Glucose and Serum Deprivation Led to Altered Proliferation, Differentiation Potential and AMPK Activation in Stem Cells from Human Deciduous Tooth
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Culture and Expansion of Stem Cells from Human Exfoliated Deciduous Teeth (SHEDs)
2.3. Experimental Groups
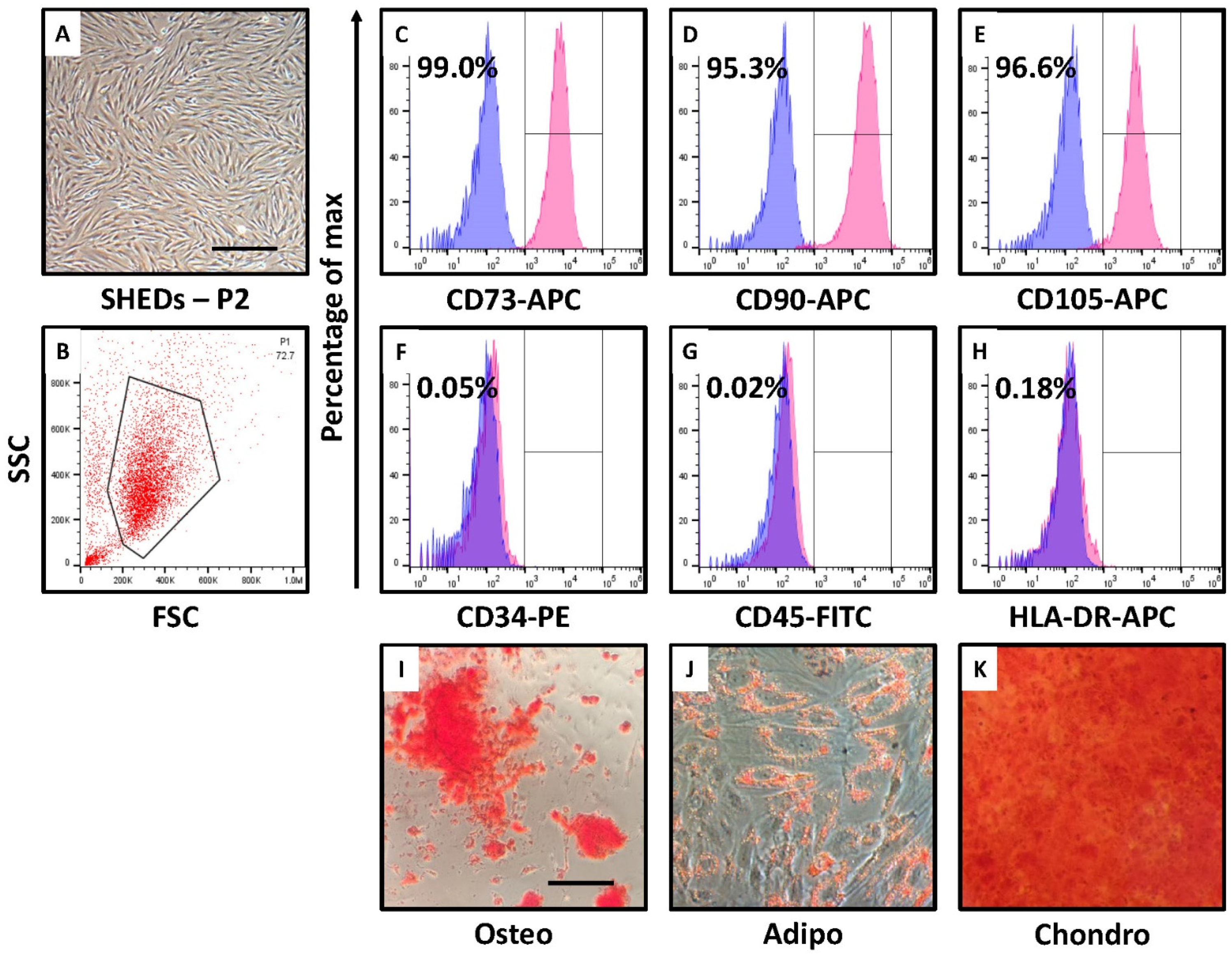
2.4. Cell Surface Marker Analysis of SHEDs by Flow Cytometry
2.5. Osteogenic Differentiation
2.6. Differentiation into Adipocytes
2.7. Chondrogenic Differentiation
2.8. Growth Curve Plotting
2.9. Analysis of Cell Cycle by Flow Cytometry Method
2.10. Quantitative Analysis of Expression of Genes by Real Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.11. Mitochondrial Potential Analysis by Flow Cytometry Method
2.12. Analysis of Cell Apoptosis Using Annexin V-FITC/PI Assay by Flow Cytometry Method
2.13. Western Blot Analysis
2.14. Statistical Analysis
3. Results
3.1. SHEDs Show MSC-like Morphology, Cell Surface Marker Expression, and Trilineage Differentiation
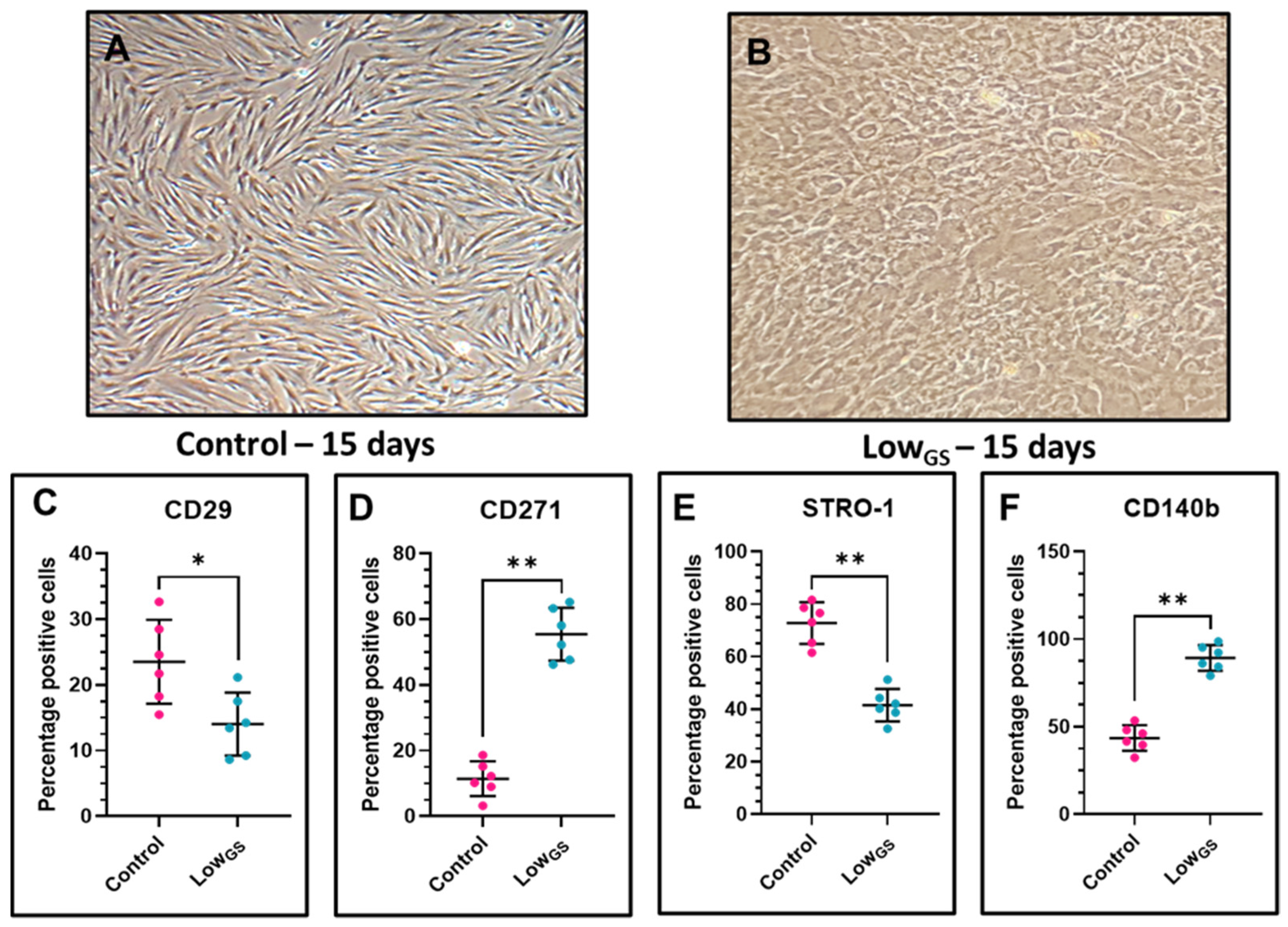
3.2. Expression of CD29, STRO-1, CD271 and CD140b in Nutrient Deprived SHEDs
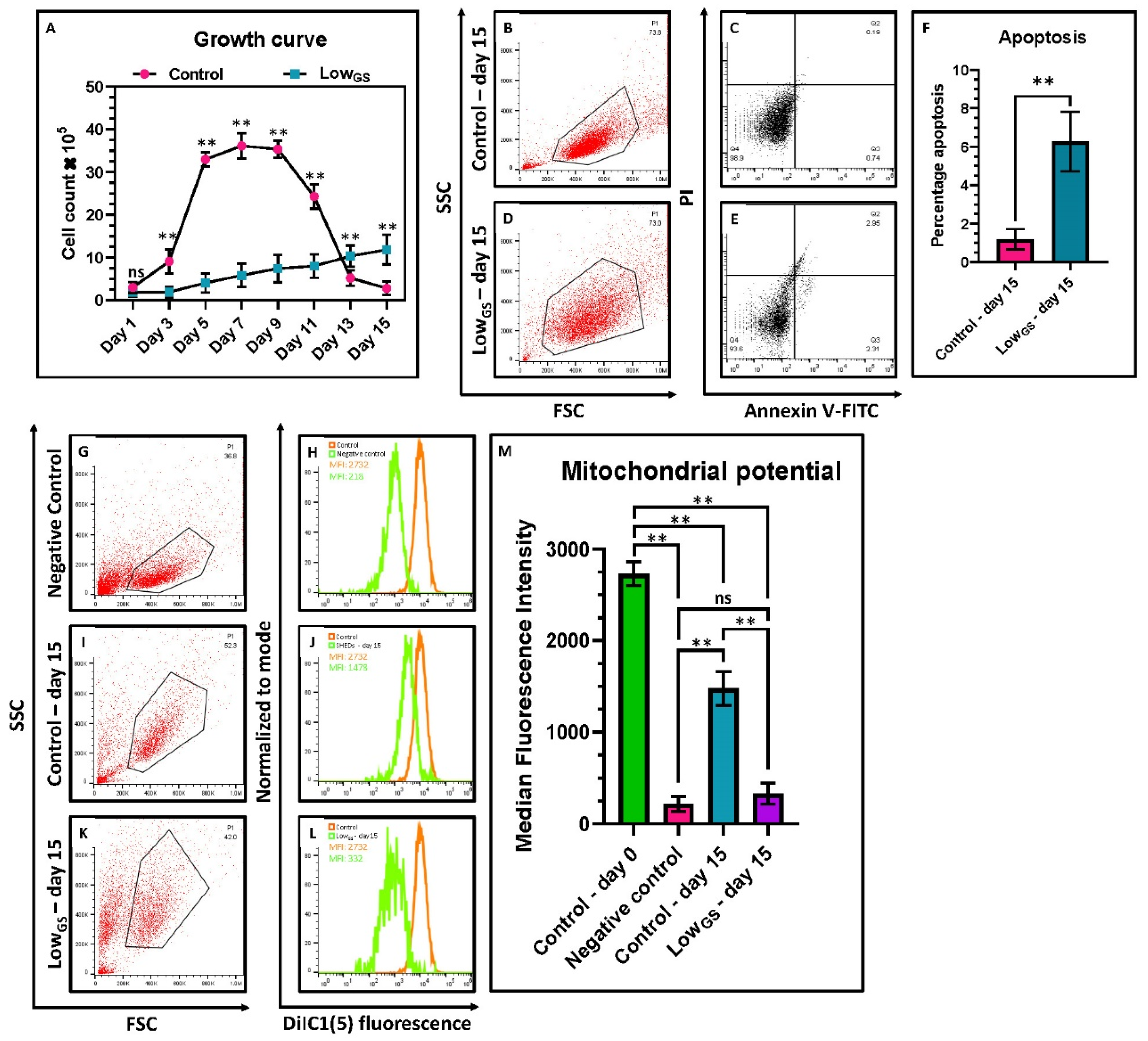
3.3. Serum Deprivation Induced Cell Cycle Arrest with Cell Cycle Synchronization in S Phase
3.4. Serum Deprived SHEDs Exhibited Decreased Proliferation, Mitochondrial Activity and Increased Apoptosis
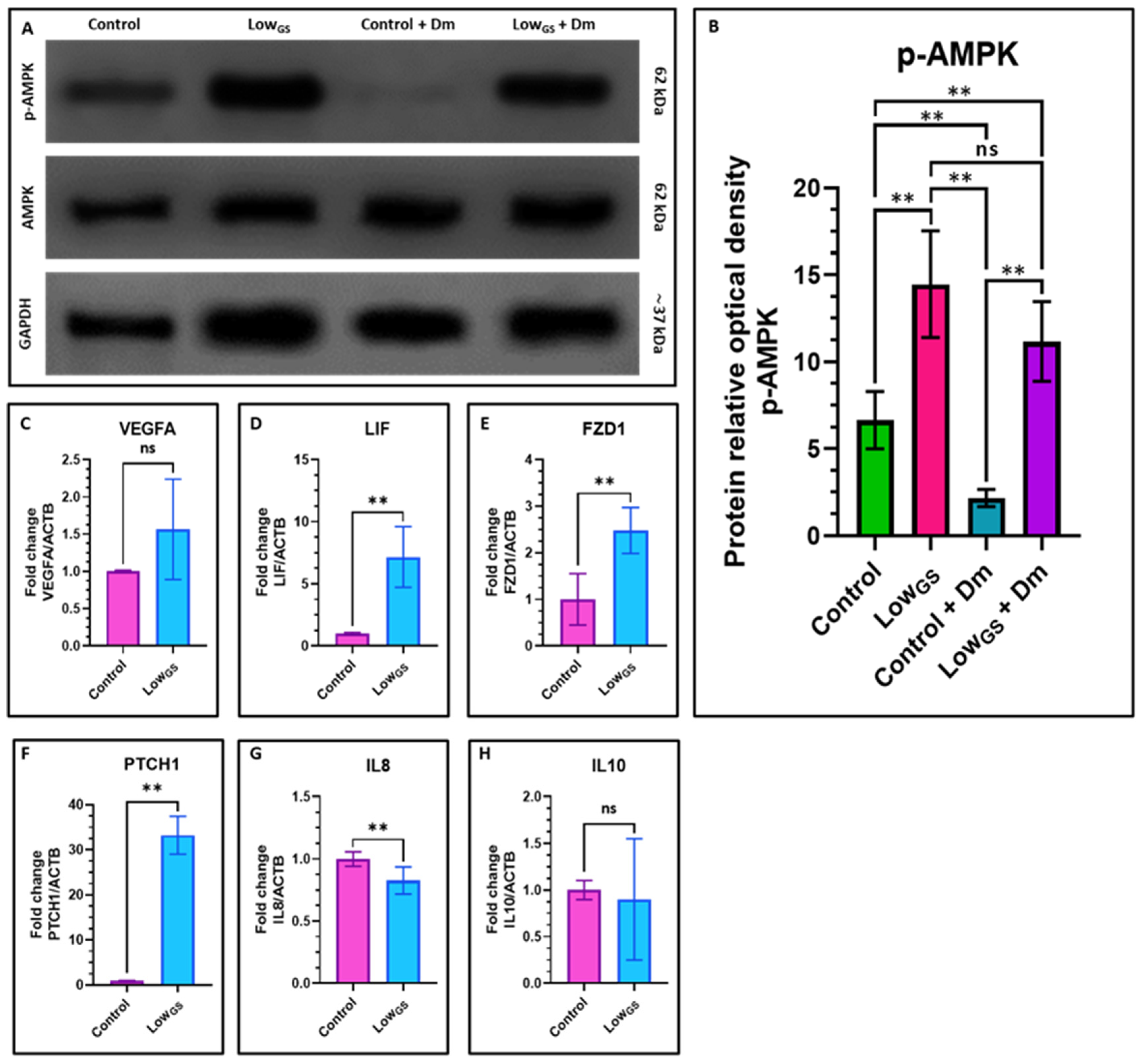
3.5. Nutrient Deprivation Leads to the Activation of AMPK in SHEDs
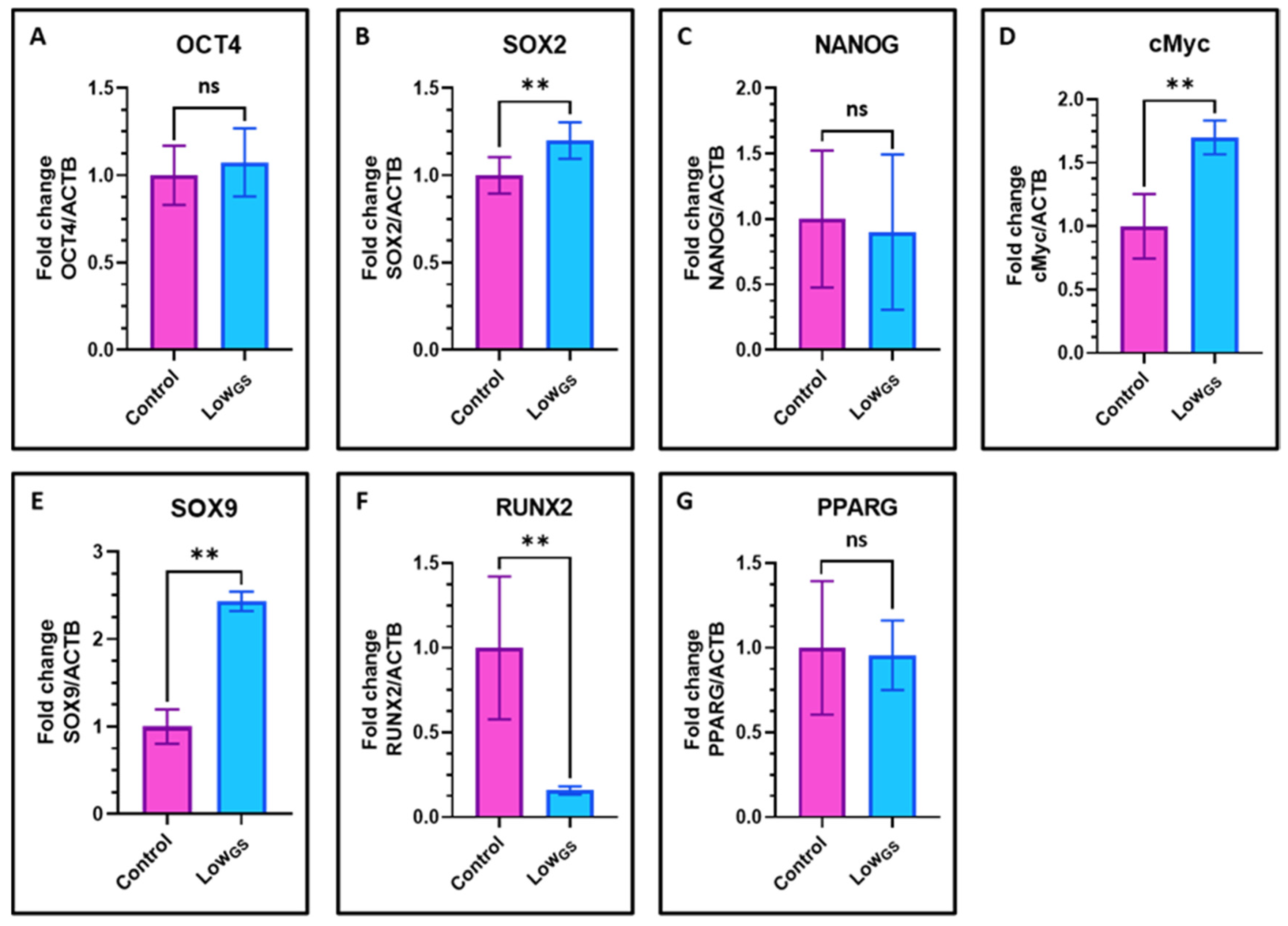
3.6. Nutrient Deprivation Elevated the Expression of Stemness Genes and Chondrogenic Differentiation in SHEDs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Sah, J.P. Challenges of Stem Cell Therapy in Developing Country. J. Stem Cell Res. Ther. 2016, 1, 96–98. [Google Scholar] [CrossRef][Green Version]
- Prockop, D.J. Marrow Stromal Cells as Stem Cells for Nonhematopoietic Tissues. Science 1997, 276, 71–74. [Google Scholar] [CrossRef]
- Weissman, I.L. Translating Stem and Progenitor Cell Biology to the Clinic: Barriers and Opportunities. Science 2000, 287, 1442–1446. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Mackay, A.M.; Beck, S.C.; Jaiswal, R.K.; Douglas, R.; Mosca, J.D.; Moorman, M.A.; Simonetti, D.W.; Craig, S.; Marshak, D.R. Multilineage Potential of Adult Human Mesenchymal Stem Cells. Science 1999, 284, 143–147. [Google Scholar] [CrossRef] [PubMed]
- Govitvattana, N.; Osathanon, T.; Taebunpakul, S.; Pavasant, P. IL-6 regulated stress-induced Rex-1 expression in stem cells from human exfoliated deciduous teeth. Oral Dis. 2012, 19, 673–682. [Google Scholar] [CrossRef]
- Sukarawan, W.; Osathanon, T. Stem Cells from Human Exfoliated Deciduous Teeth: Biology and Therapeutic Potential. In Mesenchymal Stem Cells: Isolation, Characterization and Applications; Intechopen: London, UK, 2017. [Google Scholar]
- Taguchi, T.; Yanagi, Y.; Yoshimaru, K.; Zhang, X.-Y.; Matsuura, T.; Nakayama, K.; Kobayashi, E.; Yamaza, H.; Nonaka, K.; Ohga, S.; et al. Regenerative medicine using stem cells from human exfoliated deciduous teeth (SHED): A promising new treatment in pediatric surgery. Surg. Today 2019, 49, 316–322. [Google Scholar] [CrossRef]
- Saez, D.M.; Sasaki, R.T.; da Costa Neves, A.; Da Silva, M.C. Stem Cells from Human Exfoliated Deciduous Teeth: A Growing Literature. Cells Tissues Organs 2016, 202, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Denu, R.A.; Hematti, P. Effects of Oxidative Stress on Mesenchymal Stem Cell Biology. Oxidative Med. Cell. Longev. 2016, 2016, 2989076. [Google Scholar] [CrossRef]
- Fonteneau, G.; Bony, C.; Goulabchand, R.; Maria, A.T.J.; Le Quellec, A.; Rivière, S.; Jorgensen, C.; Guilpain, P.; Noël, D. Serum-Mediated Oxidative Stress from Systemic Sclerosis Patients Affects Mesenchymal Stem Cell Function. Front. Immunol. 2017, 8, 988. [Google Scholar] [CrossRef] [PubMed]
- Anoop, M.; Datta, I. Stem Cells Derived from Human Exfoliated Deciduous Teeth (SHED) in Neuronal Disorders: A Review. Curr. Stem Cell Res. Ther. 2021, 16, 535–550. [Google Scholar] [CrossRef]
- Nakamura, S.; Yamada, Y.; Katagiri, W.; Sugito, T.; Ito, K.; Ueda, M. Stem Cell Proliferation Pathways Comparison between Human Exfoliated Deciduous Teeth and Dental Pulp Stem Cells by Gene Expression Profile from Promising Dental Pulp. J. Endod. 2009, 35, 1536–1542. [Google Scholar] [CrossRef]
- Wang, S.; Qu, X.; Zhao, R.C. Clinical applications of mesenchymal stem cells. J. Hematol. Oncol. 2012, 5, 19. [Google Scholar] [CrossRef]
- Kim, N.; Cho, S.-G. Clinical applications of mesenchymal stem cells. Korean J. Intern. Med. 2013, 28, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Yuan, Z.; Weng, J.; Pei, D.; Du, X.; He, C.; Lai, P. Challenges and advances in clinical applications of mesenchymal stromal cells. J. Hematol. Oncol. 2021, 14, 24. [Google Scholar] [CrossRef]
- Nuschke, A.; Rodrigues, M.; Stolz, D.B.; Chu, C.T.; Griffith, L.; Wells, A. Human mesenchymal stem cells/multipotent stromal cells consume accumulated autophagosomes early in differentiation. Stem Cell Res. Ther. 2014, 5, 140. [Google Scholar] [CrossRef] [PubMed]
- Farrell, M.; Shin, J.; Smith, L.; Mauck, R. Functional consequences of glucose and oxygen deprivation on engineered mesenchymal stem cell-based cartilage constructs. Osteoarthr. Cartil. 2015, 23, 134–142. [Google Scholar] [CrossRef]
- Kulkarni, G.V.; A McCulloch, C. Serum deprivation induces apoptotic cell death in a subset of Balb/c 3T3 fibroblasts. J. Cell Sci. 1994, 107 Pt 5, 1169–1179. [Google Scholar] [CrossRef]
- Wan, P.X.; Wang, B.W.; Wang, Z.C. Importance of the stem cell microenvironment for ophthalmological cell-based therapy. World J. Stem Cells 2015, 7, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhong, Q.; Yang, T.; Qi, Y.; Fu, M.; Yang, X.; Qiao, L.; Ling, Q.; Liu, S.; Zhao, Y. Comparative characterization of SHED and DPSCs during extended cultivation in vitro. Mol. Med. Rep. 2018, 17, 6551–6559. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Fu, K.Y. Serum-deprivation leads to activation-like changes in primary microglia and BV-2 cells but not astrocytes. Biomed. Rep. 2020, 13, 51. [Google Scholar] [CrossRef]
- Periasamy, R.; Elshaer, S.L.; Gangaraju, R. CD140b (PDGFRbeta) signaling in adipose-derived stem cells mediates angiogenic behavior of retinal endothelial cells. Regen. Eng. Transl. Med. 2019, 5, 1–9. [Google Scholar] [CrossRef]
- Luo, J.; Martinez, J.; Yin, X.; Sanchez, A.; Tripathy, D.; Grammas, P. Hypoxia induces angiogenic factors in brain microvascular endothelial cells. Microvasc. Res. 2012, 83, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Luo, C.; Lu, Y.; Tang, C.; Ouyang, Q. Cell cycle synchronization by nutrient modulation. Integr. Biol. 2012, 4, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Tinnemans, M.M.F.J.; Lenders, M.-H.J.H.; Velde, G.P.M.T.; Blijham, G.H.; Ramaekers, F.C.S.; Schutte, B. S-phase arrest of nutrient deprived lung cancer cells. Cytometry 1995, 19, 326–333. [Google Scholar] [CrossRef]
- Dutto, I.; Tillhon, M.; Cazzalini, O.; Stivala, L.A.; Prosperi, E. Biology of the cell cycle inhibitor p21CDKN1A: Molecular mechanisms and relevance in chemical toxicology. Arch. Toxicol. 2015, 89, 155–178. [Google Scholar] [CrossRef]
- Dávila, D.; Connolly, N.M.C.; Bonner, H.; Weisová, P.; Düssmann, H.; Concannon, C.G.; Huber, H.J.; Prehn, J.H.M. Two-step activation of FOXO3 by AMPK generates a coherent feed-forward loop determining excitotoxic cell fate. Cell Death Differ. 2012, 19, 1677–1688. [Google Scholar] [CrossRef]
- Kim, J.; Yang, G.; Kim, Y.; Kim, J.; Ha, J. AMPK activators: mechanisms of action and physiological activities. Exp. Mol. Med. 2016, 48, e224. [Google Scholar] [CrossRef]
- Mihaylova, M.M.; Shaw, R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011, 13, 1016–1023. [Google Scholar] [CrossRef] [PubMed]
- Mirouse, V.; Billaud, M. The LKB1/AMPK polarity pathway. FEBS Lett. 2010, 585, 981–985. [Google Scholar] [CrossRef]
- Ouchi, N.; Shibata, R.; Walsh, K. AMP-Activated Protein Kinase Signaling Stimulates VEGF Expression and Angiogenesis in Skeletal Muscle. Circ. Res. 2005, 96, 838–846. [Google Scholar] [CrossRef] [PubMed]
- Nylén, C.; Aoi, W.; Abdelmoez, A.M.; Lassiter, D.G.; Lundell, L.S.; Wallberg-Henriksson, H.; Naslund, E.; Pillon, N.J.; Krook, A. IL6 and LIF mRNA expression in skeletal muscle is regulated by AMPK and the transcription factors NFYC, ZBTB14, and SP1. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E995–E1004. [Google Scholar] [CrossRef]
- Yue, X.; Wu, L.; Hu, W. The Regulation of Leukemia Inhibitory Factor. Cancer Cell Microenviron. 2015, 2, e877. [Google Scholar] [CrossRef]
- Park, S.B.; Seo, K.W.; So, A.Y.; Seo, M.S.; Yu, K.R.; Kang, S.K.; Kang, K.S. SOX2 has a crucial role in the lineage determination and proliferation of mesenchymal stem cells through Dickkopf-1 and c-MYC. Cell Death Differ. 2011, 19, 534–545. [Google Scholar] [CrossRef]
- Xu, J.; Li, Z.; Hou, Y.; Fang, W. Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am. J. Transl. Res. 2015, 7, 2527–2535. [Google Scholar]

| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CCNB1 | 5′-GAC CTG TGT CAG GCT TTC TCT G-3′ | 5′-GGT ATT TTG GTC TGA CTG CTT GC-3′ |
| CCNE1 | 5′-TGT GTC CTG GAT GTT GAC TGC C-3′ | 5′-CTC TAT GTC GCA CCA CTG ATA CC-3′ |
| CCND1 | 5′-TCT ACA CCG ACA ACT CCA TCC G-3′ | 5′-TCT GGC ATT TTG GAG AGG AAG TG-3′ |
| CCNA2 | 5′-CTC TAC ACA GTC ACG GGA CAA AG-3′ | 5′-CTG TGG TGC TTT GAG GTA GGT C-3′ |
| CDKN1A | 5′-AGG TGG ACC TGG AGA CTC TCA G-3′ | 5′-TCC TCT TGG AGA AGA TCA GCC G-3′ |
| CDKN1B | 5′-ATA AGG AAG CGA CCT GCA ACC G-3′ | 5′-TTC TTG GGC GTC TGC TCC ACA G-3′ |
| FOXO1 | 5′-CTA CGA GTG GAT GGT CAA GAG C-3′ | 5′-CCA GTT CCT TCA TTC TGC ACA CG-3′ |
| FOXO3 | 5′-TCT ACG AGT GGA TGG TGC GTT G-3′ | 5′-CTC TTG CCA GTT CCC TCA TTC TG-3′ |
| VEGFA | 5′-TTG CCT TGC TGC TCT ACC TCC A-3′ | 5′-GAT GGC AGT AGC TGC GCT GAT A-3′ |
| LIF | 5′-AGA TCA GGA GCC AAC TGG CAC A-3′ | 5′-GCC ACA TAG CTT GTC CAG GTT G-3′ |
| IL8 | 5′-GTG CAG TTT TGC CAA GGA GT-3′ | 5′-TTA TGA ATT CTC AGC CCT CTT CAA-3′ |
| IL10 | 5′-TCT CCG AGA TGC CTT CAG CAG A-3′ | 5′-TCA GAC AAG GCT TGG CAA CCC A-3′ |
| PTCH1 | 5′-GCT GCA CTA CTT CAG AGA CTG G-3′ | 5′-CAC CAG GAG TTT GTA GGC AAG G-3′ |
| FZD1 | 5′-GCT TTG TGT CGC TCT TCC GCA T-3′ | 5′-TAC AGC ACG CTG AAG ACG CCA A-3′ |
| OCT3/4 | 5′-CCT GAA GCA GAA GAG GAT CAC C-3′ | 5′-AAA GCG GCA GAT GGT CGT TTG G-3′ |
| SOX2 | 5′-GCT ACA GCA TGA TGC AGG ACC A-3′ | 5′-TCT GCG AGC TGG TCA TGG AGT T-3′ |
| NANOG | 5′-CTC CAA CAT CCT GAA CCT CAG C-3′ | 5′-CGT CAC ACC ATT GCT ATT CTT CG-3′ |
| MYC | 5′-CCT GGT GCT CCA TGA GGA GAC-3′ | 5′-CA GAC TCT GAC CTT TTG CCA GG-3′ |
| SOX9 | 5′-GCC GAA AGC GGG CTC GAA AC-3′ | 5′-AAA AGT GGG GGC GCT TGC ACC-3′ |
| RUNX2 | 5′-GTG CCT AGG CGC ATT TCA-3′ | 5′-GCT CTT CTT ACT GAG AGT GGA AGG-3′ |
| PPARG | 5′-AGC CTG CGA AAG CCT TTT GGT G-3′ | 5′-GGC TTC ACA TTC AGC AAA CCT GG-3′ |
| ACTB | 5′-AGA GCT ACG AGC TGC CTG AC-3′ | 5′-AGC ACT GTG TTG GCG TAC AG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pawar, M.; Pawar, V.; Renugalakshmi, A.; Albrakati, A.; Uthman, U.S.; Dewan, H.; Mugri, M.; Sayed, M.; Bhandi, S.; Patil, V.R.; et al. Glucose and Serum Deprivation Led to Altered Proliferation, Differentiation Potential and AMPK Activation in Stem Cells from Human Deciduous Tooth. J. Pers. Med. 2022, 12, 18. https://doi.org/10.3390/jpm12010018
Pawar M, Pawar V, Renugalakshmi A, Albrakati A, Uthman US, Dewan H, Mugri M, Sayed M, Bhandi S, Patil VR, et al. Glucose and Serum Deprivation Led to Altered Proliferation, Differentiation Potential and AMPK Activation in Stem Cells from Human Deciduous Tooth. Journal of Personalized Medicine. 2022; 12(1):18. https://doi.org/10.3390/jpm12010018
Chicago/Turabian StylePawar, Madhura, Vivek Pawar, Apathsakayan Renugalakshmi, Ashraf Albrakati, Uthman S. Uthman, Harisha Dewan, Maryam Mugri, Mohammed Sayed, Shilpa Bhandi, Vikrant R. Patil, and et al. 2022. "Glucose and Serum Deprivation Led to Altered Proliferation, Differentiation Potential and AMPK Activation in Stem Cells from Human Deciduous Tooth" Journal of Personalized Medicine 12, no. 1: 18. https://doi.org/10.3390/jpm12010018
APA StylePawar, M., Pawar, V., Renugalakshmi, A., Albrakati, A., Uthman, U. S., Dewan, H., Mugri, M., Sayed, M., Bhandi, S., Patil, V. R., Reda, R., Testarelli, L., & Patil, S. (2022). Glucose and Serum Deprivation Led to Altered Proliferation, Differentiation Potential and AMPK Activation in Stem Cells from Human Deciduous Tooth. Journal of Personalized Medicine, 12(1), 18. https://doi.org/10.3390/jpm12010018











