Selecting Genetic Variants and Interactions Associated with Amyotrophic Lateral Sclerosis: A Group LASSO Approach
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Set
2.2. Genetic Model
2.3. LASSO
2.4. Group LASSO
2.5. Pairwise Interaction Model
2.6. Hierarchical Group LASSO Regularization
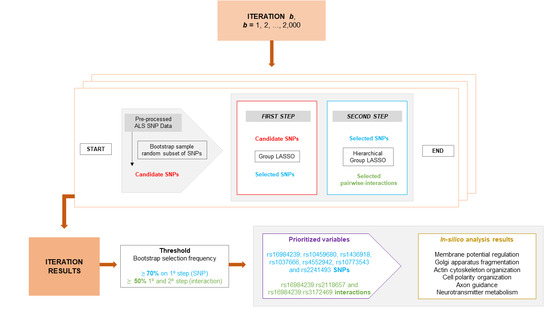
2.7. Implementation
2.8. Descriptive and Analytical Statistics
2.9. In Silico Analysis
3. Results and Discussion
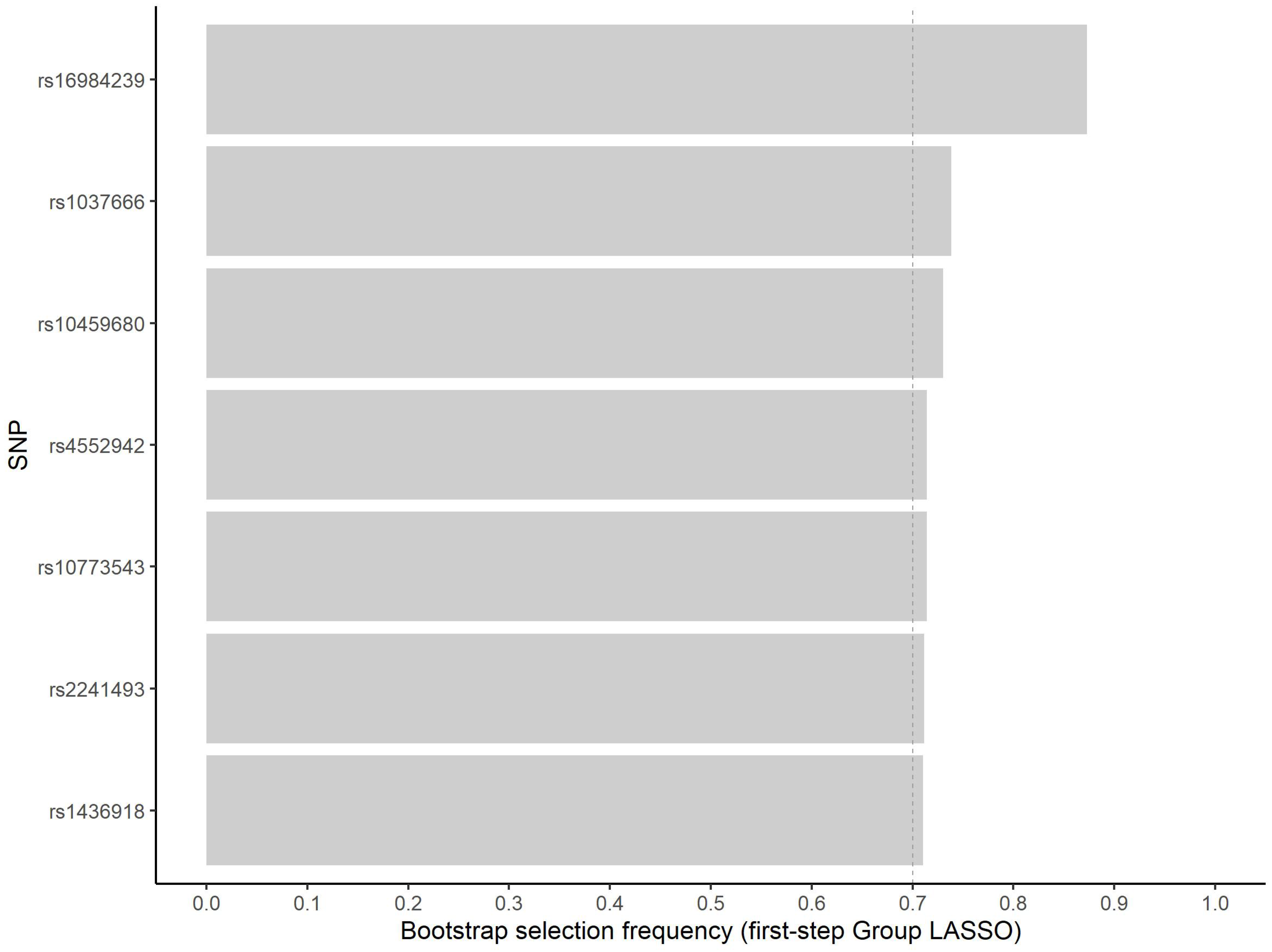
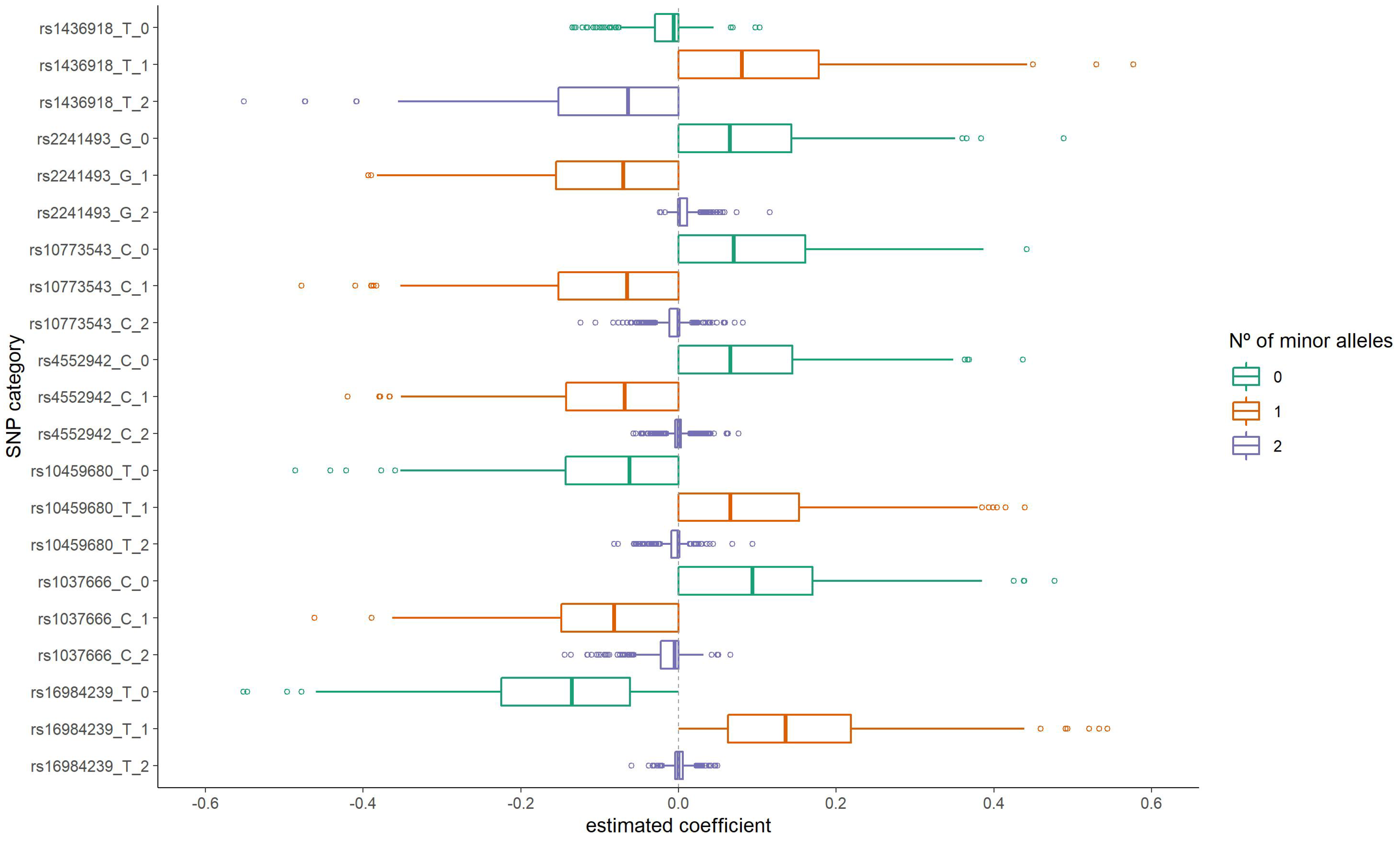
3.1. Variable Selection
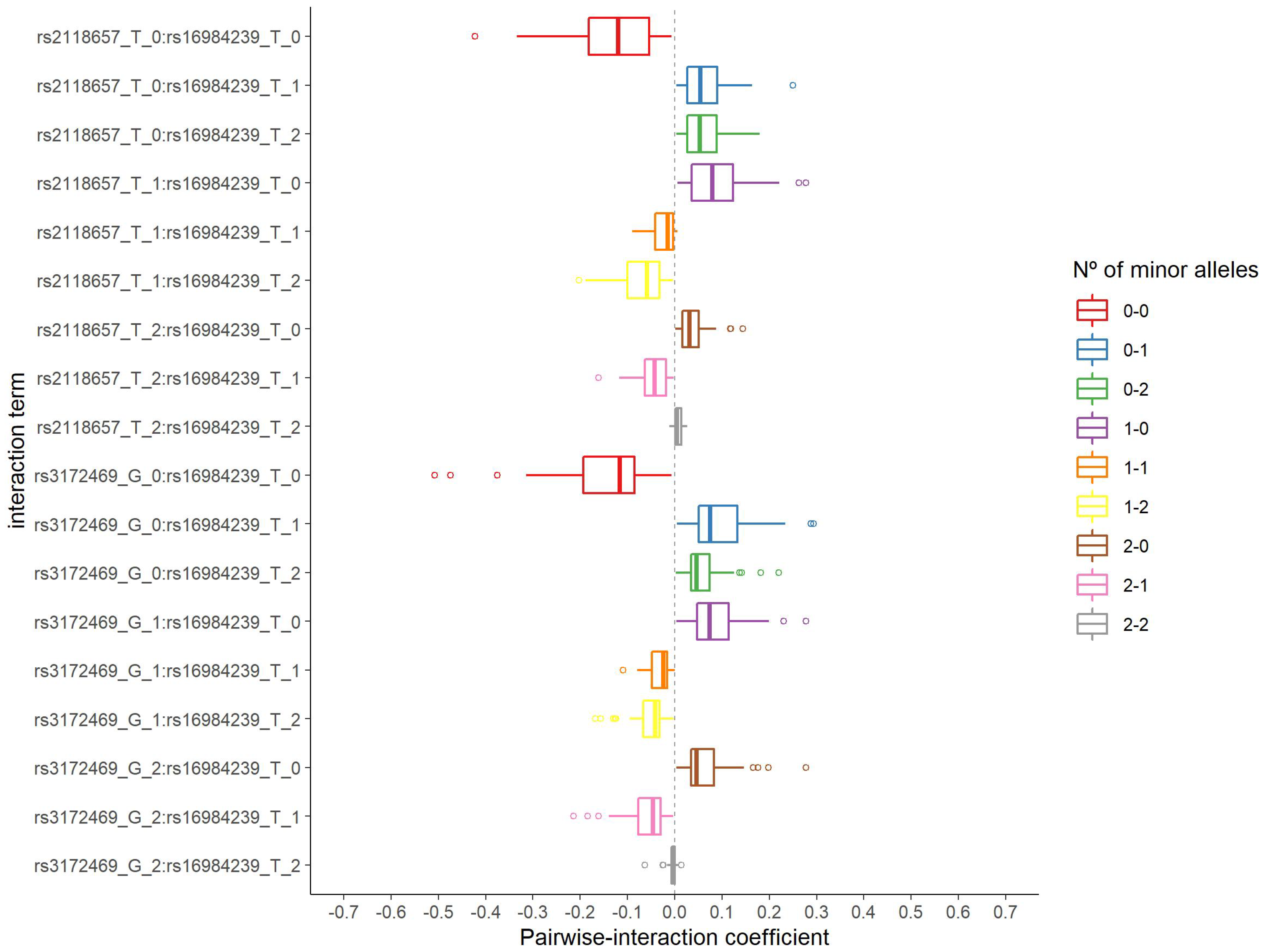
3.2. Pairwise Interaction Selection
3.3. LASSO and Related Approaches in Variable Selection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic Lateral Sclerosis |
| GWAS | Genome-Wide Association Studies |
| SNP | Single-Nucleotide Polymorphism |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| eQTL | Expression Quantitative Trait Loci |
| QC | Quality Control |
| OR | Odds Ratio |
| CI | Confidence Interval |
| LRT | Likelihood Ratio Test |
| KCNS3 | Potassium Voltage-Gated Channel Subfamily S member 3 |
| FMN2 | Formine-2 |
| RGM | Repulsive Guidance Molecules |
| RGMA | Repulsive Guidance Molecule Co-Receptor A |
| BMP | Bone Morphogenetic Protein |
| LINC02055 | Long Intergenic Non-Protein Coding RNA 2055 |
| RPS6 | Ribosomal Protein S6 |
| TMEM132C | Transmembrane Protein 132C |
| GOLGA8B | Golgin A8 Family Member B |
| TRP | Transient Receptor Potential |
| TRPM | Melastatin Transient Receptor Potential |
| ncRNA | Non-Coding RNA |
| BCHE | Butyrylcholinesterase |
| ACHE | Acetylcholinesterase |
| BCL6 | B-Cell Lymphoma 6 |
References
- Talbot, K. Motor neuron disease: The bare essentials. Pract. Neurol. 2009, 9, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; Van den Berg, L.H. Amyotrophic lateral sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef]
- Swinnen, B.; Robberecht, W. The phenotypic variability of amyotrophic lateral sclerosis. Nat. Rev. Neurol. 2014, 10, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Sabatelli, M.; Marangi, G.; Conte, A.; Tasca, G.; Zollino, M.; Lattante, S. New ALS-related genes expand the spectrum paradigm of amyotrophic lateral sclerosis. Brain Pathol. 2016, 26, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Chia, R.; Chiò, A.; Traynor, B.J. Novel genes associated with amyotrophic lateral sclerosis: Diagnostic and clinical implications. Lancet Neurol. 2018, 17, 94–102. [Google Scholar] [CrossRef]
- Brooks, B.R.; Miller, R.G.; Swash, M.; Munsat, T.L. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef]
- Van Steen, K.; Moore, J. How to increase our belief in discovered statistical interactions via large-scale association studies? Hum. Genet. 2019, 138, 293–305. [Google Scholar] [CrossRef]
- Niel, C.; Sinoquet, C.; Dina, C.; Rocheleau, G. A survey about methods dedicated to epistasis detection. Front. Genet. 2015, 6, 285. [Google Scholar] [CrossRef]
- Efron, B.; Hastie, T. Computer Age Statistical Inference; Cambridge University Press: New York, NY, USA, 2016. [Google Scholar]
- Park, M.Y.; Hastie, T. Penalized logistic regression for detecting gene interactions. Biostatistics 2008, 9, 30–50. [Google Scholar] [CrossRef]
- Wang, S.; Nan, B.; Rosset, S.; Zhu, J. Random lasso. Ann. Appl. Stat. 2011, 5, 468. [Google Scholar] [CrossRef]
- Schymick, J.C.; Scholz, S.W.; Fung, H.C.; Britton, A.; Arepalli, S.; Gibbs, J.R.; Lombardo, F.; Matarin, M.; Kasperaviciute, D.; Hernandez, D.G.; et al. Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: First stage analysis and public release of data. Lancet Neurol. 2007, 6, 322–328. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Wainwright, M. Statistical Learning with Sparsity: The Lasso and Generalizations; Chapman & Hall/CRC: Boca Raton, FL, USA, 2019. [Google Scholar]
- Agresti, A. Foundations of Linear and Generalized Linear Models; John Wiley & Sons: Hoboken, NJ, USA, 2015. [Google Scholar]
- Yang, Y.; Zou, H. A fast unified algorithm for solving group-lasso penalize learning problems. Stat. Comput. 2015, 25, 1129–1141. [Google Scholar] [CrossRef]
- Meier, L.; Van De Geer, S.; Bühlmann, P. The group lasso for logistic regression. J. R. Stat. Soc. Ser. B Stat. Methodol. 2008, 70, 53–71. [Google Scholar] [CrossRef]
- Lim, M.; Hastie, T. Learning interactions via hierarchical group-lasso regularization. J. Comput. Graph. Stat. 2015, 24, 627–654. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Imoto, S.; Miyano, S. Recursive random lasso (RRLasso) for identifying anti-cancer drug targets. PLoS ONE 2015, 10, e0141869. [Google Scholar] [CrossRef]
- Kim, Y.; Hao, J.; Mallavarapu, T.; Park, J.; Kang, M. Hi-lasso: High-dimensional lasso. IEEE Access 2019, 7, 44562–44573. [Google Scholar] [CrossRef]
- Hinrichs, A.S.; Raney, B.J.; Speir, M.L.; Rhead, B.; Casper, J.; Karolchik, D.; Kuhn, R.M.; Rosenbloom, K.R.; Zweig, A.S.; Haussler, D.; et al. UCSC data integrator and variant annotation integrator. Bioinformatics 2016, 32, 1430–1432. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The ensembl variant effect predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Ward, L.D.; Kellis, M. HaploReg: A resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012, 40, D930–D934. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef] [PubMed]
- Carithers, L.J.; Ardlie, K.; Barcus, M.; Branton, P.A.; Britton, A.; Buia, S.A.; Compton, C.C.; DeLuca, D.S.; Peter-Demchok, J.; Gelfand, E.T.; et al. A novel approach to high-quality postmortem tissue procurement: The GTEx project. Biopreserv. Biobank. 2015, 13, 311–319. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Meinshausen, N.; Meier, L.; Bühlmann, P. p-values for high-dimensional regression. J. Am. Stat. Assoc. 2009, 104, 1671–1681. [Google Scholar] [CrossRef]
- Laird, N.M.; Lang, C. The Fundamentals of Modern Statistical Genetics; Springer: New York, NY, USA, 2011. [Google Scholar]
- Sha, Q.; Zhang, Z.; Schymick, J.C.; Traynor, B.J.; Zhang, S. Genome-wide association reveals three SNPs associated with sporadic amyotrophic lateral sclerosis through a two-locus analysis. BMC Med. Genet. 2009, 10, 86. [Google Scholar] [CrossRef]
- Pan, W. Statistical tests of genetic association in the presence of gene-gene and gene-environment interactions. Hum. Hered. 2010, 69, 131–142. [Google Scholar] [CrossRef][Green Version]
- Macintyre, G.; Bailey, J.; Haviv, I.; Kowalczyk, A. is-rSNP: A novel technique for in silico regulatory SNP detection. Bioinformatics 2010, 26, i524–i530. [Google Scholar] [CrossRef]
- Han, F.; Pan, W. A composite likelihood approach to latent multivariate gaussian modeling of snp data with application to genetic association testing. Biometrics 2012, 68, 307–315. [Google Scholar] [CrossRef]
- Han, F.; Pan, W. Powerful multi-marker association tests: Unifying genomic distance-based regression and logistic regression. Genet. Epidemiol. 2010, 34, 680–688. [Google Scholar] [CrossRef]
- Audo, I.; Kohl, S.; Leroy, B.P.; Munier, F.L.; Guillonneau, X.; Mohand-Saïd, S.; Bujakowska, K.; Nandrot, E.F.; Lorenz, B.; Preising, M.; et al. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2009, 85, 720–729. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Sergouniotis, P.I.; Michaelides, M.; Mackay, D.S.; Wright, G.A.; Devery, S.; Moore, A.T.; Holder, G.E.; Robson, A.G.; Webster, A.R. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am. J. Hum. Genet. 2009, 85, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Thameem, F.; Puppala, S.; Arar, N.H.; Blangero, J.; Duggirala, R.; Abboud, H.E. Genetic variants in Transient Receptor Potential cation channel, subfamily M 1 (TRPM1) and their risk of albuminuria-related traits in Mexican Americans. Clin. Chim. Acta 2011, 412, 2058–2062. [Google Scholar] [CrossRef] [PubMed]
- Okumus, S.; Demiryürek, S.; Gürler, B.; Coskun, E.; Bozgeyik, İ.; Oztuzcu, S.; Kaydu, E.; Celik, O.; Erbagcı, İ.; Demiryürek, A.T. Association transient receptor potential melastatin channel gene polymorphism with primary open angle glaucoma. Mol. Vis. 2013, 19, 1852. [Google Scholar] [PubMed]
- Yamada, Y.; Yasukochi, Y.; Kato, K.; Oguri, M.; Horibe, H.; Fujimaki, T.; Takeuchi, I.; Sakuma, J. Identification of 26 novel loci that confer susceptibility to early-onset coronary artery disease in a Japanese population. Biomed. Rep. 2018, 9, 383–404. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, M.; Shang, Z.; Li, J.; Zhang, S.; Lian, D.; Zhang, R. Genome-wide haplotype association study identify the FGFR2 gene as a risk gene for acute myeloid leukemia. Oncotarget 2017, 8, 7891. [Google Scholar] [CrossRef]
- Van Ness, B.; Ramos, C.; Haznadar, M.; Hoering, A.; Haessler, J.; Crowley, J.; Jacobus, S.; Oken, M.; Rajkumar, V.; Greipp, P.; et al. Genomic variation in myeloma: Design, content, and initial application of the Bank on a Cure SNP Panel to detect associations with progression-free survival. BMC Med. 2008, 6, 26. [Google Scholar] [CrossRef]
- Morton, L.M.; Purdue, M.P.; Zheng, T.; Wang, S.S.; Armstrong, B.; Zhang, Y.; Menashe, I.; Chatterjee, N.; Davis, S.; Lan, Q.; et al. Risk of Non–Hodgkin Lymphoma Associated with Germline Variation in Genes that Regulate the Cell Cycle, Apoptosis, and Lymphocyte Development. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1259–1270. [Google Scholar] [CrossRef]
- Le Gall, L.; Anakor, E.; Connolly, O.; Vijayakumar, U.G.; Duddy, W.J.; Duguez, S. Molecular and cellular mechanisms affected in ALS. J. Pers. Med. 2020, 10, 101. [Google Scholar] [CrossRef]
- Stocker, M.; Kerschensteiner, D. Cloning and tissue distribution of two new potassium channel α-subunits from rat brain. Biochem. Biophys. Res. Commun. 1998, 248, 927–934. [Google Scholar] [CrossRef]
- Shepard, A.R.; Rae, J.L. Electrically silent potassium channel subunits from human lens epithelium. Am. J. Physiol.-Cell Physiol. 1999, 277, C412–C424. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, I.; Prado, Y.; Marchant, F.; Otero, C.; Eltit, F.; Cabello-Verrugio, C.; Cerda, O.; Simon, F. TRPM channels in human diseases. Cells 2020, 9, 2604. [Google Scholar] [CrossRef] [PubMed]
- Leader, B.; Leder, P. Formin-2, a novel formin homology protein of the cappuccino subfamily, is highly expressed in the developing and adult central nervous system. Mech. Dev. 2000, 93, 221–231. [Google Scholar] [CrossRef]
- Law, R.; Dixon-Salazar, T.; Jerber, J.; Cai, N.; Abbasi, A.A.; Zaki, M.S.; Mittal, K.; Gabriel, S.B.; Rafiq, M.A.; Khan, V.; et al. Biallelic truncating mutations in FMN2, encoding the actin-regulatory protein Formin 2, cause nonsyndromic autosomal-recessive intellectual disability. Am. J. Hum. Genet. 2014, 95, 721–728. [Google Scholar] [CrossRef] [PubMed]
- Mutalik, S.P. Role of the Cytoskeleton in Regulating Axonal Tension and Growth Cone Traction Dynamics. Ph.D. Thesis, Indian Institute of Science Education and Research, Pune, India, 2018. [Google Scholar]
- Sanchez-Pulido, L.; Ponting, C.P. TMEM132: An ancient architecture of cohesin and immunoglobulin domains define a new family of neural adhesion molecules. Bioinformatics 2018, 34, 721–724. [Google Scholar] [CrossRef] [PubMed]
- Severyn, C.J.; Shinde, U.; Rotwein, P. Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem. J. 2009, 422, 393–403. [Google Scholar] [CrossRef]
- Tang, J.; Zeng, X.; Li, H.; Ju, L.; Feng, J.; Yang, J. Repulsive guidance molecule-a and central nervous system diseases. BioMed Res. Int. 2021, 2021, 5532116. [Google Scholar] [CrossRef]
- Munro, S. The golgin coiled-coil proteins of the Golgi apparatus. Cold Spring Harb. Perspect. Biol. 2011, 3, a005256. [Google Scholar] [CrossRef]
- Sundaramoorthy, V.; Walker, A.K.; Yerbury, J.; Soo, K.Y.; Farg, M.A.; Hoang, V.; Zeineddine, R.; Spencer, D.; Atkin, J.D. Extracellular wildtype and mutant SOD1 induces ER–Golgi pathology characteristic of amyotrophic lateral sclerosis in neuronal cells. Cell. Mol. Life Sci. 2013, 70, 4181–4195. [Google Scholar] [CrossRef]
- Gonatas, N.; Stieber, A.; Mourelatos, Z.; Chen, Y.; Gonatas, J.; Appel, S.H.; Hays, A.; Hickey, W.; Hauw, J. Fragmentation of the Golgi apparatus of motor neurons in amyotrophic lateral sclerosis. Am. J. Pathol. 1992, 140, 731. [Google Scholar]
- Boucher, B.; Jenna, S. Genetic interaction networks: Better understand to better predict. Front. Genet. 2013, 4, 290. [Google Scholar] [CrossRef] [PubMed]
- Szklo, M.; Nieto, J. Epidemiology: Beyond the Basics; Jones & Bartlett Publishers: Burlington, MA, USA, 2014. [Google Scholar]
- Johnson, G.; Moore, S.W. Why has butyrylcholinesterase been retained? Structural and functional diversification in a duplicated gene. Neurochem. Int. 2012, 61, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Kozhemyakin, M.; Rajasekaran, K.; Kapur, J. Central cholinesterase inhibition enhances glutamatergic synaptic transmission. J. Neurophysiol. 2010, 103, 1748–1757. [Google Scholar] [CrossRef]
- Haidet-Phillips, A.M.; Hester, M.E.; Miranda, C.J.; Meyer, K.; Braun, L.; Frakes, A.; Song, S.; Likhite, S.; Murtha, M.J.; Foust, K.D.; et al. Astrocytes from familial and sporadic ALS patients are toxic to motor neurons. Nat. Biotechnol. 2011, 29, 824–828. [Google Scholar] [CrossRef] [PubMed]
- Barbour, B.; Brew, H.; Attwell, D. Electrogenic glutamate uptake in glial cells is activated by intracellular potassium. Nature 1988, 335, 433–435. [Google Scholar] [CrossRef]
- Allman, D.; Jain, A.; Dent, A.; Maile, R.R.; Selvaggi, T.; Kehry, M.R.; Staudt, L.M. BCL-6 expression during B-cell activation. Blood 1996, 87, 5257–5268. [Google Scholar] [CrossRef]
- Arlotta, P.; Molyneaux, B.J.; Chen, J.; Inoue, J.; Kominami, R.; Macklis, J.D. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 2005, 45, 207–221. [Google Scholar] [CrossRef]
- Wang, X.S.; Simmons, Z.; Liu, W.; Boyer, P.J.; Connor, J.R. Differential expression of genes in amyotrophic lateral sclerosis revealed by profiling the post mortem cortex. Amyotroph. Lateral Scler. 2006, 7, 201–216. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, C.L.; Messing, A.; Chiu, S.Y. Temperature-sensitive neuromuscular transmission in Kv1. 1 null mice: Role of potassium channels under the myelin sheath in young nerves. J. Neurosci. 1998, 18, 7200–7215. [Google Scholar] [CrossRef]
- Wu, T.T.; Chen, Y.F.; Hastie, T.; Sobel, E.; Lange, K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics 2009, 25, 714–721. [Google Scholar] [CrossRef]
- Yang, C.; Wan, X.; Yang, Q.; Xue, H.; Yu, W. Identifying main effects and epistatic interactions from large-scale SNP data via adaptive group Lasso. BMC Bioinform. 2010, 11, S18. [Google Scholar] [CrossRef]
- Shi, W.; Lee, K.E.; Wahba, G. Detecting disease-causing genes by LASSO-Patternsearch algorithm. BMC Proc. 2007, 1, S60. [Google Scholar] [CrossRef]
- Wu, C.H.; Fallini, C.; Ticozzi, N.; Keagle, P.J.; Sapp, P.C.; Piotrowska, K.; Lowe, P.; Koppers, M.; McKenna-Yasek, D.; Baron, D.M.; et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 2012, 488, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Hop, P.J.; Zwamborn, R.A.; Hannon, E.; Shireby, G.L.; Nabais, M.F.; Walker, E.M.; van Rheenen, W.; van Vugt, J.J.; Dekker, A.M.; Westeneng, H.J.; et al. Genome-wide study of DNA methylation shows alterations in metabolic, inflammatory, and cholesterol pathways in ALS. Sci. Transl. Med. 2022, 14, eabj0264. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Ou, S.; Tang, M.; Yin, Z.; Wang, Z.; Liu, G. Trauma and amyotrophic lateral sclerosis: A systematic review and meta-analysis. Amyotroph. Lateral Scler. Front. Degener. 2021, 22, 170–185. [Google Scholar] [CrossRef] [PubMed]
| Variant (Minor Allele) | Total n | ALS n (%) | OR | 95%CI | p-Value |
|---|---|---|---|---|---|
| rs16984239 (A) | |||||
| 0 | 378 | 165 (0.44) | reference | ||
| 1 | 153 | 104 (0.68) | 2.74 | 1.85; 4.10 | <0.001 |
| 2 | 13 | 7 (0.54) | 1.51 | 0.49; 4.76 | 0.469 |
| rs1037666 (C) | |||||
| 0 | 247 | 151 (0.61) | reference | ||
| 1 | 242 | 100 (0.41) | 0.45 | 0.31; 0.64 | <0.001 |
| 2 | 55 | 25 (0.45) | 0.53 | 0.29; 0.95 | 0.035 |
| rs10459680 (T) | |||||
| 0 | 281 | 118 (0.42) | reference | ||
| 1 | 230 | 142 (0.62) | 2.23 | 1.56; 3.19 | <0.001 |
| 2 | 33 | 16 (0.48) | 1.30 | 0.63; 2.69 | 0.477 |
| rs4552942 (C) | |||||
| 0 | 277 | 165 (0.60) | reference | ||
| 1 | 226 | 090 (0.40) | 0.45 | 0.31; 0.64 | <0.001 |
| 2 | 41 | 21 (0.51) | 0.71 | 0.37; 1.38 | 0.313 |
| rs10773543 (G) | |||||
| 0 | 230 | 142 (0.62) | reference | ||
| 1 | 261 | 109 (0.42) | 0.44 | 0.31; 0.64 | <0.001 |
| 2 | 53 | 25 (0.47) | 0.55 | 0.30; 1.01 | 0.054 |
| rs2241493 (C) | |||||
| 0 | 345 | 198 (0.57) | reference | ||
| 1 | 180 | 067 (0.37) | 0.44 | 0.30; 0.64 | <0.001 |
| 2 | 19 | 11 (0.58) | 1.02 | 0.40; 2.70 | 0.966 |
| rs1436918 (A) | |||||
| 0 | 129 | 061 (0.47) | reference | ||
| 1 | 288 | 171 (0.59) | 1.63 | 1.07; 2.48 | 0.022 |
| 2 | 127 | 44 (0.35) | 0.59 | 0.36; 0.98 | 0.040 |
| SNP | Chr:Location | Gene | Consequence | Phenotype | Citation |
|---|---|---|---|---|---|
| First step | |||||
| rs16984239 | 2:18053180 | - | intergenic | ALS | [12,30,31,32,33] |
| rs1037666 | 1:240195185 | FMN2 | intronic | ALS | [12] |
| rs10459680 | 15:93138241 | LOC101927025 | intronic | ALS | [12] |
| rs4552942 | 8:135862080 | LINC02055 | intronic | ALS; core binding factor acute myeloid leukemia | [12,34] |
| rs10773543 | 12:128439181 | TMEM132C | intronic | ALS | [12] |
| rs2241493 | 15:31070149 | TRPM1 | missense | Congenital stationary night blindness, type 1C | [35,36,37,38,39] |
| rs1436918 | 15:34644720 | LOC390569 | regulatory genomic region | ALS | [12,40] |
| Second step | |||||
| rs2118657 | 3:165864723 | - | intergenic | - | - |
| rs3172469 | 3:187741300 | BCL6 | intronic | Myeloma; non-Hodgkin lymphoma | [41,42] |
| Variant (Minor Allele) | Total n | ALS n (%) | OR | 95%CI | p-Value |
|---|---|---|---|---|---|
| rs2118657 (T) = 0 | |||||
| rs16984239 (A) = 0 | 189 | 61 (0.32) | reference | ||
| rs16984239 (A) = 1 | 83 | 55 (0.66) | 4.12 | 2.38; 7.13 | <0.001 |
| rs16984239 (A) = 2 | 7 | 6 (0.86) | 12.59 | 1.48; 106.89 | 0.020 |
| rs2118657 (T) = 1 | |||||
| rs16984239 (A) = 0 | 162 | 93 (0.57) | reference | ||
| rs16984239 (A) = 1 | 64 | 46 (0.72) | 1.90 | 1.01; 3.55 | 0.046 |
| rs16984239 (A) = 2 | 6 | 1 (0.17) | 0.15 | 0.02; 1.30 | 0.085 |
| rs2118657 (T) = 2 | |||||
| rs16984239 (A) = 0 | 27 | 11 (0.41) | reference | ||
| rs16984239 (A) = 1 | 6 | 3 (0.50) | 1.45 | 0.25; 8.58 | 0.679 |
| rs16984239 (A) = 2 | - | - | - | - | - |
| rs3172469 (G) = 0 | |||||
| rs16984239 (A) = 0 | 200 | 65 (0.33) | reference | ||
| rs16984239 (A) = 1 | 83 | 55 (0.66) | 4.08 | 2.37; 7.02 | <0.001 |
| rs16984239 (A) = 2 | 8 | 4 (0.50) | 2.08 | 0.50; 8.57 | 0.312 |
| rs3172469 (G) = 1 | |||||
| rs16984239 (A) = 0 | 150 | 84 (0.56) | reference | ||
| rs16984239 (A) = 1 | 63 | 45 (0.71) | 1.96 | 1.04; 3.71 | 0.037 |
| rs16984239 (A) = 2 | 5 | 3 (0.60) | 1.18 | 0.19; 7.26 | 0.859 |
| rs3172469 (G) = 2 | |||||
| rs16984239 (A) = 0 | 28 | 61 (0.47) | reference | ||
| rs16984239 (A) = 1 | 7 | 4 (0.57) | 1.00 | 0.19; 5.33 | 0.999 |
| rs16984239 (A) = 2 | - | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feronato, S.G.; Silva, M.L.M.; Izbicki, R.; Farias, T.D.J.; Shigunov, P.; Dallagiovanna, B.; Passetti, F.; dos Santos, H.G. Selecting Genetic Variants and Interactions Associated with Amyotrophic Lateral Sclerosis: A Group LASSO Approach. J. Pers. Med. 2022, 12, 1330. https://doi.org/10.3390/jpm12081330
Feronato SG, Silva MLM, Izbicki R, Farias TDJ, Shigunov P, Dallagiovanna B, Passetti F, dos Santos HG. Selecting Genetic Variants and Interactions Associated with Amyotrophic Lateral Sclerosis: A Group LASSO Approach. Journal of Personalized Medicine. 2022; 12(8):1330. https://doi.org/10.3390/jpm12081330
Chicago/Turabian StyleFeronato, Sofia Galvão, Maria Luiza Matos Silva, Rafael Izbicki, Ticiana D. J. Farias, Patrícia Shigunov, Bruno Dallagiovanna, Fabio Passetti, and Hellen Geremias dos Santos. 2022. "Selecting Genetic Variants and Interactions Associated with Amyotrophic Lateral Sclerosis: A Group LASSO Approach" Journal of Personalized Medicine 12, no. 8: 1330. https://doi.org/10.3390/jpm12081330
APA StyleFeronato, S. G., Silva, M. L. M., Izbicki, R., Farias, T. D. J., Shigunov, P., Dallagiovanna, B., Passetti, F., & dos Santos, H. G. (2022). Selecting Genetic Variants and Interactions Associated with Amyotrophic Lateral Sclerosis: A Group LASSO Approach. Journal of Personalized Medicine, 12(8), 1330. https://doi.org/10.3390/jpm12081330