An Angiogenesis-Related lncRNA Signature Is Associated with Prognosis and Tumor Immune Microenvironment in Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition and Arrangement of Raw Data
2.2. Identification of AR-lncRNAs
2.3. Definition of BC Subtypes by AR-lncRNAs
2.4. Assessment of Immune Infiltration
2.5. Analysis of Gene Set Enrichment
2.6. Evaluation and Verification of the Accuracy of Prognostic Signatures
2.7. The Creation of the Nomogram
2.8. Prediction of Response to Immune Checkpoint Inhibition (ICI) Therapy
2.9. Patients and Tissue Specimens
2.10. Cell Culture and Treatment
2.11. Quantitative Real-Time PCR
2.12. Tamoxifen-Sensitization Assays
2.13. Tube Formation Assay
2.14. Assays for Cell Migration and Invasion
2.15. Immunoblots
2.16. Statistical Analysis
3. Results
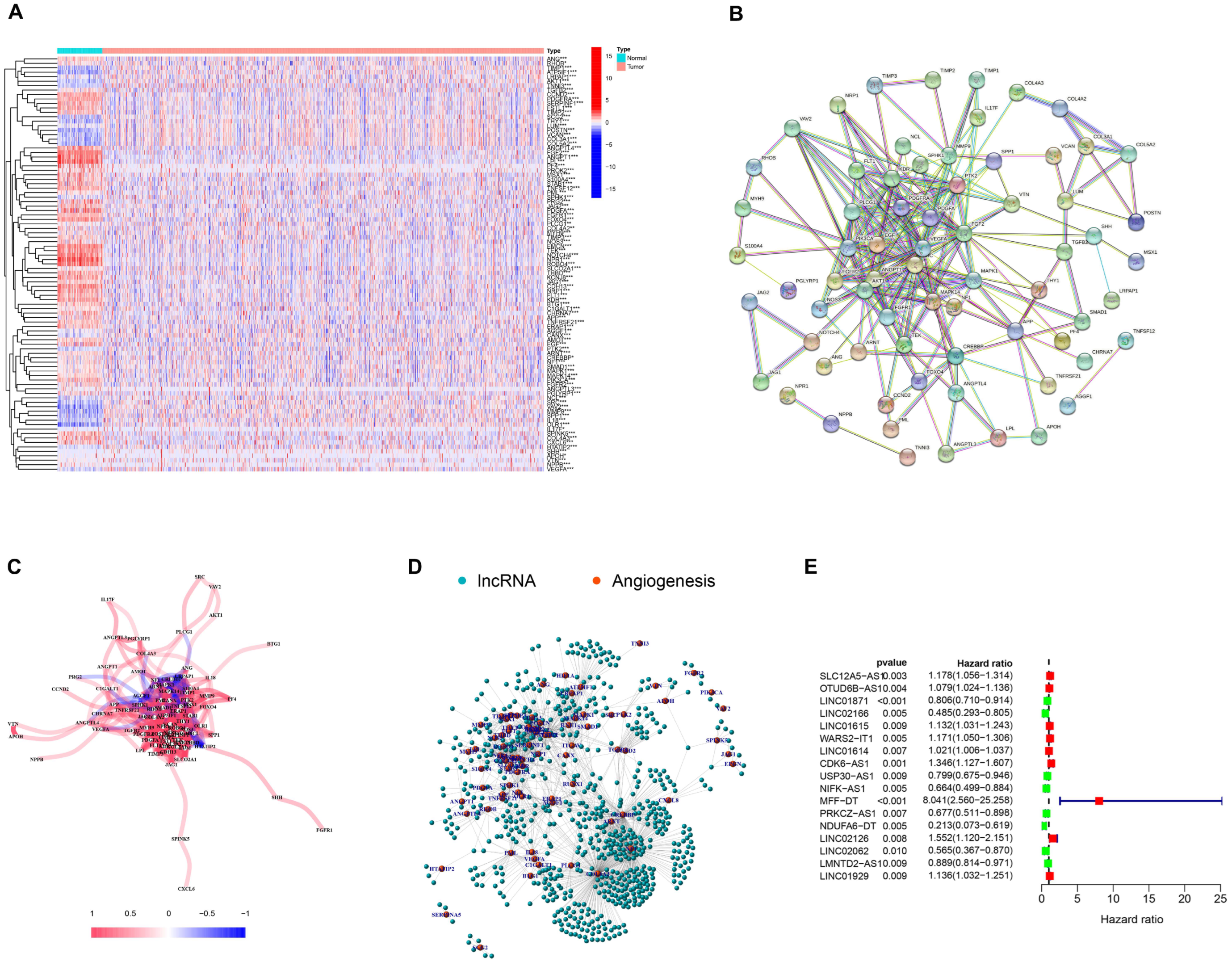
3.1. Selection of AR-lncRNAs in the Cancer Genome Atlas BC Cohort
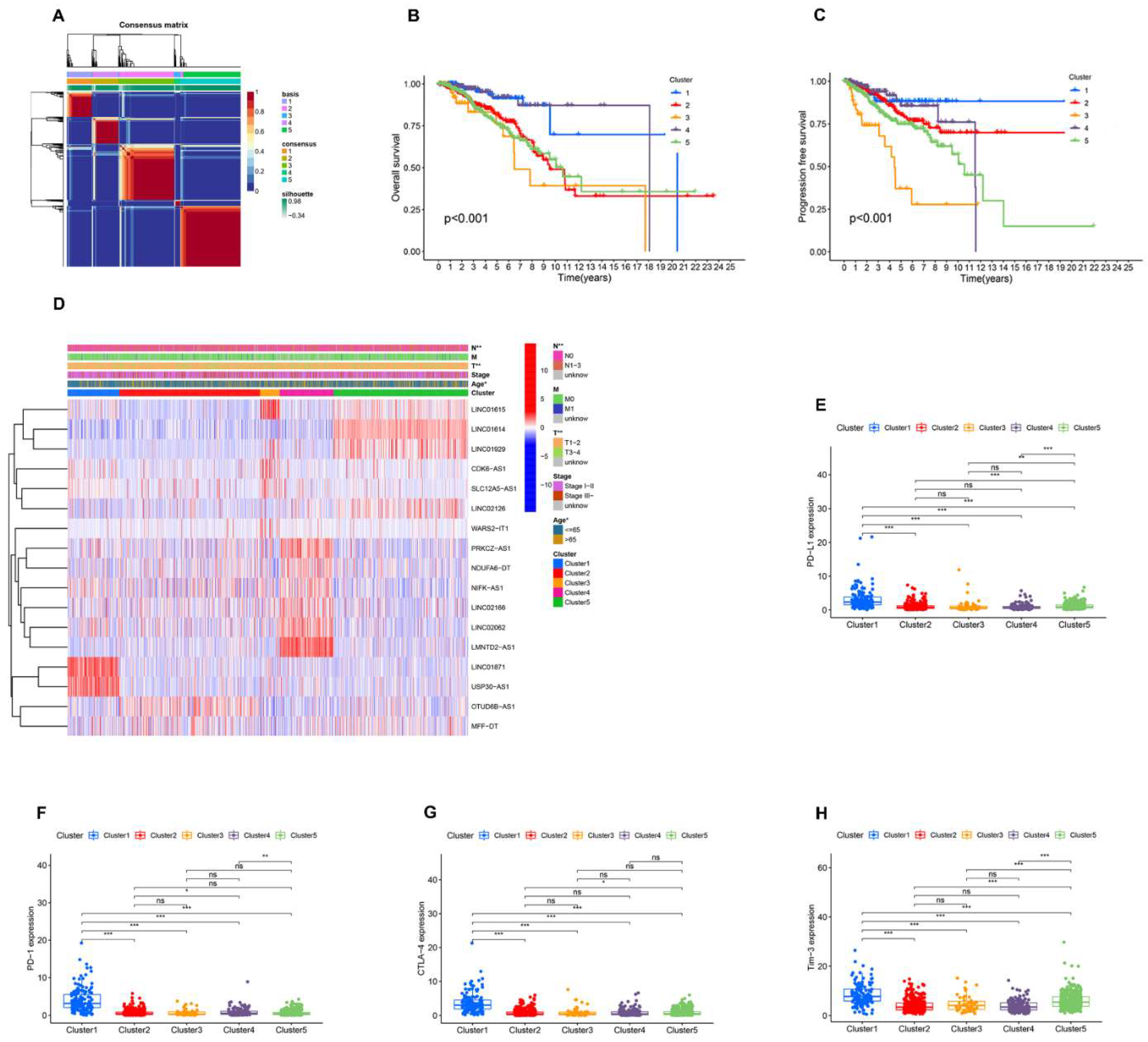
3.2. Identification of AR-lncRNAs and Immune Subtypes of BC
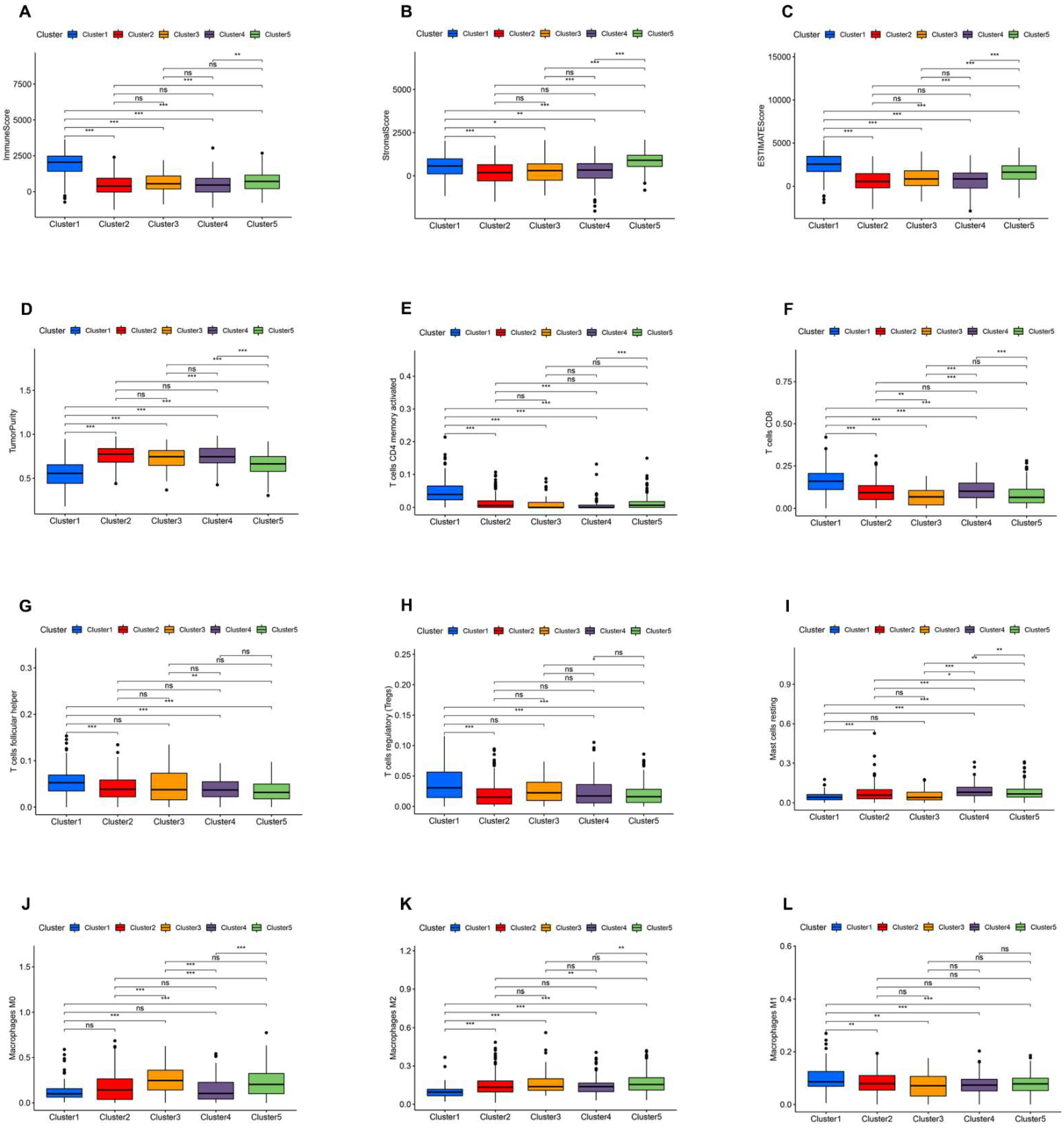
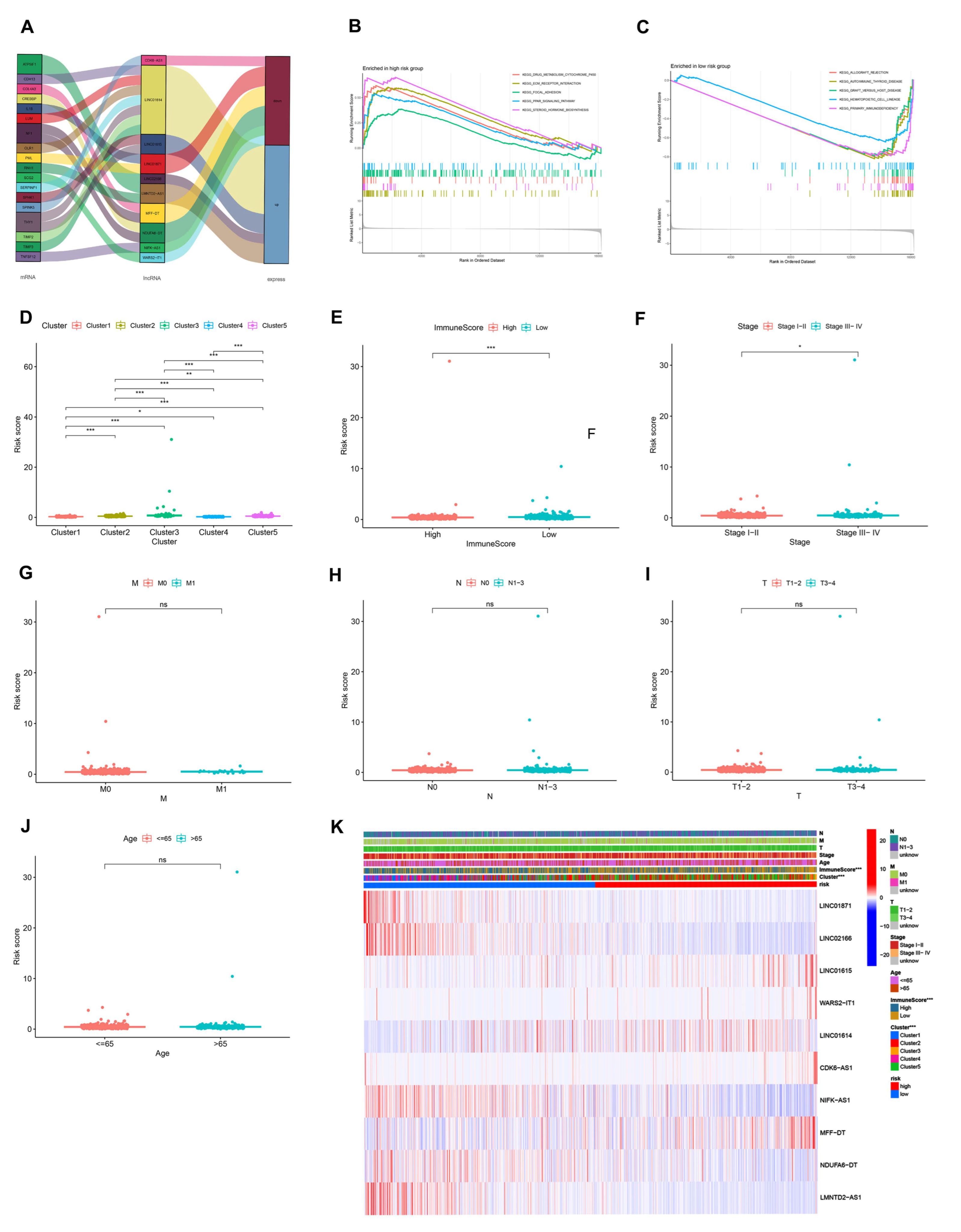
3.3. Immune Landscape of AR-lncRNA Subtypes of BC
3.4. Analysis of Gene Set Enrichment
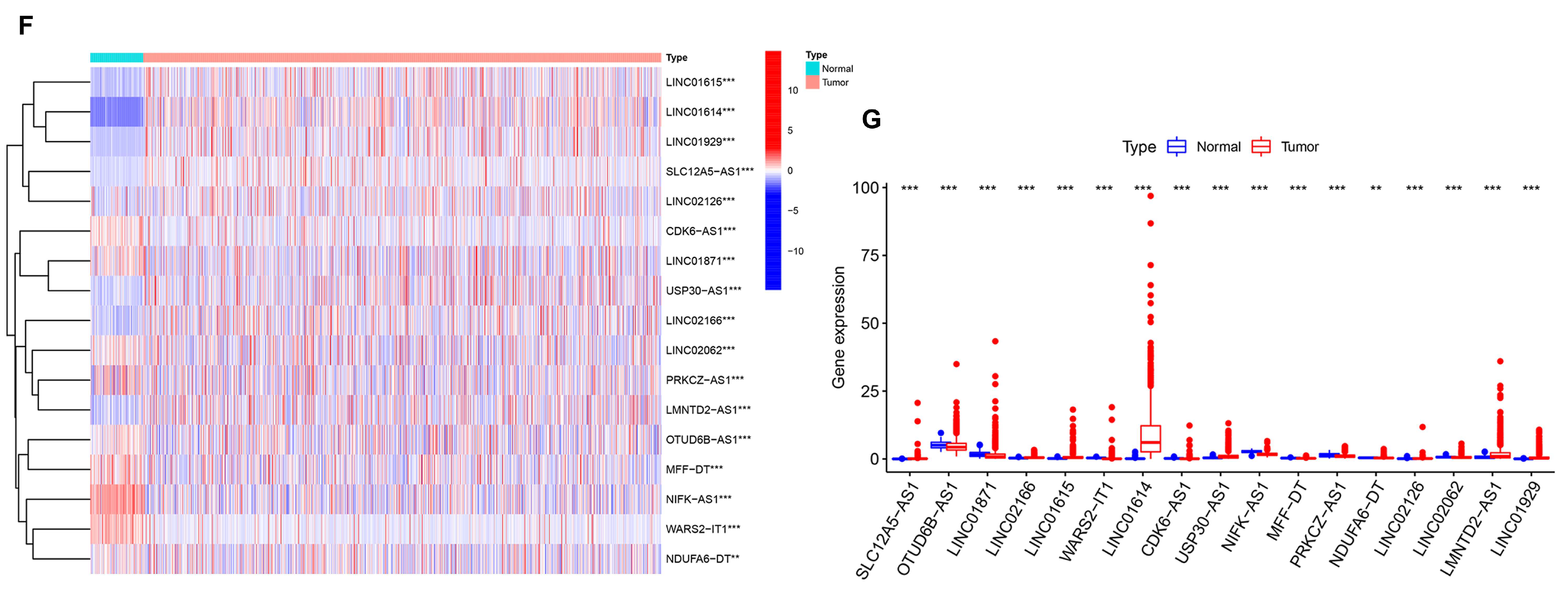
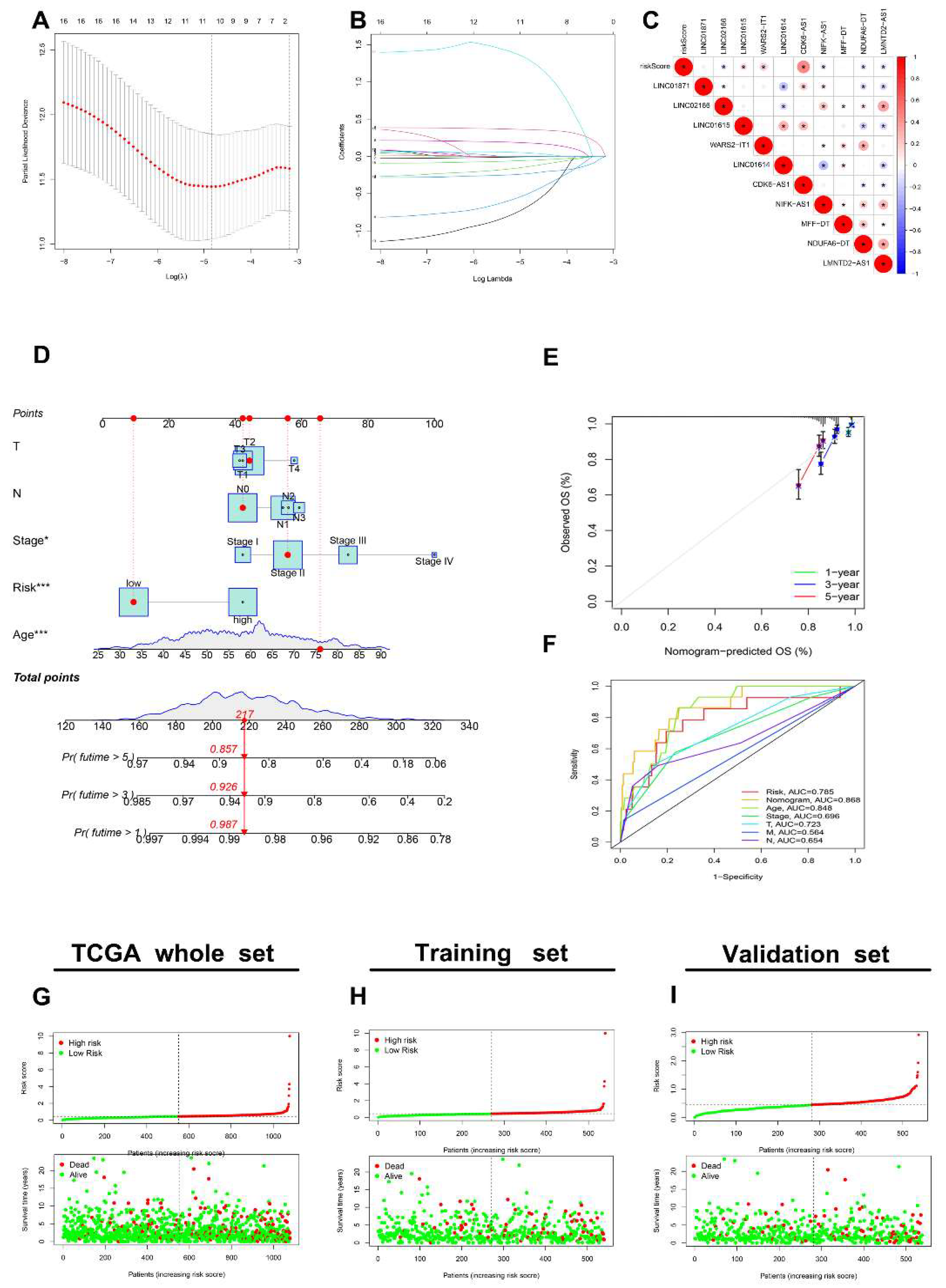
3.5. Generation of an AR-lncRNA Model for Prognosis
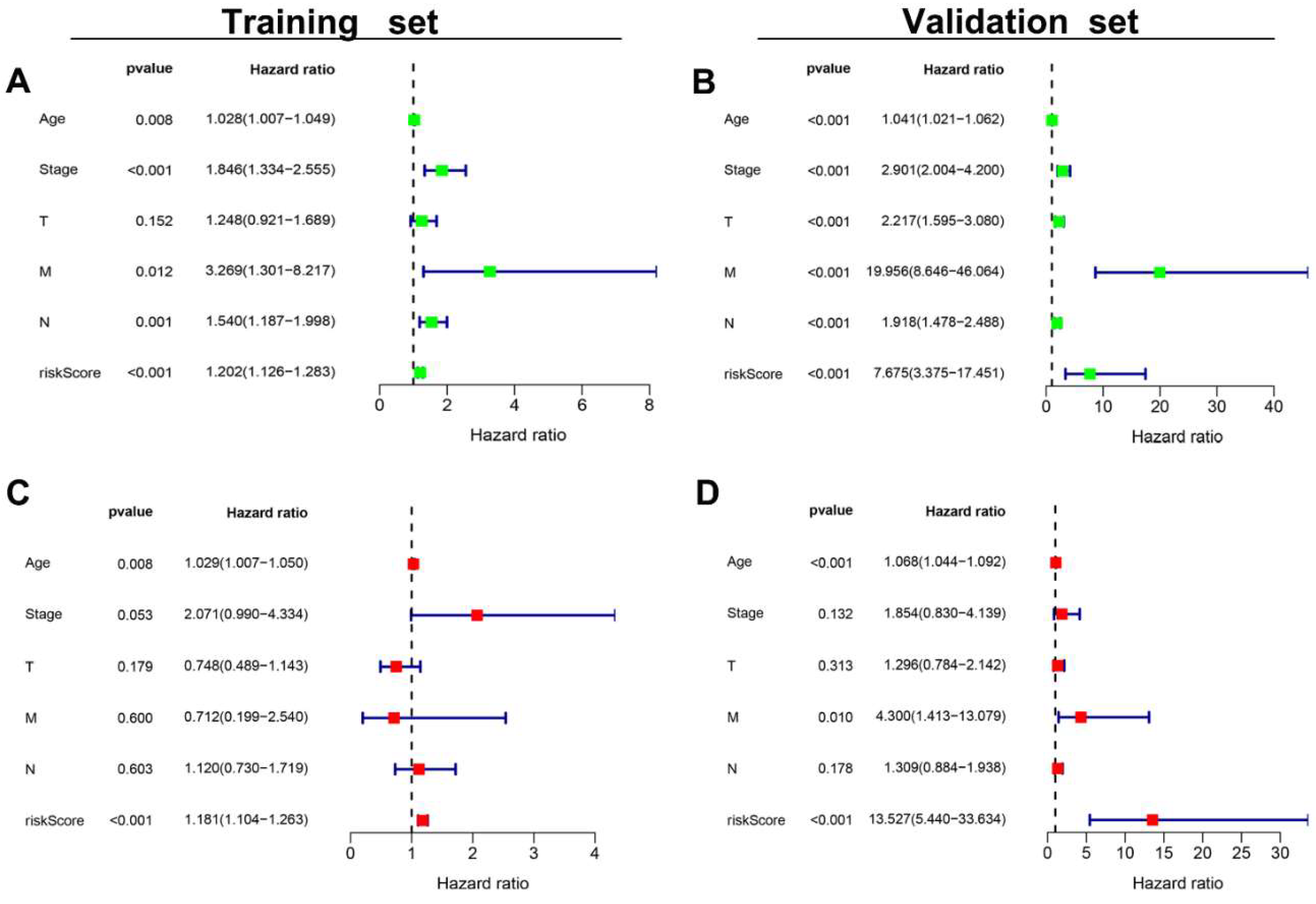
3.6. The AR-LncM Signature’s Predictive Value
3.7. Analysis of the Function and Clinical Characteristics of AR-LncM
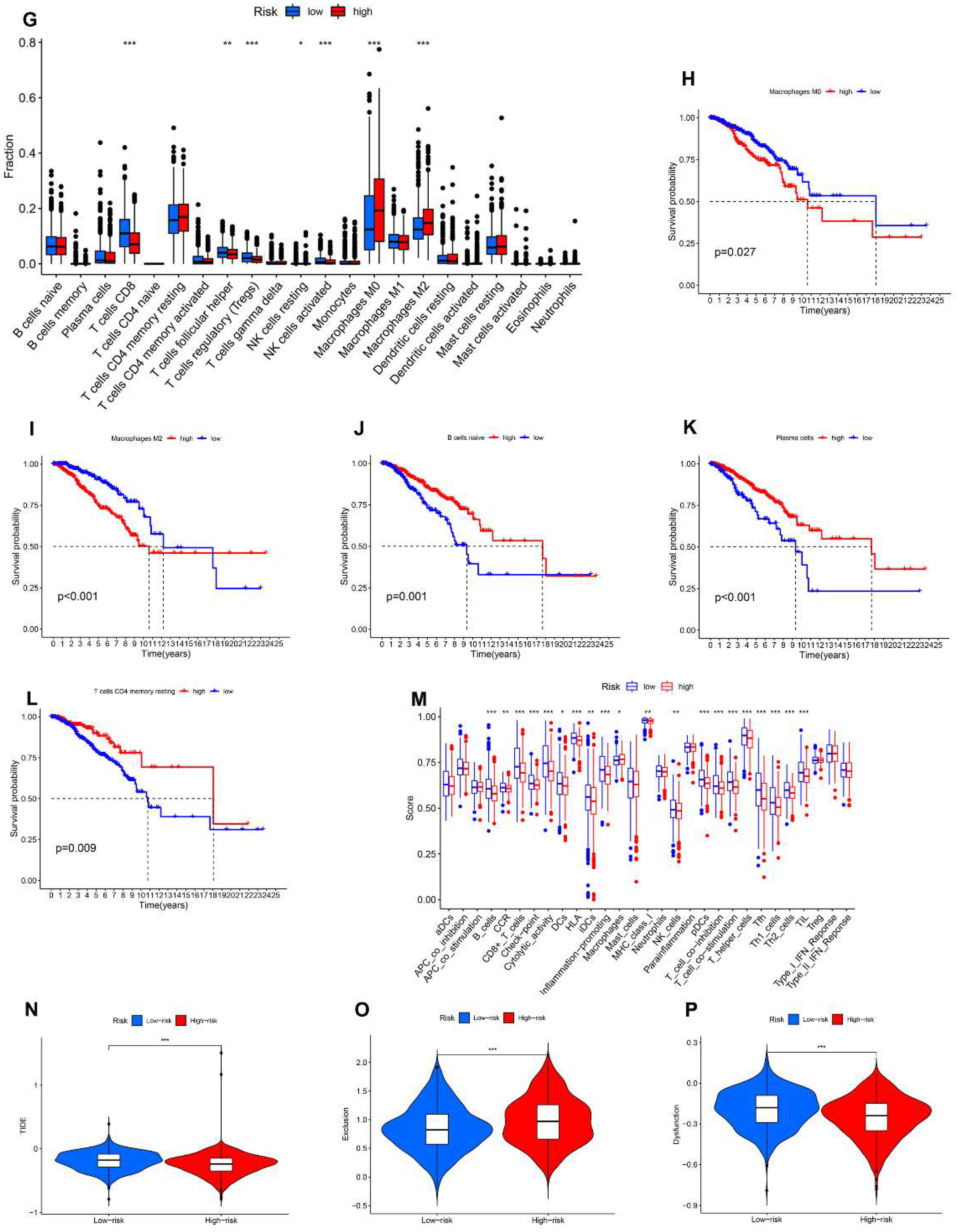
3.8. Relationship between the AR-LncM Signature and Immunotherapy and Immune Cell Infiltration
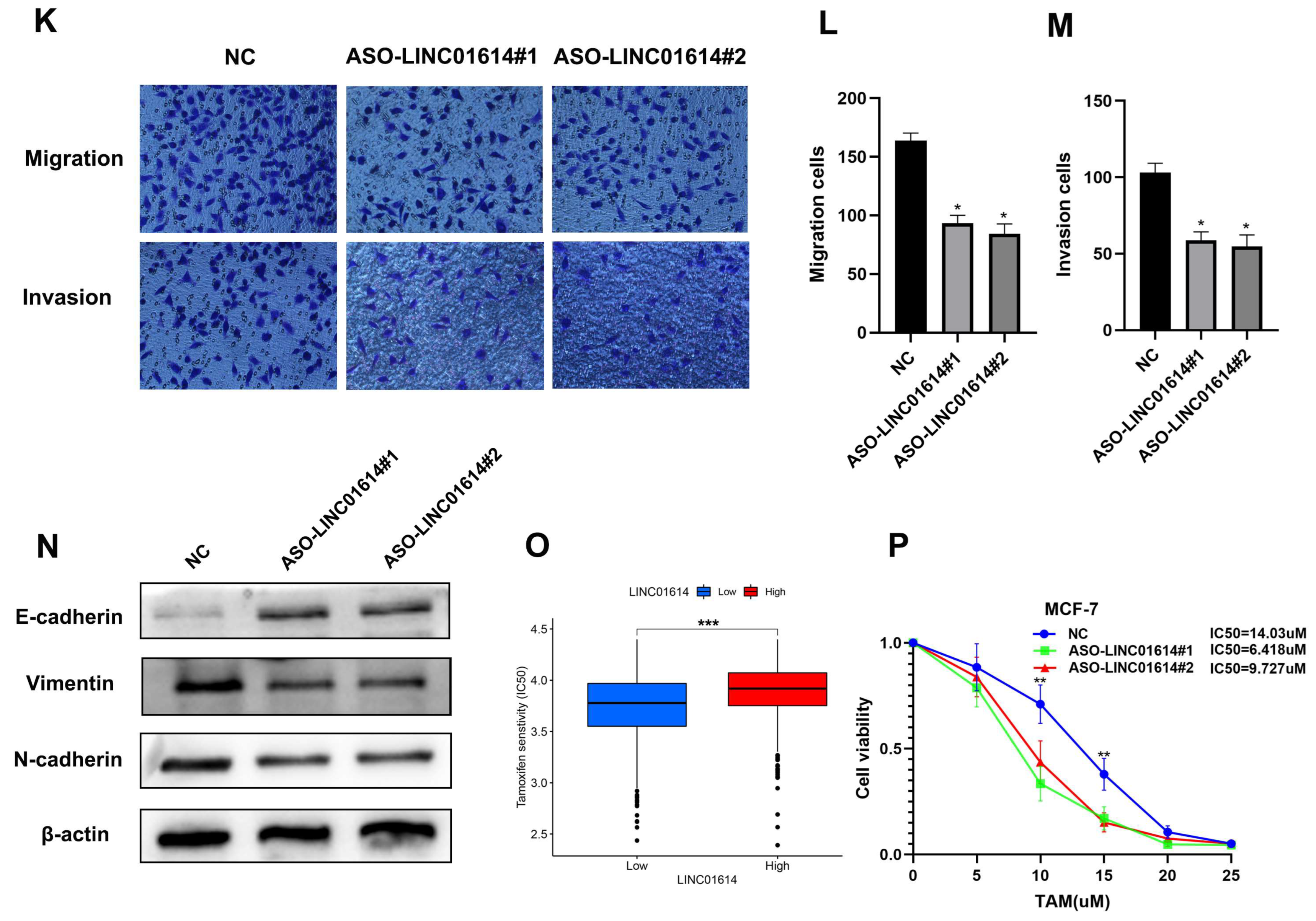
3.9. LINC01614 Is Involved in Regulating Cell Invasion, Migration, and Drug Sensitivity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Masood, S. Prognostic/predictive factors in breast cancer. Clin. Lab. Med. 2005, 25, 809–825. [Google Scholar] [CrossRef]
- Carmeliet, P. Angiogenesis in life, disease and medicine. Nature 2005, 438, 932–936. [Google Scholar] [CrossRef]
- Folkman, J. Tumor angiogenesis: A possible control point in tumor growth. Ann. Intern. Med. 1975, 82, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Valerio, G.; Casanovas, O. Angiogenesis and Metabolism: Entwined for Therapy Resistance. Trends Cancer 2017, 3, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Kuczynski, E.A.; Vermeulen, P.B.; Pezzella, F.; Kerbel, R.S.; Reynolds, A.R. Vessel co-option in cancer. Nat. Rev. Clin. Oncol. 2019, 16, 469–493. [Google Scholar] [CrossRef]
- Yi, M.; Jiao, D.; Qin, S.; Chu, Q.; Wu, K.; Li, A. Synergistic effect of immune checkpoint blockade and anti-angiogenesis in cancer treatment. Mol. Cancer 2019, 18, 60. [Google Scholar] [CrossRef]
- Han, C.; Zhang, C.; Wang, H.; Li, K.; Zhao, L. Angiogenesis-related lncRNAs predict the prognosis signature of stomach adenocarcinoma. BMC Cancer 2021, 21, 1312. [Google Scholar] [CrossRef] [PubMed]
- Lei, D.; Chen, Y.; Zhou, Y.; Hu, G.; Luo, F. An angiogenesis-related long noncoding RNA signature correlates with prognosis in patients with hepatocellular carcinoma. Biosci. Rep. 2021, 41, BSR20204442. [Google Scholar] [CrossRef] [PubMed]
- Fidler, I.J.; Ellis, L.M. The implications of angiogenesis for the biology and therapy of cancer metastasis. Cell 1994, 79, 185–188. [Google Scholar] [CrossRef]
- Schneider, B.P.; Miller, K.D. Angiogenesis of breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 1782–1790. [Google Scholar] [CrossRef]
- Banerjee, S.; Dowsett, M.; Ashworth, A.; Martin, L.A. Mechanisms of disease: Angiogenesis and the management of breast cancer. Nat. Clin. Pract. Oncol. 2007, 4, 536–550. [Google Scholar] [CrossRef] [PubMed]
- Fox, S.B.; Generali, D.G.; Harris, A.L. Breast tumour angiogenesis. Breast Cancer Res. BCR 2007, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Miles, D.W.; Diéras, V.; Cortés, J.; Duenne, A.A.; Yi, J.; O’Shaughnessy, J. First-line bevacizumab in combination with chemotherapy for HER2-negative metastatic breast cancer: Pooled and subgroup analyses of data from 2447 patients. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 2773–2780. [Google Scholar] [CrossRef]
- Hegde, P.S.; Karanikas, V.; Evers, S. The Where, the When, and the How of Immune Monitoring for Cancer Immunotherapies in the Era of Checkpoint Inhibition. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Yagi, T.; Baba, Y.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Watanabe, M.; Baba, H. PD-L1 Expression, Tumor-infiltrating Lymphocytes, and Clinical Outcome in Patients With Surgically Resected Esophageal Cancer. Ann. Surg. 2019, 269, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Teppola, P.; Taavitsainen, V.M. Parsimonious and robust multivariate calibration with rational function Least Absolute Shrinkage and Selection Operator and rational function Elastic Net. Anal. Chim. Acta 2013, 768, 57–68. [Google Scholar] [CrossRef]
- Kamarudin, A.N.; Cox, T.; Kolamunnage-Dona, R. Time-dependent ROC curve analysis in medical research: Current methods and applications. BMC Med. Res. Methodol. 2017, 17, 53. [Google Scholar] [CrossRef]
- Blanche, P.; Dartigues, J.F.; Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef] [PubMed]
- Fu J, Li K, Zhang W; et al. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef]
- Viallard, C.; Larrivée, B. Tumor angiogenesis and vascular normalization: Alternative therapeutic targets. Angiogenesis 2017, 20, 409–426. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. CR 2019, 38, 173. [Google Scholar] [CrossRef]
- Wu, S.; Guo, B.; Zhang, L.; Zhu, X.; Zhao, P.; Deng, J.; Zheng, J.; Li, F.; Wang, Y.; Zhang, S.; et al. A micropeptide XBP1SBM encoded by lncRNA promotes angiogenesis and metastasis of TNBC via XBP1s pathway. Oncogene 2022, 41, 2163–2172. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Camacho, C.V.; Setlem, R.; Kim, K.; Malladi, S.; Hou, T.Y.; Nandu, T.; Gadad, S.S.; Kraus, W.L. Functional Characterization of lncRNA152 as an Angiogenesis-Inhibiting Tumor Suppressor in Triple-Negative Breast Cancers. Mol. Cancer Res. MCR 2022, 20, 1623–1635. [Google Scholar] [CrossRef]
- Zeng, H.; Hou, Y.; Zhou, X.; Lang, L.; Luo, H.; Sun, Y.; Wan, X.; Yuan, T.; Wang, R.; Liu, Y.; et al. Cancer-associated fibroblasts facilitate premetastatic niche formation through lncRNA SNHG5-mediated angiogenesis and vascular permeability in breast cancer. Theranostics 2022, 12, 7351–7370. [Google Scholar] [CrossRef] [PubMed]
- Harry, J.A.; Ormiston, M.L. Novel Pathways for Targeting Tumor Angiogenesis in Metastatic Breast Cancer. Front. Oncol. 2021, 11, 772305. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.; Wang, Y.; Zhang, X.; Wang, C.; Yang, D.; Chen, J.; Long, Y.; Jiang, Y.; Zhou, X.; Zhang, N. Identification of Angiogenesis-Related Prognostic Biomarkers Associated with Immune Cell Infiltration in Breast Cancer. Front. Cell Dev. Biol. 2022, 10, 853324. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Li, J.; Li, Y.; Duan, W.; Qi, Q.; Wang, T.; Yang, Q.; Du, L.; Mao, H.; Wang, C. A novel long non-coding RNA AC073352.1 promotes metastasis and angiogenesis via interacting with YBX1 in breast cancer. Cell Death Dis. 2021, 12, 670. [Google Scholar] [CrossRef]
- Xu, Y.; Peng, Y.; Shen, M.; Liu, L.; Lei, J.; Gao, S.; Wang, Y.; Lan, A.; Li, H.; Liu, S. Construction and Validation of Angiogenesis-Related Prognostic Risk Signature to Facilitate Survival Prediction and Biomarker Excavation of Breast Cancer Patients. J. Oncol. 2022, 2022, 1525245. [Google Scholar] [CrossRef] [PubMed]
- Teng, M.W.; Galon, J.; Fridman, W.H.; Smyth, M.J. From mice to humans: Developments in cancer immunoediting. J. Clin. Investig. 2015, 125, 3338–3346. [Google Scholar] [CrossRef]
- Spill, F.; Reynolds, D.S.; Kamm, R.D.; Zaman, M.H. Impact of the physical microenvironment on tumor progression and metastasis. Curr. Opin. Biotechnol. 2016, 40, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, A.; Schioppa, T.; Tiberio, L.; Stabile, H.; Sozzani, S. Leukocyte trafficking in tumor microenvironment. Curr. Opin. Pharmacol. 2017, 35, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hu, Y.; Liu, W. Pyroptosis-Mediated Molecular Subtypes are Characterized by Distinct Tumor Microenvironment Infiltration Characteristics in Breast Cancer. J. Inflamm. Res. 2022, 15, 345–362. [Google Scholar] [CrossRef]
- Jiang, T.; Wang, Y.; Chen, X.; Xia, W.; Xue, S.; Gu, L.; Guo, L.; Lin, H. Neutrophil extracellular traps (NETs)-related lncRNAs signature for predicting prognosis and the immune microenvironment in breast cancer. Front. Cell Dev. Biol. 2023, 11, 1117637. [Google Scholar] [CrossRef]
- Zhang, Z.; Qiu, X.; Yan, Y.; Liang, Q.; Cai, Y.; Peng, B.; Xu, Z.; Xia, F. Evaluation of Ferroptosis-related Gene AKR1C1 as a Novel Biomarker Associated with the Immune Microenvironment and Prognosis in Breast Cancer. Int. J. Gen. Med. 2021, 14, 6189–6200. [Google Scholar] [CrossRef]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Medrek, C.; Pontén, F.; Jirström, K.; Leandersson, K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer 2012, 12, 306. [Google Scholar] [CrossRef]
- Fortis, S.P.; Sofopoulos, M.; Sotiriadou, N.N.; Haritos, C.; Vaxevanis, C.K.; Anastasopoulou, E.A.; Janssen, N.; Arnogiannaki, N.; Ardavanis, A.; Pawelec, G.; et al. Differential intratumoral distributions of CD8 and CD163 immune cells as prognostic biomarkers in breast cancer. J. Immunother. Cancer 2017, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.; Ferlazzo, G.; Albini, A.; Noonan, D.M. A think tank of TINK/TANKs: Tumor-infiltrating/tumor-associated natural killer cells in tumor progression and angiogenesis. J. Natl. Cancer Inst. 2014, 106, dju200. [Google Scholar] [CrossRef] [PubMed]
- Solis-Castillo, L.A.; Garcia-Romo, G.S.; Diaz-Rodriguez, A.; Reyes-Hernandez, D.; Tellez-Rivera, E.; Rosales-Garcia, V.H.; Mendez-Cruz, A.R.; Jimenez-Flores, J.R.; Villafana-Vazquez, V.H.; Pedroza-Gonzalez, A. Tumor-infiltrating regulatory T cells, CD8/Treg ratio, and cancer stem cells are correlated with lymph node metastasis in patients with early breast cancer. Breast Cancer J. Jpn. Breast Cancer Soc. 2020, 27, 837–849. [Google Scholar] [CrossRef] [PubMed]






| HALLMARK_ANGIOGENESIS | WP_ANGIOGENESIS | ANGIOGENESIS |
|---|---|---|
| APOH, APP, CCND2, COL3A1, COL5A2, CXCL6, FGFR1, FSTL1, ITGAV, JAG1, JAG2, KCNJ8, LPL, LRPAP1, LUM, MSX1, NRP1, OLR1, DGFA, PF4, PGLYRP1, POSTN, PRG2, PTK2, S100A4, SERPINA5, SLCO2A1, SPP1, STC1, THBD, TIMP1, TNFRSF21, VAV2, VCAN, VEGFA, VTN | AKT1, ANGPT1, ARNT, CREBBP, FGF2, FGFR2, FLT1, HIF1A, KDR, MAPK1, MAPK14, MMP9, NOS3, PDGFB, PDGFRA, PIK3CA, PLCG1, PTK2, SMAD1, SRC, TEK, TIMP2, TIMP3, EGFA | ACVRL1, AGGF1, AMOT, ANG, ANGPTL3, ANGPTL4, ATP5IF1, BTG1, C1GALT1, CANX, CDH13, CHRNA7, COL4A2, COL4A3, CXCL8, EGF, EMCN, EPGN, ERAP1, FOXO4, HTATIP2, IL17F, IL18, MYH9, NCL, NF1, NOTCH4, NPPB, NPR1, PF4, PLG, PML, PROK2, RHOB, RNH1, ROBO4, RUNX1, SCG2, SERPINF1, SHH, SPHK1, SPINK5, STAB1, TGFB2, THY1, TNFSF12, TNNI3, VEGFA |
| Cas NO. | Sex | Age, Years | Diagnosis | Tumor Location | Tumor Size (cm) | TNM Stage |
|---|---|---|---|---|---|---|
| 1 | Female | 69 | IBC | Left | 3.0 × 3.0 | T2N0M0 |
| 2 | Female | 49 | IBC | Right | 3.2 × 1.7 | T2N1M0 |
| 3 | Female | 61 | IBC | Right | 1.9 × 1.7 | T1N1M0 |
| 4 | Female | 55 | IBC | Right | 2.5 × 2.1 | T2N0M0 |
| 5 | Female | 46 | IBC | Right | 2.0 × 1.0 | T1N1M0 |
| 6 | Female | 48 | IBC | Left | 9.0 × 6.0 | T3N2M1 |
| 7 | Female | 41 | IBC | Left | 2.0 × 1.0 | T2NOM0 |
| 8 | Female | 38 | IBC | Right | 2.6 × 0.5 | T2N1M0 |
| 9 | Female | 63 | IBC | Left | 2.1 × 1.6 | T2NOM0 |
| 10 | Female | 61 | IBC | Left | 2.4 × 1.4 | T2NOM0 |
| 11 | Female | 64 | IBC | Right | 2.9 × 1.7 | T2NOM0 |
| 12 | Female | 55 | IBC | Left | 1.9 × 1.2 | T1N1M0 |
| GENE | HR | p Value | Coeffient |
|---|---|---|---|
| LINC01871 | 0.805696513 | 0.000786757 | −0.139091012 |
| LINC02166 | 0.485364461 | 0.005052064 | −0.504430065 |
| LINC01615 | 1.131750977 | 0.009372695 | 0.046593523 |
| WARS2-IT1 | 1.170943297 | 0.00455321 | 0.180717203 |
| LINC01614 | 1.021172679 | 0.006761123 | −0.005591609 |
| CDK6-AS1 | 1.346135832 | 0.001023502 | 0.359540355 |
| NIFK-AS1 | 0.664026974 | 0.005039487 | −0.231976683 |
| MFF-DT | 8.040952442 | 0.000357726 | 1.310004855 |
| NDUFA6-DT | 0.212778995 | 0.004502171 | −0.67551201 |
| LMNTD2-AS1 | 0.889106279 | 0.008835066 | −0.05439316 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, S.; Wang, Y.; Xu, Y.; Liu, S. An Angiogenesis-Related lncRNA Signature Is Associated with Prognosis and Tumor Immune Microenvironment in Breast Cancer. J. Pers. Med. 2023, 13, 513. https://doi.org/10.3390/jpm13030513
Gao S, Wang Y, Xu Y, Liu S. An Angiogenesis-Related lncRNA Signature Is Associated with Prognosis and Tumor Immune Microenvironment in Breast Cancer. Journal of Personalized Medicine. 2023; 13(3):513. https://doi.org/10.3390/jpm13030513
Chicago/Turabian StyleGao, Shun, Yuan Wang, Yingkun Xu, and Shengchun Liu. 2023. "An Angiogenesis-Related lncRNA Signature Is Associated with Prognosis and Tumor Immune Microenvironment in Breast Cancer" Journal of Personalized Medicine 13, no. 3: 513. https://doi.org/10.3390/jpm13030513
APA StyleGao, S., Wang, Y., Xu, Y., & Liu, S. (2023). An Angiogenesis-Related lncRNA Signature Is Associated with Prognosis and Tumor Immune Microenvironment in Breast Cancer. Journal of Personalized Medicine, 13(3), 513. https://doi.org/10.3390/jpm13030513






