 5′-AGA ATT CCC AAC CTC CTG CC-3′) were designed to produce a PCR product of 216 bp. For these two variants, the PCR reaction was processed within a thermal cycler comprising 25 µL DreamTaq Green PCR Master Mix (2×) (Thermo Fisher Scientific), 1 µM forward primer, 1 µM reverse primer, 50 µL nuclease-free water, and 1 µg purified template DNA. The amplification reaction was adjusted with an initial denaturation at 94 °C for 5 min, followed by 35 cycles involving denaturation at 94 °C for 30 s, annealing at 60 °C for 60 s, and extension at 72 °C for 60 s, with a final extension at 72 °C for 7 min. The amplified products of the XPO5 (rs34324334; c.722G>A) variant were subjected to endonuclease restriction enzyme Tsp45I (NmuCI) (New England Biolabs, Ipswich, MA, USA) and incubated at 65 °C for an hour, while the RAN (rs14035; c.*770C>T) variant was incubated with endonuclease restriction enzyme BseLI (Bs1I) (New England Biolabs) at 55 °C for 15 min. The digested fragments were electrophoresed using 2.5% agarose gel with ethidium bromide to allow for visualization.
2.3. Bioinformatic Analysis
Various online databases were explored for the identification of the functional roles of XPO5 and RAN along with their potential proteins, including the Ensembl database for chromosomal screening (https://www.ensembl.org/), National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/gene/), compartments for localization of specific proteins inside the cell (https://www.proteinatlas.org/), Kaplan–Meier plotter database for survival analysis (https://kmplot.com/analysis/), protein data bank for the crystal structure of the specific proteins (https://www.rcsb.org/), string database for protein–protein interactions and gene ontology (https://string-db.org/), and GeneMANIA database for gene prediction (https://genemania.org/). All these electronic databases were accessed on 28 February 2023.
2.4. Statistical Analysis
Statistical analysis was conducted using STATA software version 17.0 (Stata Corporation, College Station, TX, USA). The detection of the degree of normal distribution was assessed by applying the Shapiro–Wilk test. Continuous data were assessed using the Wilcoxon rank-sum test, while categorical findings were evaluated using Fisher’s exact test and odds ratio (OR) with a 95% confidence interval [38,39]. The Hardy–Weinberg equilibrium for the XPO5 (rs34324334; c.722G>A) and RAN (rs14035; c.*770C>T) variants was executed to identify the level of equilibrium among observed and expected values of HCC patients and cancer-free subjects [40]. Genetic association models were carried out using the electronic online tool for SNP analysis (www.snpstats.net, accessed on 25 October 2022). Multivariate analysis was performed and processed using R software version 4.2.2 with R studio version 2023.03 Build 386. Principal component analysis (PCA) was carried out using the “FactoMineR” and “Factoextra” packages. The statistical level was adjusted as a p-value of not more than 0.05.
3. Results
3.1. The Fundamental Characteristics of the Study Population
Generally, HCC patients showed a significantly higher correlation with tobacco smoking and weight (p-value < 0.05) compared to cancer-free controls. Of the HCC patients, 79 (73.8%) were cirrhotic, while 72 (67.3%) were ascitic. Biochemical, serological, and hematological measurements differed significantly between HCC patients and controls (p-value < 0.05) (Table 1).

Table 1.
The main demographic, clinical, and laboratory variables of the study participants.
3.2. XPO5*rs34324334 and RAN*rs14035 Variants with Susceptibility to HCC
The XPO5*rs34324334 and RAN*rs14035 variants approached Hardy–Weinberg equilibrium among cancer-free controls (p-value > 0.05). For the XPO5*rs34324334 variant, the most prevalent genotype (A/A genotype) in HCC patients was 43%, compared to 3.2% in cancer-free controls (Figure 1A). The XPO5*rs34324334 and RAN*rs14035 variants among healthy subjects from the 1000 genome project dataset (https://www.internationalgenome.org/data, accessed on 28 February 2023) are summarized in Figure 1B,D. Principal component analysis (PCA) diagrams for HCC patients with XPO5*rs34324334 and RAN*rs14035 variants represented no distinct demarcation among different genotypes (Figure 1E,F). PCA categorized HCC patients into three clusters based on the genotype variants. Axis one and two for these tests accounted for 6.7% and 10.2% for the XPO5 genotypes and 7.1% and 10.2% for the RAN genotypes.
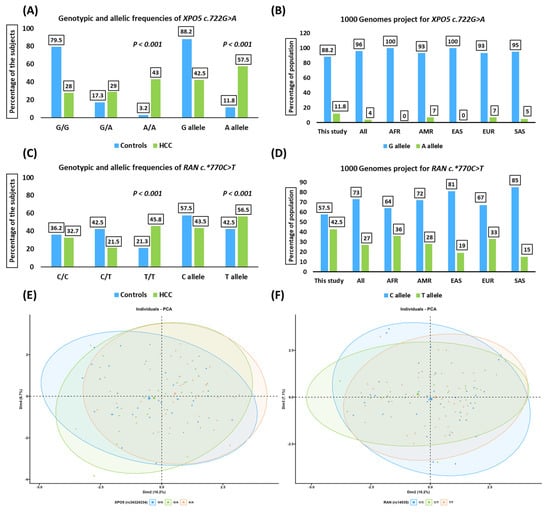
Figure 1.
Genotypic and allelic frequencies of the study population. (A) Genotypic and allelic distribution of the XPO5 (rs34324334; c.722G>A) variant among HCC patients compared to cancer-free controls. (B) The 1000 genome project for the XPO5 (rs34324334; c.722G>A) variant. (C) Genotypic and allelic distribution of the RAN (rs14035; c.*770C>T) variant among HCC patients compared to cancer-free controls. (D) The 1000 genome project for the RAN (rs14035; c.*770C>T) variant. (E,F) Principal component analysis (PCA) of the XPO5 (rs34324334; c.722G>A) and RAN (rs14035; c.*770C>T) variants among HCC patients. These figures present no distinct demarcation of the HCC patients with different genotypes. AFR, Africa; AMR, America; EAS, East Asia; EUR, Europe; SAS, South Asia; HCC, hepatocellular carcinoma.
The minor allele frequency (A allele) was 57.5% among HCC patients compared to 11.8% among cancer-free controls (OR = 10.09, 95% CI = 6.32–16.11, p-value < 0.001; Table 2). The most prevalent genotype (T/T genotype) for the RAN*rs14035 variant in HCC patients was 45.8%, compared to 21.3% in cancer-free controls (Figure 1C). The minor allele frequency was 56.5% in HCC patients and 42.5% in controls (OR = 1.76, 95% CI = 1.22–2.54, p-value < 0.001) (Table 2).

Table 2.
Genotypic and allelic frequencies of the XPO5*rs34324334 and RAN*rs14035 variants among the study population.
XPO5*rs34334334 was significantly associated with HCC status among different genetic models, including heterozygote (OR = 4.73, 95% CI = 2.38–9.39, p-value < 0.001), homozygote (OR = 39.9, 95% CI = 13.2–121.1, p-value < 0.001), dominant (OR = 10.1, 95% CI = 5.46–18.4, p-value < 0.001), and recessive (OR = 24.1, 95% CI = 8.23–70.8, p-value < 0.001) (Table 3). The RAN*rs14035 variant showed a significant association with elevated risk of HCC compared to cancer-free controls among different hereditary models, including homozygote (OR = 2.44, 95% CI = 1.27–4.68, p-value < 0.001) and recessive (OR = 3.27, 95% CI = 1.83–5.83, p-value < 0.001) (Table 3).

Table 3.
Genetic association models of the XPO5*rs34324334 and RAN*rs14035 variants with the risk of hepatocellular carcinoma.
3.3. XPO5*rs34324334 and RAN*rs14035 Variants Stratified by Clinical and Laboratory Measurements among HCC Patients
The XPO5*rs34324334 variant (A/A genotype) presented a significant association with hepatitis C cirrhosis (p-value = 0.012), ascites (p-value = 0.003), and higher levels of alpha-fetoproteins (p-value = 0.011) (Supplementary Materials: Table S1), while the RAN*rs14035 variant (T/T genotype) failed to achieve statistical significance with any clinical or laboratory parameters (p-value > 0.05) (Figure 2).
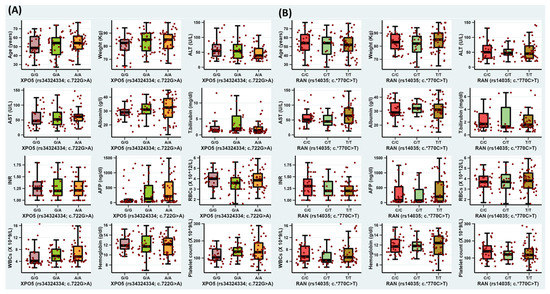
Figure 2.
The impact of XPO5*rs34324334 and RAN*14035 variants on serological, biochemical, and hematological variables. (A) Box plot for the XPO5*rs34324334 variant among HCC patients; (B) box plot for the RAN*14035 variant among HCC patients.
3.4. In Silico Data Analysis
Computational bioinformatic analyses of the exportin 5 (XPO5) and ras-related nuclear protein (RAN) genes are summarized in Figure 3. The XPO5 gene has specific properties that indicate its coding protein comprising 1204 amino acid residues. This encoded protein is a major component of the nuclear export receptor complex along with RAN. The RAN gene encodes a specific protein with 216 amino acid residues. The XPO5*rs34324334 (c.722G>A; p.Ser241Asn) variant is located on the seventh exon, while the RAN*rs14035 (c.*770C>T) variant is situated on the last exon. Protein–protein networks indicated that XPO5 and RanGTP binding proteins were involved in various miRNA biological functions, including pre-miRNA export from the nucleus, miRNA metabolic processes, RNA transport, and the miRNA biogenesis pathway. Data provided from the GeneMANIA database for gene–gene interactions revealed the role of the XPO5-RanGTP complex in post-transcriptional gene silencing by RNA, miRNA gene silencing, nuclear export, and RNA localization from the nucleus. The protein atlas database identified the presence of these proteins in higher abundance in the nuclear membrane. The Kaplan–Meier plotter database detected low and high mRNA expression levels of XPO5 and RAN among patients with hepatic carcinoma, respectively.
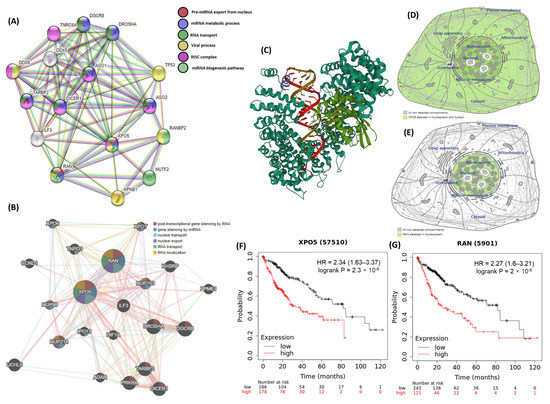
Figure 3.
The bioinformatic framework analysis of the XPO5 and RAN genes. (A) protein–protein interactions of the XPO5 and RanGTP proteins using the STRING database. (B) gene–gene interactions using the GeneMANIA database. (C) the crystal structure of the XPO5:RanGTP:pre-miRNA complex. (D) subcellular localization of the XPO5 protein, with darker colors indicating more abundance. (E) subcellular localization of the RanGTP protein. (F,G) survival analysis data for high and low XPO5 and RAN expression. (Data source: The Human Protein Atlas, GeneMANIA, STRING version 11.0, and Kaplan–Meier plotter database).
4. Discussion
MicroRNAs (miRNAs) are a crucial category of noncoding RNAs, with about 22 nucleotides that are involved in the regulation mechanisms of hereditary and epigenetic pathways [41]. Each miRNA can bind to several mRNA targets, causing instability, degradation, and inhibition [42]. Generally, miRNAs regulate gene expression by attaching to the 3′ untranslated region (3′ UTR) of target mRNAs, but any alteration in expression of synthesized miRNA could affect hundreds of genes [43]. miRNA biogenesis genes and their variants are involved in the pathogenesis of several diseases, including HCC [12,44,45]. To the best of our knowledge, this work is the first to study the association of the XPO5*rs34324334 and RAN*rs14035 variants with increased risk of HCC among Egyptian subjects. The XPO5 gene belongs to the karyopherin β family, which plays a crucial role in shuttling from nucleus to cytoplasm in a RANGTP-dependent manner. The XPO5*rs34324334 variant could represent a missense variant (XPO5*p.Ser241Asn) modulating the protein structure, which could reflect a change in its behavior associated with pre-miRNA nuclear export and a subsequent regulatory role in HCC.
In the present study, the XPO5*rs34324334 (c.722G>A) variant showed a significant association according to the allelic model (OR = 10.09 and p-value < 0.001). Testing the inheritance model with frequency of genotypes revealed a significant effect on the risk of HCC in dominant and recessive models, with odds ratios of 10.1 and 24.1, respectively. We also found significant associations between the XPO5*rs34324334 variant and cirrhotic liver and ascites status among HCC patients. These findings were consistent with a study that revealed a strong association between the XPO5*rs34324334 variant and elevated risk of breast carcinoma in postmenopausal women with breast cancer (dominant model: OR = 1.76, p-value < 0.05) [46]. The XPO5*rs34324334 variant is thought to be in a highly conserved exportin-1/importin-b-like region and may affect the development of carcinomas. To test and predict the changes in protein stability upon mutation, we used DynaMut (https://biosig.lab.uq.edu.au/dynamut/; accessed on 20 December 2022) to analyze the modification in the XPO5 (rs34324334; p.Ser241Asn) missense variant and identified a destabilization effect (ΔΔG = −0.236 kcal/mol) on the functional mechanism of the XPO5 protein. Lack of XPO5 expression and mutated forms of XPO5 are both highly correlated with reduced levels of miRNAs and their inhibitory targets; restoring functional XPO5 seemed to act as a tumor suppressor. XPO5 mutations subsequently disrupt its role as a tumor suppressor gene and modulator for cancer through attenuated production of miRNAs that, in turn, promote growth regulatory gene TGGBR2 as well as proapoptotic gene BAX. Moreover, the aberration of XPO5 is linked to a global reduction in miRNAs and subsequent altered regulation [30,47].
The RAN gene acts as a powerhouse for the pre-miRNA nuclear shuttling process via exportin 5. Our RAN*rs14035 variant data suggest an association within the allelic model (T allele vs. C allele; odds ratio 1.76 and p-value < 0.05). Upon testing the inheritance model, this 3′ UTR variant was associated with a significantly elevated risk of HCC under homozygote and recessive comparisons (OR = 2.44 and 3.27, respectively). Similar results were found in a study among Asian patients diagnosed with HCC, which revealed a strong correlation with elevated risk of hepatic cancer under a recessive model (OR = 2.54) [32]. Another report, among Korean patients with colorectal cancer (CRC), indicated that the RAN*rs14035 variant was significantly correlated with a decreased risk of CRC (OR = 0.690; p-value = 0.016) [48]. Several meta-analysis reports addressed the contribution of the RAN*rs14035 variant to cancer development among various ethnic populations [49,50,51]. Another study revealed a significant association between the RAN*rs14035 variant and an increased risk of end-stage renal disease among Egyptian subjects (OR = 5.18) [44], whereas the RAN*rs14035 variant showed no association with elevated risk of various types of carcinomas, including hepatocellular carcinoma [45] and renal cell carcinoma [52].
Testing the functional role of the regulatory non-coding region within the RAN*rs14035 variant using the RegulomeDB electronic tool (https://regulomedb.org/, accessed on 20 December 2022) resulted in a score of 0.59 with a moderate shift toward 1, suggesting that alterations within the RAN*rs14035 variant may modify the stability of various mRNAs, particularly miR-575 and miR-182, as has been previously suggested for larynx cancer [53]. Our results were consistent with a comprehensive meta-analysis that confirmed the contribution of RAN*rs14035 to cancer risk [54]. Additionally, knocking down XPO5 expression resulted in a reduction in global miRNA levels, thus enhancing carcinogenesis [55].
Another tool for exploring annotations of noncoding variants, HaploReg, revealed overlap between RAN*rs14035 and nearby variants as well as interactions with PLZF, ZEB1, and ZBTB12 motifs (Supplementary Materials: Table S2). The promyelocytic leukemia zinc finger protein (PLZF) contributes to tumor formation by regulating cell growth, differentiation, and apoptosis, in addition to its role regulating cytokine production to ameliorate cancer progression [56,57,58]. Zinc finger E-box-binding homeobox 1 (ZEB1) is involved in epithelial-to-mesenchymal transition (EMT) and is significantly associated with poorer cancer prognosis, including for HCC [59]. RAN*rs14035 may contribute to changes in the expression of RAN itself, binding of miRNAs, and interaction with network motifs, all of which can lead to oncogenesis.
5. Conclusions
Evidence suggests that the genes XPO5 and RAN may contribute to the development of tumors, including HCC, and may be involved in the mechanism by which microRNAs (miRNAs) are synthesized in cancer. This is the first work studying the association of XPO5*rs34324334 and RAN*rs14035 variants with elevated risk of HCC among Egyptian subjects. We found evidence suggesting that these genetic variants are associated with a variety of indicators of poor liver health, including HCC. Our data strongly suggests that XPO5*rs34324334 and RAN*rs14035 variants could represent independent risk factors for developing HCC. Genetic testing for these variants in HCC patients may be a useful tool for predicting which patients are at a higher risk of aggressive characteristics and poorer outcomes, allowing clinicians to choose more personalized and effective treatments early in the course of the disease when treatment is most likely to be effective.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jpm13060959/s1: Table S1, genotypic frequencies of XPO5 (rs34324334; c.722G>A) and RAN (rs14035; c.*770C>T) variants stratified by the demographic, clinical, and laboratory variables of HCC patients; Table S2, impact and linkage disequilibrium of the studied RAN*rs14035 variant with other variants (r2 ≥ 0.8).
Author Contributions
Conceptualization, A.F.S. and R.M.E.; methodology, M.I.E., A.F.S. and R.M.E.; software, M.I.E., T.D., E.T. and R.M.E.; validation, T.D., A.F.S. and M.I.E.; formal analysis, M.I.E., E.T. and R.M.E.; investigation, T.D., M.I.E. and M.G.; resources, A.F.S. and M.G.; data curation, M.I.E., M.G. and R.M.E.; writing—original draft preparation, M.I.E. and R.M.E.; writing—review and editing, T.D., M.I.E., A.F.S., M.G. and R.M.E.; visualization, M.I.E., E.T. and R.M.E.; supervision, A.F.S. and R.M.E.; project administration, A.F.S., M.G. and R.M.E. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of Tanta University, Egypt, (protocol code 34230/11/20, 2 November 2020). Patient data were obtained from hospital medical records, which were anonymized and de-identified before analysis.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Data Availability Statement
Data are available from the corresponding author upon reasonable request. Sources of in silico data analysis sections are available in public repositories through the links provided in the manuscript.
Acknowledgments
A sincere thank you to Loula Burton from Tulane’s Research Proposal Development Office for her diligent editing and proofreading of this paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yang, C.; Huang, X.; Liu, Z.; Qin, W.; Wang, C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 2020, 14, 896–913. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Saad, A.M.; Abdel-Megied, A.E.S.; Elbaz, R.A.; Hassab El-Nabi, S.E.; Elshazli, R.M. Genetic variants of APEX1 p.Asp148Glu and XRCC1 p.Gln399Arg with the susceptibility of hepatocellular carcinoma. J. Med. Virol. 2021, 93, 6278–6291. [Google Scholar] [CrossRef] [PubMed]
- Amer, T.; El-Baz, R.; Mokhtar, A.R.; El-Shaer, S.; Elshazli, R.; Settin, A. Genetic polymorphisms of IL-23R (rs7517847) and LEP (rs7799039) among Egyptian patients with hepatocellular carcinoma. Arch. Physiol. Biochem. 2017, 123, 279–285. [Google Scholar] [CrossRef]
- Lehman, E.M.; Soliman, A.S.; Ismail, K.; Hablas, A.; Seifeldin, I.A.; Ramadan, M.; El-Hamzawy, H.; Shoushtari, C.S.; Wilson, M.L. Patterns of hepatocellular carcinoma incidence in Egypt from a population-based cancer registry. Hepatol. Res. 2008, 38, 465–473. [Google Scholar] [CrossRef] [PubMed]
- Ezzat, R.; Eltabbakh, M.; El Kassas, M. Unique situation of hepatocellular carcinoma in Egypt: A review of epidemiology and control measures. World J. Gastrointest. Oncol. 2021, 13, 1919–1938. [Google Scholar] [CrossRef]
- Sagnelli, E.; Macera, M.; Russo, A.; Coppola, N.; Sagnelli, C. Epidemiological and etiological variations in hepatocellular carcinoma. Infection 2020, 48, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Margini, C.; Dufour, J.F. The story of HCC in NAFLD: From epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016, 36, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.S.; Zheng, H.; Huang, Z.P.; Hong, Y.G.; Ou, Y.L.; Tao, Y.P.; Wang, M.C.; Wang, Z.G.; Yang, Y.; Zhou, W.P. MicroRNA-197-3p acts as a prognostic marker and inhibits cell invasion in hepatocellular carcinoma. Oncol. Lett. 2019, 17, 2317–2327. [Google Scholar] [CrossRef]
- Chu, R.; Mo, G.; Duan, Z.; Huang, M.; Chang, J.; Li, X.; Liu, P. miRNAs affect the development of hepatocellular carcinoma via dysregulation of their biogenesis and expression. Cell Commun. Signal. 2014, 12, 45. [Google Scholar] [CrossRef]
- Sun, H.L.; Cui, R.; Zhou, J.; Teng, K.Y.; Hsiao, Y.H.; Nakanishi, K.; Fassan, M.; Luo, Z.; Shi, G.; Tili, E.; et al. ERK Activation Globally Downregulates miRNAs through Phosphorylating Exportin-5. Cancer Cell 2016, 30, 723–736. [Google Scholar] [CrossRef]
- Elshazli, R.M.; Toraih, E.A.; Hussein, M.H.; Ruiz, E.M.; Kandil, E.; Fawzy, M.S. Pan-Cancer Study on Variants of Canonical miRNA Biogenesis Pathway Components: A Pooled Analysis. Cancers 2023, 15, 338. [Google Scholar] [CrossRef]
- Oura, K.; Morishita, A.; Masaki, T. Molecular and Functional Roles of MicroRNAs in the Progression of Hepatocellular Carcinoma—A Review. Int. J. Mol. Sci. 2020, 21, 8362. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Pasha, T.; Zatorska, A.; Sharipov, D.; Rogelj, B.; Hortobágyi, T.; Hirth, F. Karyopherin abnormalities in neurodegenerative proteinopathies. Brain 2021, 144, 2915–2932. [Google Scholar] [CrossRef]
- Quan, Y.; Ji, Z.-L.; Wang, X.; Tartakoff, A.M.; Tao, T. Evolutionary and Transcriptional Analysis of Karyopherin β Superfamily Proteins. Mol. Cell. Proteom. 2008, 7, 1254–1269. [Google Scholar] [CrossRef]
- Wu, K.; He, J.; Pu, W.; Peng, Y. The Role of Exportin-5 in MicroRNA Biogenesis and Cancer. Genom. Proteom. Bioinform. 2018, 16, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes. Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Matchett, K.B.; McFarlane, S.; Hamilton, S.E.; Eltuhamy, Y.S.; Davidson, M.A.; Murray, J.T.; Faheem, A.M.; El-Tanani, M. Ran GTPase in nuclear envelope formation and cancer metastasis. Adv. Exp. Med. Biol. 2014, 773, 323–351. [Google Scholar] [CrossRef]
- El-Tanani, M.; Dakirel, H.; Raynor, B.; Morgan, R. Mechanisms of Nuclear Export in Cancer and Resistance to Chemotherapy. Cancers 2016, 8, 35. [Google Scholar] [CrossRef]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef]
- Bao, X.; Liu, H.; Liu, X.; Ruan, K.; Zhang, Y.; Zhang, Z.; Hu, Q.; Liu, Y.; Akram, S.; Zhang, J.; et al. Mitosis-specific acetylation tunes Ran effector binding for chromosome segregation. J. Mol. Cell Biol. 2018, 10, 18–32. [Google Scholar] [CrossRef]
- Sanz-García, M.; López-Sánchez, I.; Lazo, P.A. Proteomics identification of nuclear Ran GTPase as an inhibitor of human VRK1 and VRK2 (vaccinia-related kinase) activities. Mol. Cell. Proteom. 2008, 7, 2199–2214. [Google Scholar] [CrossRef]
- Hayder, H.; O’Brien, J.; Nadeem, U.; Peng, C. MicroRNAs: Crucial regulators of placental development. Reproduction 2018, 155, R259–R271. [Google Scholar] [CrossRef]
- Zeng, Y.; Cullen, B.R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 2004, 32, 4776–4785. [Google Scholar] [CrossRef] [PubMed]
- Muqbil, I.; Bao, B.; Abou-Samra, A.B.; Mohammad, R.M.; Azmi, A.S. Nuclear export mediated regulation of microRNAs: Potential target for drug intervention. Curr. Drug. Targets 2013, 14, 1094–1100. [Google Scholar] [CrossRef]
- Lee, E.J.; Baek, M.; Gusev, Y.; Brackett, D.J.; Nuovo, G.J.; Schmittgen, T.D. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA 2008, 14, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Fu, Z.; Yang, G.; Gao, D.; Wang, T.; Liu, Z.; Li, G.; Wang, Y. Exportin-5 SUMOylation promotes hepatocellular carcinoma progression. Exp. Cell Res. 2020, 395, 112219. [Google Scholar] [CrossRef]
- Melo, S.A.; Moutinho, C.; Ropero, S.; Calin, G.A.; Rossi, S.; Spizzo, R.; Fernandez, A.F.; Davalos, V.; Villanueva, A.; Montoya, G.; et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 2010, 18, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Esteller, M. A precursor microRNA in a cancer cell nucleus: Get me out of here! Cell Cycle 2011, 10, 922–925. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Song, C.H.; Yang, W.J.; Dai, L.P.; Wang, P.; Shi, J.X.; Zhang, J.Y.; Wang, K.J. [Correlation between tag single nucleotide polymorphisms of microRNA regulatory genes and the genetic susceptibility of primary liver cancer]. Zhonghua Yu Fang Yi Xue Za Zhi 2012, 46, 533–537. [Google Scholar]
- Gomaa, A.; Allam, N.; Elsharkawy, A.; El Kassas, M.; Waked, I. Hepatitis C infection in Egypt: Prevalence, impact and management strategies. Hepat. Med. 2017, 9, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Marrero, J.A.; Kulik, L.M.; Sirlin, C.B.; Zhu, A.X.; Finn, R.S.; Abecassis, M.M.; Roberts, L.R.; Heimbach, J.K. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018, 68, 723–750. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Galal, A.A.; Abd Elmajeed, A.A.; Elbaz, R.A.; Wafa, A.M.; Elshazli, R.M. Association of Apolipoprotein E gene polymorphism with the risk of T2DM and obesity among Egyptian subjects. Gene 2021, 769, 145223. [Google Scholar] [CrossRef]
- Elsaid, A.; Elshazli, R.; El-Tarapely, F.; Darwish, H.; Abdel-Malak, C. Association of monoallelic MUTYH mutation among Egyptian patients with colorectal cancer. Fam. Cancer 2017, 16, 83–90. [Google Scholar] [CrossRef]
- Yahia, S.; Hammad, A.; El-Gilany, A.H.; El-Assmy, M.; El-Tanbouly, R.; Elsaid, A.M.; Elmoursi, L.Z.; Elshazli, R.M.; Shoaib, R.M. Genetic variant in the 5′ untranslated region of endothelin1 (EDN1) gene in children with primary nephrotic syndrome. J. Biochem. Mol. Toxicol. 2022, 36, e22963. [Google Scholar] [CrossRef]
- Alghamdi, S.A.; Kattan, S.W.; Toraih, E.A.; Alrowaili, M.G.; Fawzy, M.S.; Elshazli, R.M. Association of AIRE (rs2075876), but not CTLA4 (rs231775) polymorphisms with systemic lupus erythematosus. Gene 2021, 768, 145270. [Google Scholar] [CrossRef] [PubMed]
- Pajares, M.J.; Alemany-Cosme, E.; Goñi, S.; Bandres, E.; Palanca-Ballester, C.; Sandoval, J. Epigenetic Regulation of microRNAs in Cancer: Shortening the Distance from Bench to Bedside. Int. J. Mol. Sci. 2021, 22, 7350. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef]
- Deng, Y.; Wang, C.C.; Choy, K.W.; Du, Q.; Chen, J.; Wang, Q.; Li, L.; Chung, T.K.H.; Tang, T. Therapeutic potentials of gene silencing by RNA interference: Principles, challenges, and new strategies. Gene 2014, 538, 217–227. [Google Scholar] [CrossRef]
- Fawzy, M.S.; Abu AlSel, B.T.; Toraih, E.A. Analysis of microRNA processing machinery gene (DROSHA, DICER1, RAN, and XPO5) variants association with end-stage renal disease. J. Clin. Lab. Anal. 2020, 34, e23520. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.N.; Kim, J.O.; Lee, S.M.; Park, H.; Lee, J.H.; Rim, K.S.; Hwang, S.G.; Kim, N.K. Variation in the Dicer and RAN Genes Are Associated with Survival in Patients with Hepatocellular Carcinoma. PLoS ONE 2016, 11, e0162279. [Google Scholar] [CrossRef]
- Leaderer, D.; Hoffman, A.E.; Zheng, T.; Fu, A.; Weidhaas, J.; Paranjape, T.; Zhu, Y. Genetic and epigenetic association studies suggest a role of microRNA biogenesis gene exportin-5 (XPO5) in breast tumorigenesis. Int. J. Mol. Epidemiol. Genet. 2011, 2, 9–18. [Google Scholar] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef]
- Cho, S.H.; Ko, J.J.; Kim, J.O.; Jeon, Y.J.; Yoo, J.K.; Oh, J.; Oh, D.; Kim, J.W.; Kim, N.K. 3′-UTR Polymorphisms in the MiRNA Machinery Genes DROSHA, DICER1, RAN, and XPO5 Are Associated with Colorectal Cancer Risk in a Korean Population. PLoS ONE 2015, 10, e0131125. [Google Scholar] [CrossRef]
- Shao, Y.; Shen, Y.; Zhao, L.; Guo, X.; Niu, C.; Liu, F. Association of microRNA biosynthesis genes XPO5 and RAN polymorphisms with cancer susceptibility: Bayesian hierarchical meta-analysis. J. Cancer 2020, 11, 2181–2191. [Google Scholar] [CrossRef]
- Song, K.; Yi, J.; Shen, X.; Cai, Y. Genetic polymorphisms of glutathione S-transferase genes GSTM1, GSTT1 and risk of hepatocellular carcinoma. PLoS ONE 2012, 7, e48924. [Google Scholar] [CrossRef]
- Gholami, M.; Larijani, B.; Sharifi, F.; Hasani-Ranjbar, S.; Taslimi, R.; Bastami, M.; Atlasi, R.; Amoli, M.M. MicroRNA-binding site polymorphisms and risk of colorectal cancer: A systematic review and meta-analysis. Cancer Med. 2019, 8, 7477–7499. [Google Scholar] [CrossRef] [PubMed]
- Horikawa, Y.; Wood, C.G.; Yang, H.; Zhao, H.; Ye, Y.; Gu, J.; Lin, J.; Habuchi, T.; Wu, X. Single Nucleotide Polymorphisms of microRNA Machinery Genes Modify the Risk of Renal Cell Carcinoma. Clin. Cancer Res. 2008, 14, 7956–7962. [Google Scholar] [CrossRef] [PubMed]
- Osuch-Wojcikiewicz, E.; Bruzgielewicz, A.; Niemczyk, K.; Sieniawska-Buccella, O.; Nowak, A.; Walczak, A.; Majsterek, I. Association of Polymorphic Variants of miRNA Processing Genes with Larynx Cancer Risk in a Polish Population. BioMed. Res. Int. 2015, 2015, 298378. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, F.; Xing, C. A Systematic Review and Meta-Analysis for the Association of Gene Polymorphisms in RAN with Cancer Risk. Dis. Markers 2020, 2020, 9026707. [Google Scholar] [CrossRef]
- Wang, C.; Dong, H.; Fan, H.; Wu, J.; Wang, G. Genetic polymorphisms of microRNA machinery genes predict overall survival of esophageal squamous carcinoma. J. Clin. Lab. Anal. 2018, 32, e22170. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, X.; Li, Y.; Chen, S.; Shen, X.; Dong, X.; Song, Y.; Zhang, X.; Huang, K. Expression of the PTEN/FOXO3a/PLZF signalling pathway in pancreatic cancer and its significance in tumourigenesis and progression. Investig. New Drugs 2020, 38, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Zhu, S.; Zhou, W.; Li, J.; Liu, C.; Xuan, H.; Yan, J.; Zheng, L.; Zhou, L.; Yu, J.; et al. PLZF mediates the PTEN/AKT/FOXO3a signaling in suppression of prostate tumorigenesis. PLoS ONE 2013, 8, e77922. [Google Scholar] [CrossRef]
- Shen, H.; Zhan, M.; Zhang, Y.; Huang, S.; Xu, S.; Huang, X.; He, M.; Yao, Y.; Man, M.; Wang, J. PLZF inhibits proliferation and metastasis of gallbladder cancer by regulating IFIT2. Cell Death Dis. 2018, 9, 71. [Google Scholar] [CrossRef]
- Qin, Y.; Yu, J.; Zhang, M.; Qin, F.; Lan, X. ZEB1 promotes tumorigenesis and metastasis in hepatocellular carcinoma by regulating the expression of vimentin. Mol. Med. Rep. 2019, 19, 2297–2306. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).