Associations between the Levels of Estradiol-, Progesterone-, and Testosterone-Sensitive MiRNAs and Main Clinicopathologic Features of Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Tissue Samples
2.3. MicroRNA Isolation
2.4. MiRNA Reverse Transcription and Real-Time PCR (RT-PCR)
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Selection of Estradiol-, Progesterone-, Testosterone-Sensitive miRNAs
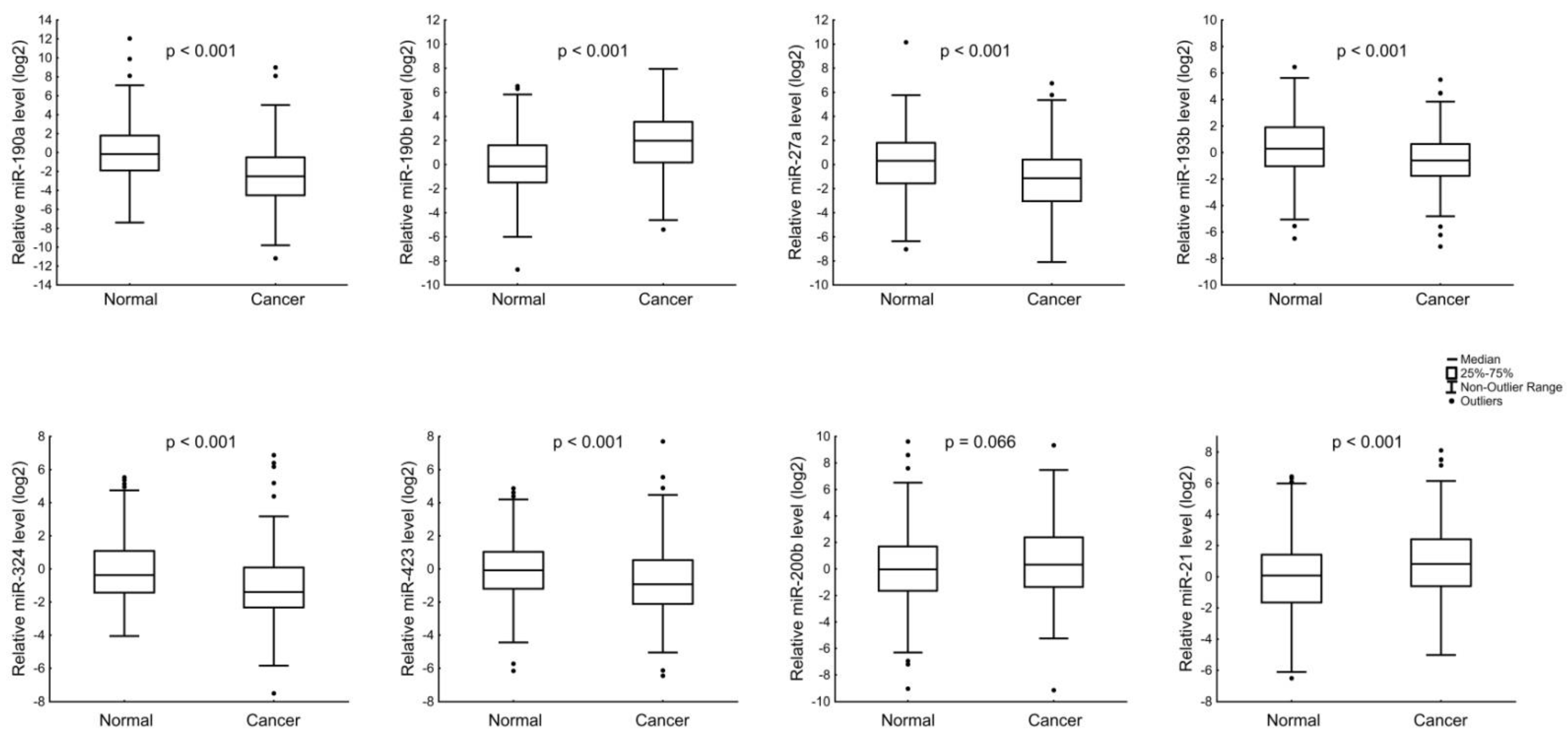
3.2. Analysis of the Hormone-Sensitive MiRNAs Expression in Breast Cancer
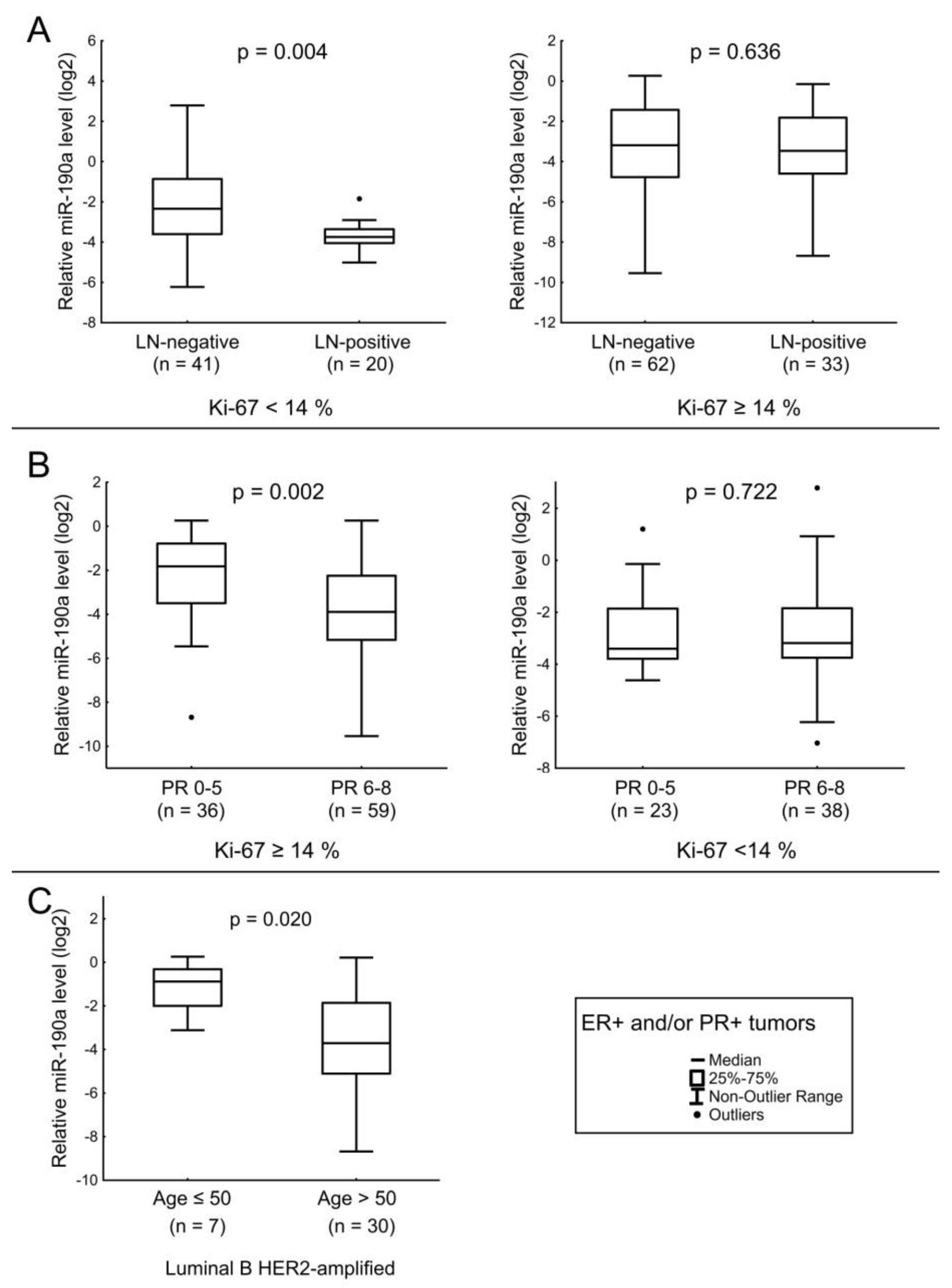
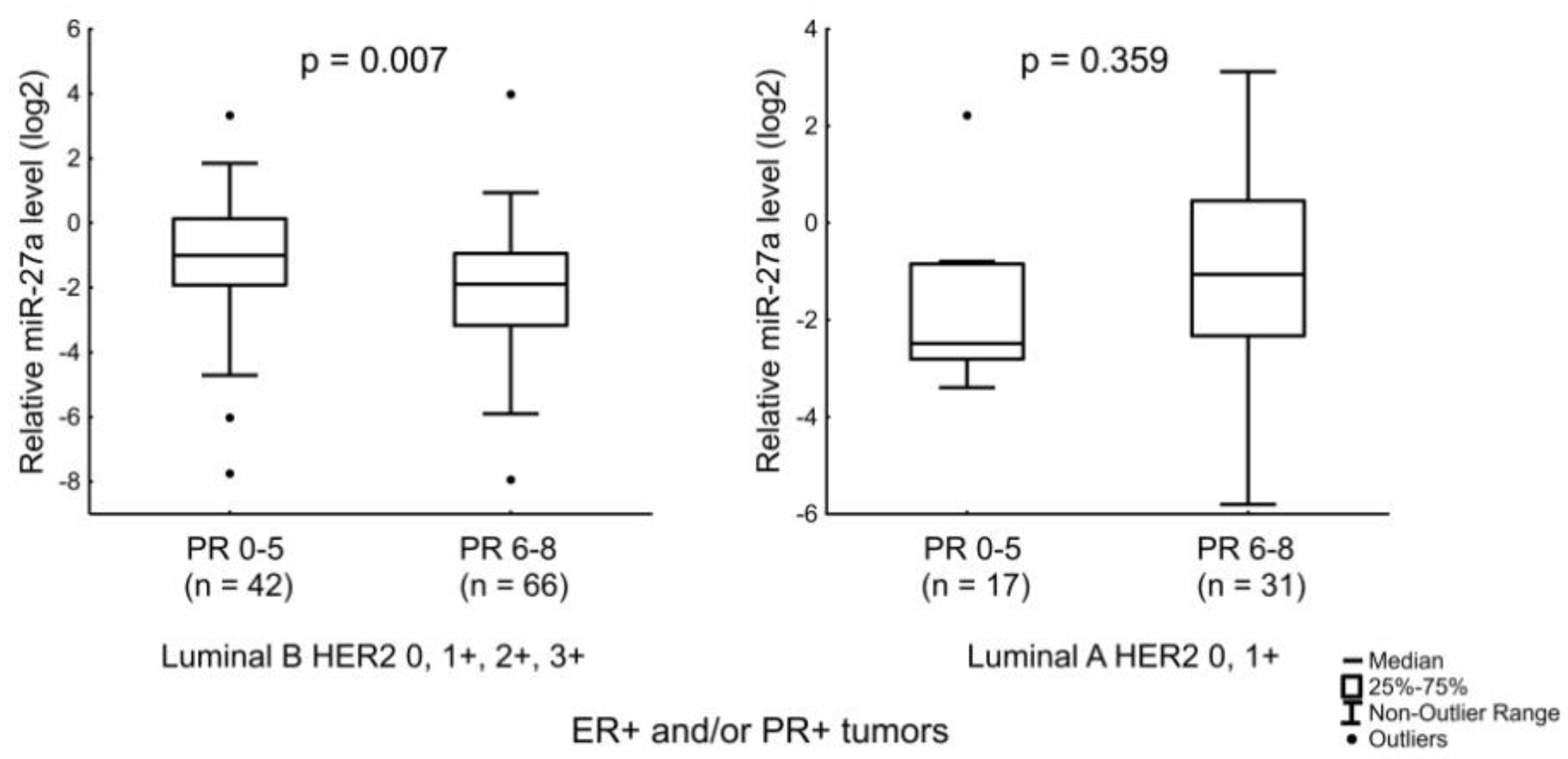
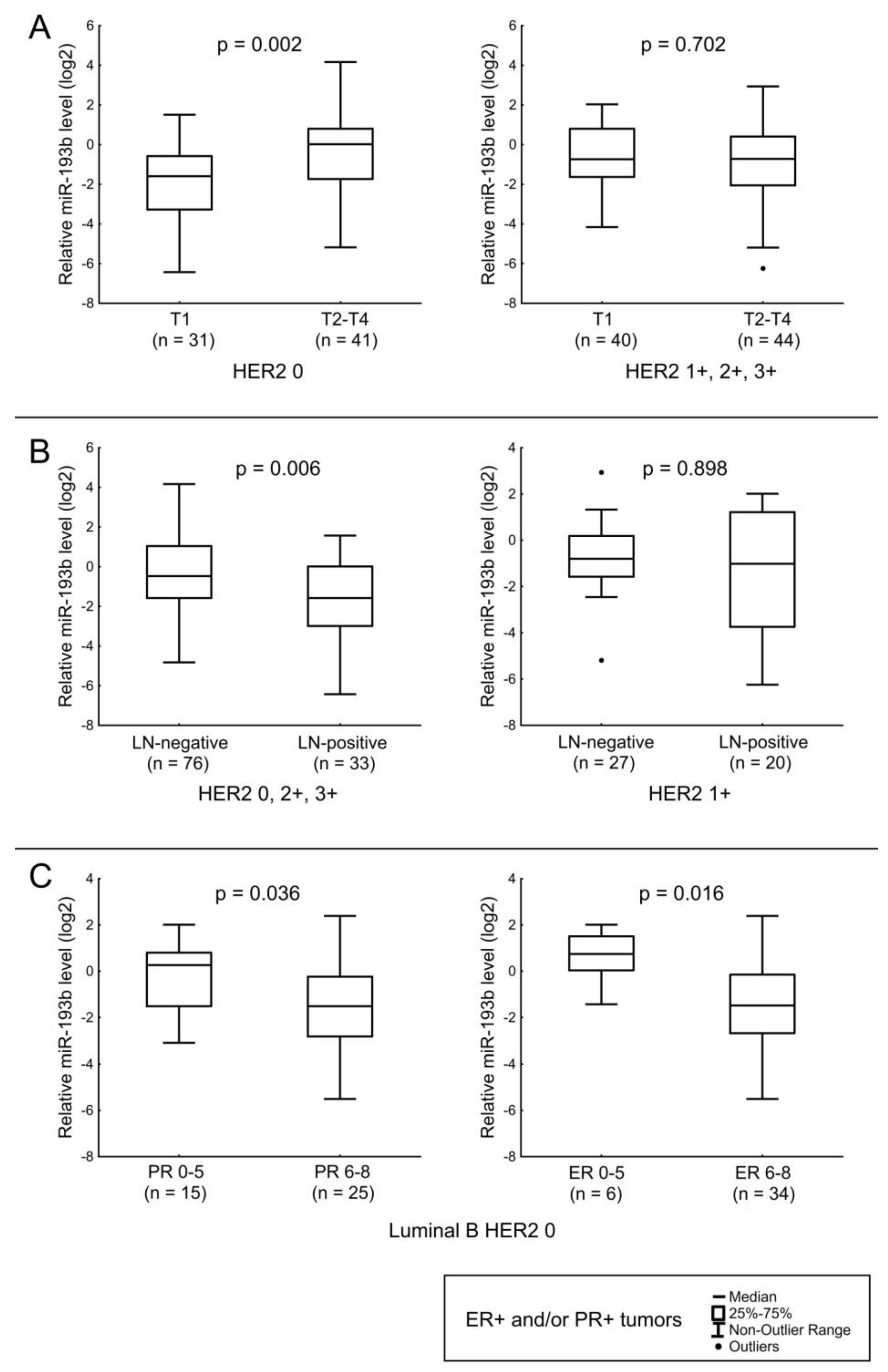
3.3. Expression of MiR-190a, MiR-190b, MiR-27a, MiR-193b, MiR-324, MiR-423, MiR-200b, and MiR-21 in Relation to Clinicopathologic Features of ER- and/or PR-Positive BC
3.4. Expression of MiR-190a, MiR-190b, MiR-27a, MiR-193b, MiR-324, MiR-423, MiR-200b, and MiR-21 in Relation to Clinicopathologic Features of ER- and PR-Negative BC
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gown, A.M. Current issues in ER and HER2 testing by IHC in breast cancer. Mod. Pathol. 2008, 21, S8–S15. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Downs, B.M.; Cope, L.M.; Fackler, M.J.; Zhang, X.; Song, C.G.; VandenBussche, C.; Zhang, K.; Han, Y.; Liu, Y.; et al. Automated and rapid detection of cancer in suspicious axillary lymph nodes in patients with breast cancer. NPJ Breast Cancer 2021, 7, 89. [Google Scholar] [CrossRef]
- Li, H.; Jun, Z.; Zhi-Cheng, G.; Xiang, Q. Factors that affect the false negative rate of sentinel lymph node mapping with methylene blue dye alone in breast cancer. J. Int. Med. Res. 2019, 47, 4841–4853. [Google Scholar] [CrossRef]
- Shah, R.; Rosso, K.; Nathanson, S.D. Pathogenesis, prevention, diagnosis and treatment of breast cancer. World J. Clin. Oncol. 2014, 5, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Dey, S.; Nath, S. Steroid hormone receptors: Links with cell cycle machinery and breast cancer progression. Front. Oncol. 2021, 11, 620214. [Google Scholar] [CrossRef]
- Amaral, J.D.; Solá, S.; Steer, C.J.; Rodrigues, C.M. Role of nuclear steroid receptors in apoptosis. Curr. Med. Chem. 2009, 16, 3886–3902. [Google Scholar] [CrossRef] [PubMed]
- Jansen, J.; Greither, T.; Behre, H.M. Androgen-regulated microRNAs (AndroMiRs) as novel players in adipogenesis. Int. J. Mol. Sci. 2019, 20, 5767. [Google Scholar] [CrossRef] [PubMed]
- Klinge, C.M. Estrogen Regulation of MicroRNA Expression. Curr Genomics. 2009, 10, 169–183. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.J.; Panel members. Strategies for subtypes--dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Harvey, J.M.; Clark, G.M.; Osborne, C.K.; Allred, D.C. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J. Clin. Oncol. 1999, 17, 1474–1481. [Google Scholar] [CrossRef]
- Kalinina, T.S.; Kononchuk, V.V.; Yakovleva, A.K.; Alekseenok, E.Y.; Sidorov, S.V.; Gulyaeva, L.F. Association between lymph node status and expression levels of androgen receptor, miR-185, miR-205, and miR-21 in breast cancer subtypes. Int. J. Breast Cancer 2020, 2020, 3259393. [Google Scholar] [CrossRef]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Ovchinnikov, V.Y.; Antonets, D.V.; Gulyaeva, L.F. The search of CAR, AhR, ESRs binding sites in promoters of intronic and intergenic microRNAs. J. Bioinform. Comput. Biol. 2018, 16, 1750029. [Google Scholar] [CrossRef] [PubMed]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef]
- Mathelier, A.; Fornes, O.; Arenillas, D.J.; Chen, C.Y.; Denay, G.; Lee, J.; Shi, W.; Shyr, C.; Tan, G.; Worsley-Hunt, R.; et al. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016, 44, D110–D115. [Google Scholar] [CrossRef] [PubMed]
- Pagès, H.; Aboyoun, P.; Gentleman, R.; DebRoy, S. Biostrings: String Objects Representing Biological Sequences, and Matching Algorithms. R Package Version 2.44.2. 2017. Available online: https://bioconductor.org/packages/release/bioc/html/Biostrings.html (accessed on 25 October 2021).
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stähler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877. [Google Scholar] [CrossRef]
- Ribas, J.; Ni, X.; Haffner, M.; Wentzel, E.A.; Salmasi, A.H.; Chowdhury, W.H.; Kudrolli, T.A.; Yegnasubramanian, S.; Luo, J.; Rodriguez, R.; et al. miR-21: An androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009, 69, 7165–7169. [Google Scholar] [CrossRef]
- Casaburi, I.; Cesario, M.G.; Donà, A.; Rizza, P.; Aquila, S.; Avena, P.; Lanzino, M.; Pellegrino, M.; Vivacqua, A.; Tucci, P.; et al. Androgens downregulate miR-21 expression in breast cancer cells underlining the protective role of androgen receptor. Oncotarget 2016, 7, 12651–12661. [Google Scholar] [CrossRef]
- Wickramasinghe, N.S.; Manavalan, T.T.; Dougherty, S.M.; Riggs, K.A.; Li, Y.; Klinge, C.M. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009, 37, 2584–2595. [Google Scholar] [CrossRef]
- Cizeron-Clairac, G.; Lallemand, F.; Vacher, S.; Lidereau, R.; Bieche, I.; Callens, C. MiR-190b, the highest up-regulated miRNA in ERα-positive compared to ERα-negative breast tumors, a new biomarker in breast cancers? BMC Cancer 2015, 15, 499. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.V.; Veliceasa, D.; Vinokour, E.; Volpert, O.V. miR-200b inhibits prostate cancer EMT, growth and metastasis. PLoS ONE 2013, 8, e83991. [Google Scholar] [CrossRef]
- Manavalan, T.T.; Teng, Y.; Litchfield, L.M.; Muluhngwi, P.; Al-Rayyan, N.; Klinge, C.M. Reduced expression of miR-200 family members contributes to antiestrogen resistance in LY2 human breast cancer cells. PLoS ONE 2013, 8, e62334. [Google Scholar]
- Fletcher, C.E.; Dart, D.A.; Sita-Lumsden, A.; Cheng, H.; Rennie, P.S.; Bevan, C.L. Androgen-regulated processing of the oncomir miR-27a, which targets Prohibitin in prostate cancer. Hum. Mol. Genet. 2012, 21, 3112–3127. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.Y.; Wong, D.K.; Seto, W.K.; Lai, C.L.; Yuen, M.F. Estradiol induces apoptosis via activation of miRNA-23a and p53: Implication for gender difference in liver cancer development. Oncotarget 2015, 6, 34941–34952. [Google Scholar] [CrossRef]
- He, Y.J.; Wu, J.Z.; Ji, M.H.; Ma, T.; Qiao, E.Q.; Ma, R.; Tang, J.H. miR-342 is associated with estrogen receptor-α expression and response to tamoxifen in breast cancer. Exp. Ther. Med. 2013, 5, 813–818. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.W.; Cheng, C.W.; Chou, W.C.; Hu, L.Y.; Wang, H.W.; Hsiung, C.N.; Hsu, H.M.; Wu, P.E.; Hou, M.F.; Shen, C.Y.; et al. A novel estrogen receptor-microRNA 190a-PAR-1-pathway regulates breast cancer progression, a finding initially suggested by genome-wide analysis of loci associated with lymph-node metastasis. Hum. Mol. Genet. 2014, 23, 355–367. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, S.; Wang, T.; Song, W.; Jiang, T.; Zhang, F.; Yin, Y.; Jiang, S.W.; Wu, K.; Yu, Z.; Wang, C.; et al. The inhibitory effects of AR/miR-190a/YB-1 negative feedback loop on prostate cancer and underlying mechanism. Sci. Rep. 2015, 5, 13528. [Google Scholar] [CrossRef]
- Lyu, S.; Liu, H.; Liu, X.; Liu, S.; Wang, Y.; Yu, Q.; Niu, Y. Interrelation of androgen receptor and miR-30a and miR-30a function in ER-, PR-, AR+ MDA-MB-453 breast cancer cells. Oncol. Lett. 2017, 14, 4930–4936. [Google Scholar] [CrossRef]
- Milevskiy, M.J.G.; Gujral, U.; Del Lama Marques, C.; Stone, A.; Northwood, K.; Burke, L.J.; Gee, J.M.W.; Nephew, K.; Clark, S.; Brown, M.A. MicroRNA-196a is regulated by ER and is a prognostic biomarker in ER+ breast cancer. Br. J. Cancer 2019, 120, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Leivonen, S.K.; Mäkelä, R.; Ostling, P.; Kohonen, P.; Haapa-Paananen, S.; Kleivi, K.; Enerly, E.; Aakula, A.; Hellström, K.; Sahlberg, N.; et al. Protein lysate microarray analysis to identify microRNAs regulating estrogen receptor signaling in breast cancer cell lines. Oncogene 2009, 28, 3926–3936. [Google Scholar] [CrossRef]
- Cui, W.; Li, Q.; Feng, L.; Ding, W. MiR-126-3p regulates progesterone receptors and involves development and lactation of mouse mammary gland. Mol. Cell. Biochem. 2011, 355, 17–25. [Google Scholar] [CrossRef]
- Lambein, K.; Van Bockstal, M.; Vandemaele, L.; Geenen, S.; Rottiers, I.; Nuyts, A.; Matthys, B.; Praet, M.; Denys, H.; Libbrecht, L. Distinguishing score 0 from score 1+ in HER2 immunohistochemistry-negative breast cancer: Clinical and pathobiological relevance. Am. J. Clin. Pathol. 2013, 140, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Schettini, F.; Chic, N.; Brasó-Maristany, F.; Paré, L.; Pascual, T.; Conte, B.; Martínez-Sáez, O.; Adamo, B.; Vidal, M.; Barnadas, E.; et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer 2021, 7, 1. [Google Scholar] [CrossRef] [PubMed]
- Kalinina, T.; Kononchuk, V.; Alekseenok, E.; Obukhova, D.; Sidorov, S.; Strunkin, D.; Gulyaeva, L. Expression of estrogen receptor- and progesterone receptor-regulating microRNAs in breast cancer. Genes (Basel) 2021, 12, 582. [Google Scholar] [CrossRef]
- You, K.; Park, S.; Ryu, J.M.; Kim, I.; Lee, S.K.; Yu, J.; Kim, S.W.; Nam, S.J.; Lee, J.E. Comparison of core needle biopsy and surgical specimens in determining intrinsic biological subtypes of breast cancer with immunohistochemistry. J. Breast Cancer 2017, 20, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Petrović, N.; Mandušić, V.; Dimitrijević, B.; Roganović, J.; Lukić, S.; Todorović, L.; Stanojević, B. Higher miR-21 expression in invasive breast carcinomas is associated with positive estrogen and progesterone receptor status in patients from Serbia. Med. Oncol. 2014, 31, 977. [Google Scholar] [CrossRef]
- Yu, Q.; Niu, Y.; Liu, N.; Zhang, J.Z.; Liu, T.J.; Zhang, R.J.; Wang, S.L.; Ding, X.M.; Xiao, X.Q. Expression of androgen receptor in breast cancer and its significance as a prognostic factor. Ann. Oncol. 2011, 22, 1288–1294. [Google Scholar] [CrossRef] [PubMed]
- Li, X.F.; Yan, P.J.; Shao, Z.M. Downregulation of miR-193b contributes to enhance urokinase-type plasminogen activator (uPA) expression and tumor progression and invasion in human breast cancer. Oncogene 2009, 28, 3937–3948. [Google Scholar] [CrossRef]
- Yu, Y.; Luo, W.; Yang, Z.J.; Chi, J.R.; Li, Y.R.; Ding, Y.; Ge, J.; Wang, X.; Cao, X.C. miR-190 suppresses breast cancer metastasis by regulation of TGF-β-induced epithelial-mesenchymal transition. Mol. Cancer 2018, 17, 70. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Chung, H.F.; Dobson, A.J.; Pandeya, N.; Giles, G.G.; Bruinsma, F.; Brunner, E.J.; Kuh, D.; Hardy, R.; Avis, N.E.; et al. Age at natural menopause and risk of incident cardiovascular disease: A pooled analysis of individual patient data. Lancet Public Health 2019, 4, e553–e564. [Google Scholar] [CrossRef]




| Characteristics | ER- and/or PR-Positive (n = 156) | ER- and PR-Negative (n = 40) | |
|---|---|---|---|
| Age (mean and range, year) | 61 (27–90) | 55 (38–76) | |
| T stage | T1 | 71 | 18 |
| T2 | 81 | 20 | |
| T3 | 2 | 1 | |
| T4 | 2 | 1 | |
| N stage | N0 | 103 | 27 |
| N1 | 37 | 7 | |
| N2 | 11 | 6 | |
| N3 | 5 | - | |
| ER score | 0–2 | 3 | 40 |
| 3–5 | 7 | - | |
| 6–8 | 146 | - | |
| PR score | 0–2 | 25 | 40 |
| 3–5 | 34 | - | |
| 6–8 | 97 | - | |
| HER2 score | 0 | 72 | 18 |
| 1 | 47 | 5 | |
| 2–3 | 37 | 17 | |
| miRNA | ESR1 and ESR2 Binding Sites in Promoter According to ChipSeq Data (Homo Sapiens) [13] | ESR1 and ESR2 Binding Sites in Promoter According to Position Weight Matrix (Homo Sapiens) [13] | ESR1 and ESR2 Binding Sites in Promoter According to Position Weight Matrix (Mus Musculus and Rattus Norvegicus) [13] | AR/PR Binding Sites in Promoter According to Position Weight Matrix (Homo Sapiens) | AR/PR Binding Sites in Promoter According to Position Weight Matrix (Rattus Norvegicus) | Comments |
|---|---|---|---|---|---|---|
| hsa-mir-21 | + | + | only mouse | + | + | It was demonstrated that androgen induced AR binding to the miR-21 promoter; MiR-21 expression was induced by R1881 in LNCaP and LAPC-4 cells [19]. Mibolerone inhibited basal expression of miR-21 in MCF-7 breast cancer cells [20]. Estradiol inhibited miR-21 expression in MCF-7 cells [21]. |
| hsa-mir-190b | + | + | + | + | - | MiR-190b is the highest up-regulated miRNA in ER+ breast cancers compared to ER− tumors. Did not observe an increase of miR-190b expression levels in MCF-7 or in T-47D treated by estradiol (1 nM for MCF-7 and 10 nM for T-47D, 6 h, 18 h, and 4 days) [22]. |
| hsa-mir-200a/ hsa-mir-200b/ hsa-mir-429 | + | + | + | - | - | MiR-200b showed the highest fold change under the influence of R1881 among androgen-sensitive miRNAs (PC3-AR cells) [23]. MiR-200b expression in MCF-7 cells decreased after 6 h of incubation with 10 nM estradiol [24]. |
| hsa-mir-23a/ hsa-mir-24-2/ hsa-mir-27a | + | - | + | - | - | AR is able to associate transiently with the miR-23a/27a/24-2 promoter in response to androgen to initiate cluster transcription. The highest-fold change was observed for miR-27a and miR-23a (LNCaP cells treated with mibolerone) [25]. Estrogen induced miR-23a expression in SNU-387 cells [26]. |
| hsa-mir-342 | + | - | + | + | - | MiR-342 expression is positively correlated with ERα mRNA expression in human BC [27]. |
| hsa-mir-190a | - | - | - | + | + | The promoter region of miR-190a contains half of an estrogen response element. ERα binds directly to this promoter [28]. Androgen inhibits miR-190a expression through direct binding to the half-site of ARE in miR-190a promoter (LNCaP cells) [29]. |
| hsa-mir-378a | - | + | + | + | + | |
| hsa-mir-324 | - | + | + | + | + | |
| hsa-mir-423 | - | + | only rat | + | + | |
| hsa-mir-149 | - | + | only rat | + | + | |
| hsa-mir-365b | + | + | only rat | + | - | |
| hsa-mir-574 | + | - | only mouse | + | - | |
| hsa-mir-30a | + | - | only mouse | + | - | AR does not target the miR-30a promoter; AR activating signal may indirectly downregulate miR-30a (MDA-MB-453 cells) [30]. |
| hsa-mir-10a | + | + | + | - | - | |
| hsa-mir-483 | + | + | + | - | - | |
| hsa-let-7a-3/ hsa-let-7b | + | - | + | - | - | |
| hsa-mir-196a-2 | + | + | - | - | - | MiR-196a expression is regulated by the estrogen receptor [31]. |
| hsa-mir-33b | + | - | - | - | - | |
| hsa-miR-193b | - | + | + | + | - | Targets ER [32]. |
| hsa-miR-126 | - | + | + | + | - | Targets PR (regulation confirmed using mouse mammary epithelial cells) [33]. |
| miRNA | Time, h | Relative Level of miRNA | |||||
|---|---|---|---|---|---|---|---|
| Estradiol | Testosterone | Progesterone | |||||
| 10 nM | 100 nM | 10 nM | 100 nM | 10 nM | 100 nM | ||
| miR-23a | 6 | 1.12 | 1.08 | 0.96 | 0.86 | 0.84 | 0.84 |
| 24 | 0.89 | 0.95 | 0.88 | 1.00 | 0.92 | 0.88 | |
| 48 | 0.89 | 0.85 | 1.02 | 0.94 | 1.11 | 0.92 | |
| miR-27a | 6 | 1.12 | 0.88 | 0.92 | 0.76 * | 0.89 | 0.93 |
| 24 | 1.07 | 1.10 | 1.09 | 1.08 | 0.93 | 0.92 | |
| 48 | 0.92 | 0.95 | 1.13 | 1.09 | 1.01 | 1.16 | |
| miR-190b | 6 | 0.97 | 0.75 ** | 0.96 | 0.92 | 0.93 | 1.10 |
| 24 | 1.35 * | 1.37 ** | 1.13 | 1.09 | 1.02 | 1.03 | |
| 48 | 1.12 | 1.09 | 1.05 | 1.15 | 1.14 | 1.29 * | |
| miR-190a | 6 | 1.05 | 1.01 | 0.91 | 0.88 | 1.01 | 0.90 |
| 24 | 1.09 | 1.02 | 0.90 | 0.99 | 0.79 | 0.75 * | |
| 48 | 1.08 | 1.17 | 1.24 * | 1.38 * | 0.90 | 0.96 | |
| miR-200b | 6 | 1.01 | 0.79 ** | 1.00 | 0.88 | 1.02 | 0.98 |
| 24 | 0.98 | 1.01 | 1.10 | 0.99 | 0.91 | 0.96 | |
| 48 | 1.01 | 1.11 | 1.24 * | 1.32 ** | 0.95 | 1.07 | |
| miR-21 | 6 | 1.03 | 0.98 | 0.92 | 0.62 ** | 1.17 | 1.36 * |
| 24 | 0.97 | 0.98 | 0.89 | 0.91 | 0.92 | 0.90 | |
| 48 | 1.07 | 1.08 | 1.36 ** | 1.40 ** | 1.11 | 1.08 | |
| miR-126 | 6 | 1.01 | 1.03 | 0.90 | 1.05 | 0.87 | 0.95 |
| 24 | 0.94 | 0.88 | 1.06 | 0.94 | 1.01 | 1.12 | |
| 48 | 1.13 | 1.11 | 1.18 | 0.99 | 0.93 | 1.07 | |
| miR-378 | 6 | 1.09 | 1.00 | 0.88 | 0.94 | 0.90 | 1.08 |
| 24 | 1.05 | 1.05 | 1.11 | 0.97 | 0.95 | 1.04 | |
| 48 | 1.00 | 1.17 | 1.15 | 1.19 | 0.94 | 1.12 | |
| miR-423 | 6 | 0.98 | 0.90 | 0.89 | 0.93 | 0.91 | 1.09 |
| 24 | 1.04 | 1.06 | 0.99 | 0.87 | 0.96 | 0.99 | |
| 48 | 0.97 | 0.99 | 1.50 ** | 1.42 ** | 0.90 | 0.97 | |
| miR-149 | 6 | 1.25 | 1.01 | 0.95 | 0.91 | 0.96 | 1.06 |
| 24 | 0.98 | 0.94 | 0.99 | 0.95 | 0.90 | 0.94 | |
| 48 | 0.89 | 1.05 | 1.05 | 0.89 | 1.02 | 1.18 | |
| miR-193b | 6 | 1.07 | 1.00 | 0.93 | 0.95 | 0.92 | 1.07 |
| 24 | 1.02 | 1.10 | 1.05 | 1.08 | 1.00 | 0.89 | |
| 48 | 1.24 | 1.30 ** | 1.30 ** | 1.40 ** | 1.07 | 1.08 | |
| miR-324 | 6 | 1.35 * | 1.21 | 0.91 | 0.95 | 1.61 ** | 2.20 ** |
| 24 | 0.99 | 1.06 | 1.06 | 1.05 | 1.05 | 1.04 | |
| 48 | 1.08 | 1.07 | 1.15 | 1.33 ** | 0.94 | 1.09 | |
| miR-342 | 6 | 0.83 | 0.96 | 0.89 | 1.10 | 0.92 | 1.12 |
| 24 | 0.96 | 1.02 | 1.15 | 1.10 | 0.89 | 0.94 | |
| 48 | 1.07 | 1.08 | 1.17 | 1.19 | 0.90 | 1.18 | |
| Characteristics | n | Relative Level * of miRNA and p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miR-190a | p-Value | miR-190b | p-Value | miR-27a | p-Value | miR-193b | p-Value | |||
| ER and PR status | ER+ and/or PR+ | 156 | 0.10 | 0.165 | 4.84 | <0.001 | 0.33 | 0.443 | 0.58 | 0.168 |
| ER− and PR− | 40 | 0.33 | 1.13 | 0.44 | 0.36 | |||||
| HER2 status | HER2+ | 52 | 0.10 | 0.728 | 3.86 | 0.985 | 0.34 | 0.388 | 0.72 | 0.193 |
| HER2− | 144 | 0.12 | 3.76 | 0.39 | 0.47 | |||||
| Ki-67 index (%) | <14 | 65 | 0.11 | 0.753 | 3.89 | 0.941 | 0.49 | 0.394 | 0.49 | 0.567 |
| ≥14 | 131 | 0.11 | 3.64 | 0.35 | 0.53 | |||||
| Age | ≤50 | 48 | 0.21 | 0.107 | 2.79 | 0.119 | 0.48 | 0.045 | 0.56 | 0.861 |
| >50 | 148 | 0.11 | 4.26 | 0.33 | 0.52 | |||||
| N stage | N0 | 130 | 0.17 | 0.103 | 3.47 | 0.592 | 0.37 | 0.834 | 0.65 | 0.022 |
| N1-N3 | 66 | 0.09 | 4.21 | 0.42 | 0.37 | |||||
| miR-324 | p-Value | miR-423 | p-Value | miR-200b | p-Value | miR-21 | p-Value | |||
| ER and PR status | ER+ and/or PR+ | 156 | 0.48 | 0.129 | 0.65 | 0.800 | 1.77 | 0.676 | 1.78 | 0.004 |
| ER− and PR− | 40 | 0.68 | 0.74 | 1.53 | 3.48 | |||||
| HER2 status | HER2+ | 52 | 0.60 | 0.180 | 0.80 | 0.004 | 2.99 | 0.024 | 1.62 | 0.342 |
| HER2− | 144 | 0.47 | 0.55 | 1.46 | 1.95 | |||||
| Ki-67 index (%) | <14 | 65 | 0.39 | 0.257 | 0.56 | 0.038 | 1.26 | 0.030 | 2.02 | 0.947 |
| ≥14 | 131 | 0.60 | 0.71 | 2.04 | 1.88 | |||||
| Age | ≤50 | 48 | 0.60 | 0.103 | 0.69 | 0.958 | 1.68 | 0.598 | 3.52 | 0.003 |
| >50 | 148 | 0.48 | 0.65 | 1.50 | 1.68 | |||||
| N stage | N0 | 130 | 0.55 | 0.210 | 0.68 | 0.068 | 1.51 | 0.491 | 1.84 | 0.445 |
| N1-N3 | 66 | 0.48 | 0.52 | 1.78 | 2.13 | |||||
| Characteristics | n | Relative Level * of miRNA and p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miR-190a | p-Value | miR-190b | p-Value | miR-27a | p-Value | miR-193b | p-Value | |||
| ER+ and/or PR+ | ||||||||||
| T stage | T1 | 71 | 0.11 | 0.783 | 4.80 | 0.995 | 0.30 | 0.248 | 0.43 | 0.070 |
| T2–T4 | 85 | 0.09 | 4.93 | 0.46 | 0.74 | |||||
| N stage | N0 | 103 | 0.13 | 0.058 | 4.57 | 0.477 | 0.37 | 0.391 | 0.69 | 0.013 |
| N1–N3 | 53 | 0.08 | 5.41 | 0.52 | 0.38 | |||||
| Ki-67 index (%) | <M ** | 81 | 0.17 | 0.071 | 4.59 | 0.231 | 0.47 | 0.261 | 0.56 | 0.316 |
| ≥M ** | 75 | 0.09 | 5.42 | 0.32 | 0.66 | |||||
| ER score | 6–8 | 146 | 0.09 | 0.475 | 5.04 | 0.078 | 0.32 | 0.113 | 0.54 | 0.013 |
| 0–5 | 10 | 0.19 | 2.12 | 0.52 | 1.74 | |||||
| PR score | 6–8 | 97 | 0.09 | 0.004 | 4.71 | 0.840 | 0.29 | 0.048 | 0.49 | 0.258 |
| 0–5 | 59 | 0.21 | 6.41 | 0.49 | 0.81 | |||||
| Age | ≤50 | 30 | 0.12 | 0.226 | 5.41 | 0.893 | 0.48 | 0.227 | 0.90 | 0.360 |
| >50 | 126 | 0.10 | 4.80 | 0.31 | 0.55 | |||||
| miR-324 | p-Value | miR-423 | p-Value | miR-200b | p-Value | miR-21 | p-Value | |||
| ER+ and/or PR+ | ||||||||||
| T stage | T1 | 71 | 0.43 | 0.216 | 0.59 | 0.290 | 2.09 | 0.109 | 1.68 | 0.247 |
| T2–T4 | 85 | 0.50 | 0.67 | 1.36 | 1.81 | |||||
| N stage | N0 | 103 | 0.53 | 0.313 | 0.68 | 0.039 | 1.64 | 0.832 | 1.74 | 0.802 |
| N1–N3 | 53 | 0.46 | 0.49 | 1.77 | 1.95 | |||||
| Ki-67 index (%) | <M ** | 81 | 0.44 | 0.694 | 0.57 | 0.055 | 1.53 | 0.253 | 1.84 | 0.494 |
| ≥M ** | 75 | 0.49 | 0.68 | 2.04 | 1.69 | |||||
| ER score | 6–8 | 146 | 0.48 | 0.467 | 0.62 | 0.756 | 1.88 | 0.006 | 1.69 | 0.106 |
| 0–5 | 10 | 0.89 | 0.88 | 0.28 | 2.09 | |||||
| PR score | 6–8 | 97 | 0.45 | 0.211 | 0.63 | 0.600 | 1.85 | 0.858 | 1.76 | 0.336 |
| 0–5 | 59 | 0.61 | 0.65 | 1.50 | 1.83 | |||||
| Age | ≤50 | 30 | 0.57 | 0.232 | 0.71 | 0.741 | 1.72 | 0.729 | 2.28 | 0.085 |
| >50 | 126 | 0.46 | 0.62 | 1.73 | 1.65 | |||||
| Characteristics | n | Relative Level * of miRNA and p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| miR-190a | p-Value | miR-190b | p-Value | miR-27a | p-Value | miR-193b | p-Value | |||
| T stage | T1 | 18 | 0.33 | 0.372 | 0.90 | 0.629 | 0.40 | 0.240 | 0.57 | 0.176 |
| T2–T4 | 22 | 0.37 | 1.26 | 0.57 | 0.35 | |||||
| N stage | N0 | 27 | 0.37 | 0.824 | 1.07 | 0.835 | 0.39 | 0.183 | 0.44 | 0.725 |
| N1–N3 | 13 | 0.24 | 1.14 | 0.52 | 0.35 | |||||
| Ki-67 index (%) | ≤M ** | 19 | 0.39 | 0.162 | 1.45 | 0.191 | 0.45 | 0.779 | 0.47 | 0.272 |
| >M ** | 21 | 0.08 | 1.02 | 0.44 | 0.35 | |||||
| Age | ≤50 | 18 | 0.34 | 0.228 | 0.64 | 0.275 | 0.55 | 0.091 | 0.37 | 0.617 |
| >50 | 22 | 0.26 | 1.24 | 0.39 | 0.43 | |||||
| miR-324 | p-Value | miR-423 | p-Value | miR-200b | p-Value | miR-21 | p-Value | |||
| T stage | T1 | 18 | 0.65 | 0.987 | 0.80 | 0.729 | 1.11 | 0.036 | 3.87 | 0.703 |
| T2–T4 | 26 | 0.75 | 0.51 | 2.04 | 3.47 | |||||
| N stage | N0 | 27 | 0.69 | 0.391 | 0.70 | 0.969 | 1.33 | 0.349 | 3.33 | 0.101 |
| N1–N3 | 13 | 0.62 | 0.80 | 2.04 | 6.68 | |||||
| Ki-67 index (%) | ≤M ** | 19 | 0.89 | 0.101 | 0.79 | 0.546 | 1.34 | 0.706 | 3.48 | 0.582 |
| >M ** | 21 | 0.56 | 0.49 | 2.07 | 3.47 | |||||
| Age | ≤50 | 18 | 0.65 | 0.565 | 0.55 | 0.617 | 1.78 | 0.266 | 6.15 | 0.019 |
| >50 | 22 | 0.68 | 0.80 | 1.32 | 1.95 | |||||
| miRNA | Regulation of miRNA Expression According to Bioinformatic Analysis | Observed Changes | ||
|---|---|---|---|---|
| MCF-7 | ||||
| Estradiol | Testosterone | Progesterone | ||
| miR-27a | ER *, AR * | - | down (6 h) | - |
| miR-190b | ER *, AR, PR | down (6 h) up (24 h) | - | up (48 h) |
| miR-190a | ER *, AR, PR | - | up (48 h) | down (24 h) |
| miR-200b | ER *, AR ** | down (6 h) | up (48 h) | - |
| miR-21 | ER *, AR *, PR | - | down (6 h) up (48 h) | up (6 h) |
| miR-423 | ER, AR, PR | - | up (48 h) | - |
| miR-193b | ER, PR, AR | up (48 h) | up (48 h) | - |
| miR-324 | ER, PR, AR | up (6 h) | up (48 h) | up (6 h) |
| miRNA | Associated Tumor Characteristics |
|---|---|
| miR-27a | Age of the patients and the level of PR expression. |
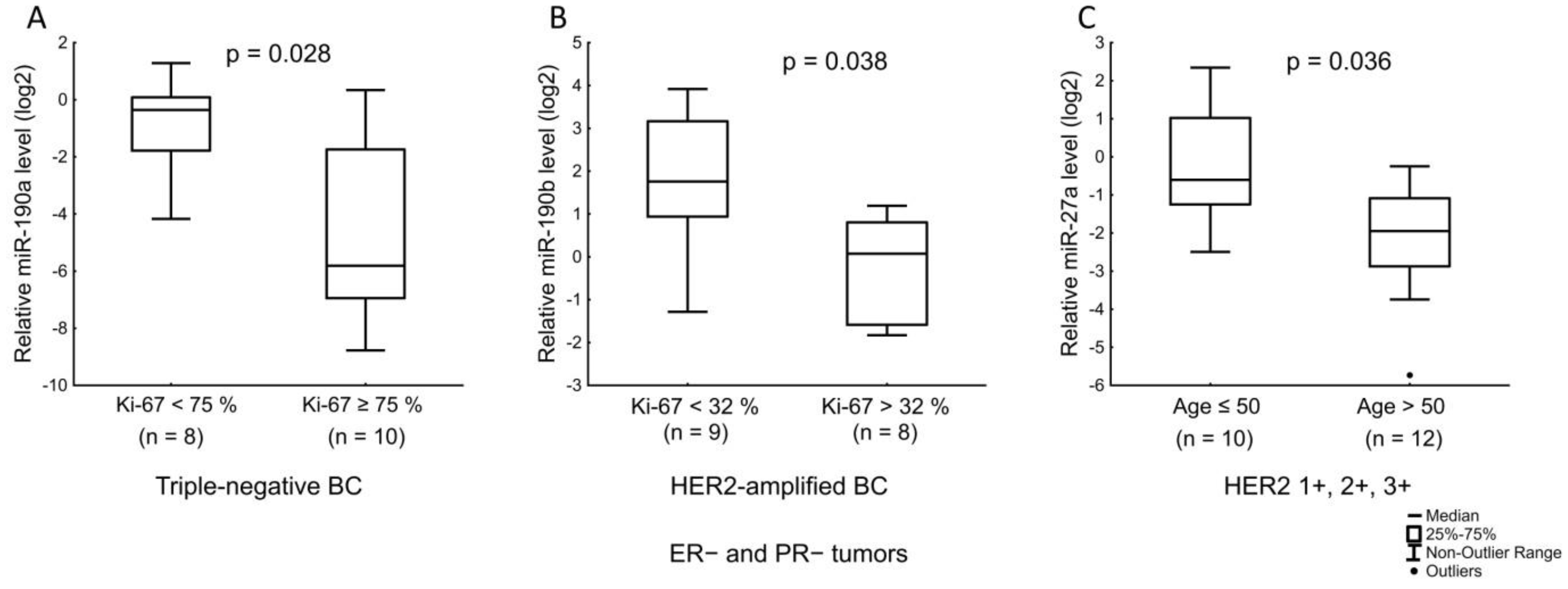
| miR-190b | ER receptor status. Ki-67 index in ER+ and/or PR+ HER2-expressing, and Ki-67 index in ER− and PR− HER2-amplified tumors. |
| miR-190a | Expression level of PR. LN status in ER+ and/or PR+ tumors with low Ki-67. Ki-67 index in triple-negative tumors. |
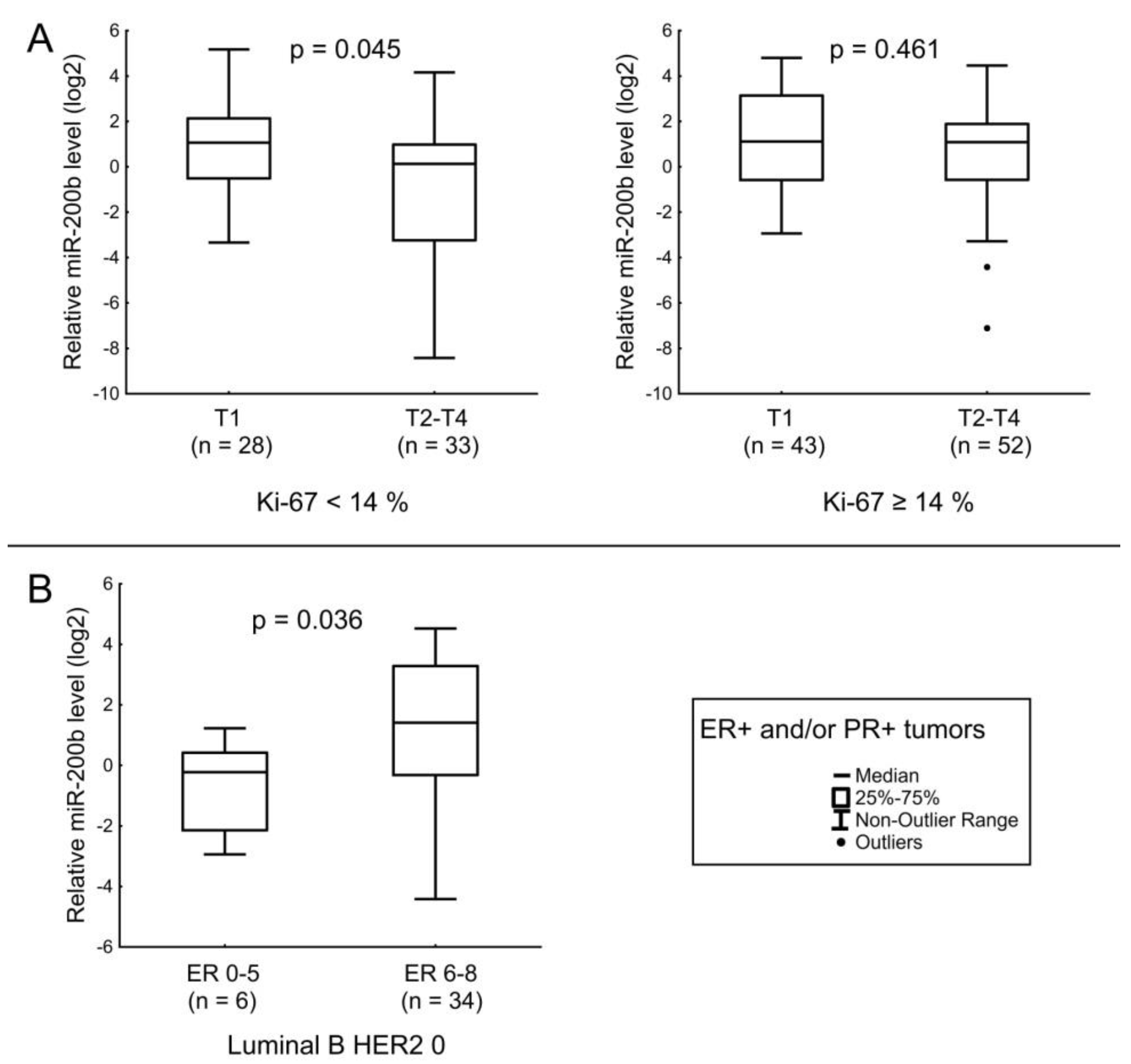
| miR-200b | HER2 status and Ki-67 index. Tumor size in ER+ and/or PR+ tumors with low Ki-67. Level of ER expression in Luminal B tumors not expressing HER2. |
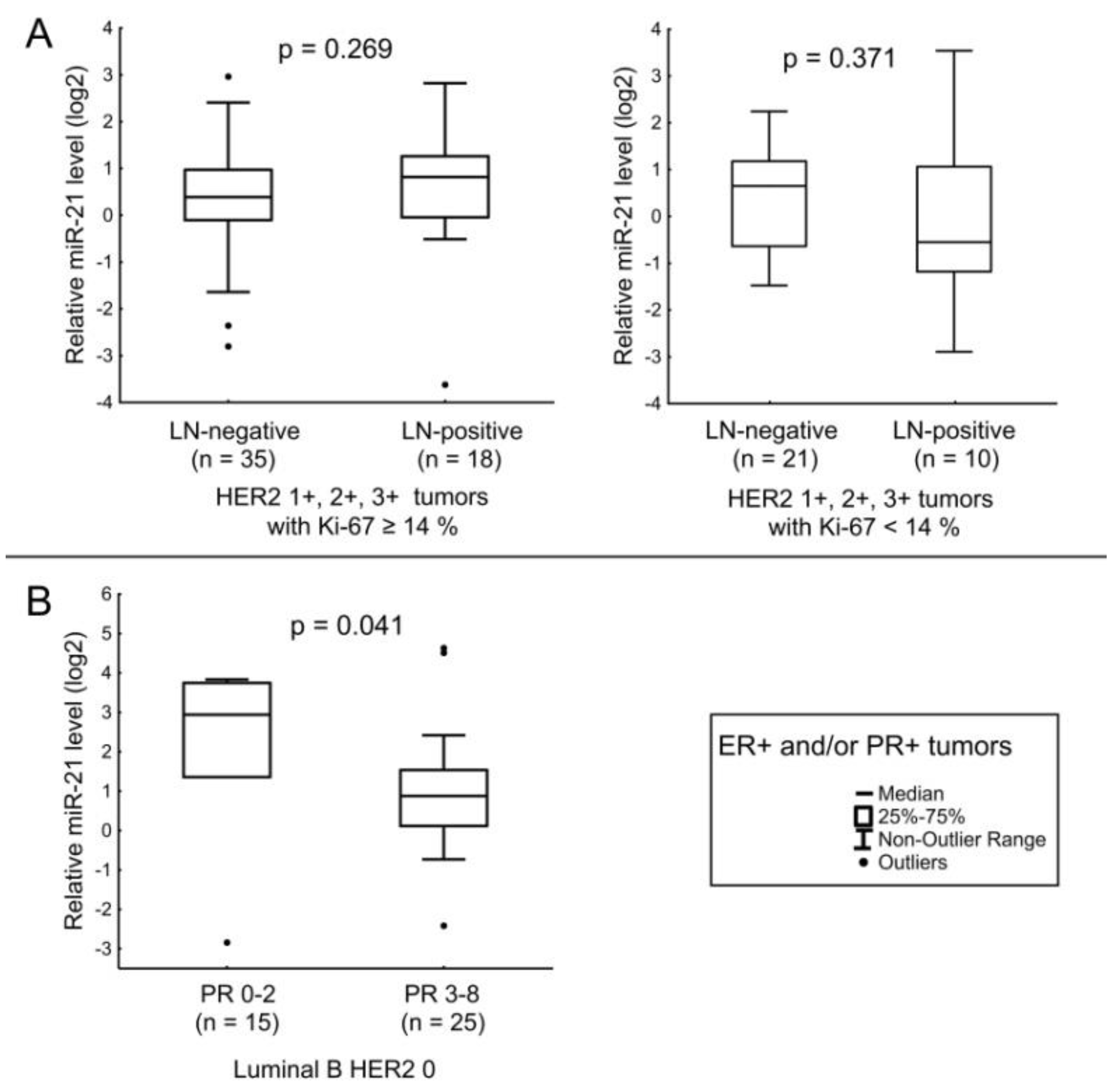
| miR-21 | ER receptor status and age of the patients. Expression level of HER2. |
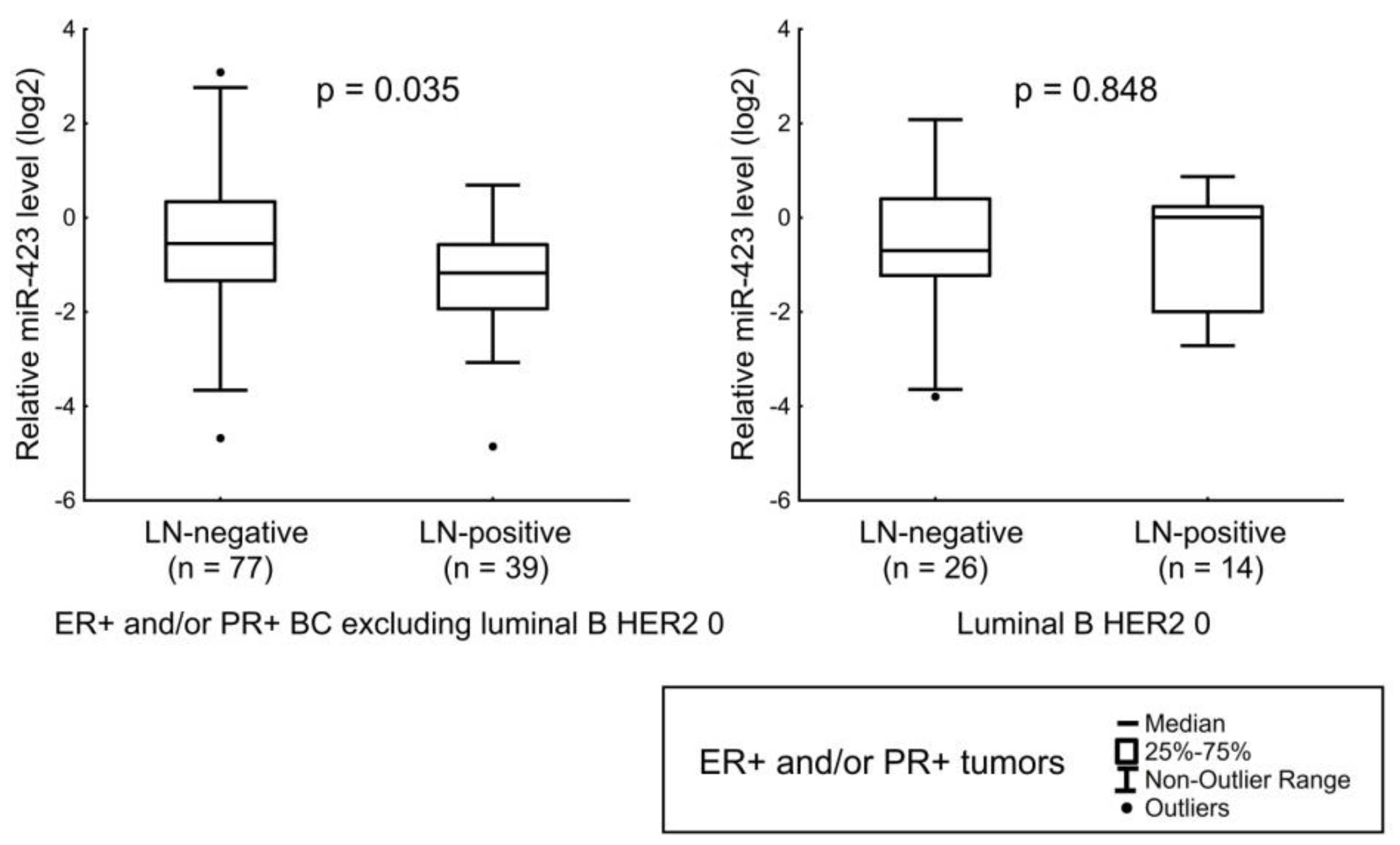
| miR-423 | HER2 status and Ki-67 index. LN status in luminal A, luminal B HER2-amplified, and luminal B HER2 1+ tumors. |
| miR-193b | LN status in ER+ and/or PR+ tumors (except HER2 1+ cases). Tumor size in ER+ and/or PR+ HER2-non-expressing tumors. Expression level of ER in Luminal B HER2 0 tumors. |
| miR-324 | Tumor size in Luminal B HER2-non-amplified tumors. Age of the patients and HER2 status in ER+ and/or PR+ tumors with low Ki-67. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalinina, T.; Kononchuk, V.; Alekseenok, E.; Abdullin, G.; Sidorov, S.; Ovchinnikov, V.; Gulyaeva, L. Associations between the Levels of Estradiol-, Progesterone-, and Testosterone-Sensitive MiRNAs and Main Clinicopathologic Features of Breast Cancer. J. Pers. Med. 2022, 12, 4. https://doi.org/10.3390/jpm12010004
Kalinina T, Kononchuk V, Alekseenok E, Abdullin G, Sidorov S, Ovchinnikov V, Gulyaeva L. Associations between the Levels of Estradiol-, Progesterone-, and Testosterone-Sensitive MiRNAs and Main Clinicopathologic Features of Breast Cancer. Journal of Personalized Medicine. 2022; 12(1):4. https://doi.org/10.3390/jpm12010004
Chicago/Turabian StyleKalinina, Tatiana, Vladislav Kononchuk, Efim Alekseenok, Grigory Abdullin, Sergey Sidorov, Vladimir Ovchinnikov, and Lyudmila Gulyaeva. 2022. "Associations between the Levels of Estradiol-, Progesterone-, and Testosterone-Sensitive MiRNAs and Main Clinicopathologic Features of Breast Cancer" Journal of Personalized Medicine 12, no. 1: 4. https://doi.org/10.3390/jpm12010004
APA StyleKalinina, T., Kononchuk, V., Alekseenok, E., Abdullin, G., Sidorov, S., Ovchinnikov, V., & Gulyaeva, L. (2022). Associations between the Levels of Estradiol-, Progesterone-, and Testosterone-Sensitive MiRNAs and Main Clinicopathologic Features of Breast Cancer. Journal of Personalized Medicine, 12(1), 4. https://doi.org/10.3390/jpm12010004







