Abstract
Amelogenesis imperfecta (AI) is a rare genetic condition affecting the quantity and/or quality of tooth enamel. Hypomaturation AI is characterized by brownish-yellow discoloration with increased opacity and poorly mineralized enamel prone to fracture and attrition. We recruited three families affected by hypomaturation AI and performed whole exome sequencing with selected individuals in each family. Bioinformatic analysis and Sanger sequencing identified and confirmed mutations and segregation in the families. Family 1 had a novel homozygous frameshift mutation in GPR68 gene (NM_003485.3:c.78_83delinsC, p.(Val27Cysfs*146)). Family 2 had a novel homozygous nonsense mutation in SLC24A4 gene (NM_153646.4:c.613C>T, NP_705932.2:p.(Arg205*)). Family 3 also had a homozygous missense mutation in SLC24A4 gene which was reported previously (c.437C>T, p.(Ala146Val)). This report not only expands the mutational spectrum of the AI-causing genes but also improves our understanding of normal and pathologic amelogenesis.
1. Introduction
Tooth enamel is the hardest tissue in the human body, covering the crown of the tooth not only to protect the underlying dentin–pulp complex but also to enhance esthetics and facilitate masticatory function. The formation of enamel begins with the ameloblasts’ secretion of enamel matrix proteins on the predentin, which triggers the growth and elongation of enamel crystallites [1]. The process continues until the full thickness of the enamel is achieved although only 30% of mineral by weight is produced. When the ameloblasts enter the maturation stage, they cycle through the ruffle-ended and smooth-ended phases during which matrix proteins are further degraded and removed and rapid appositional growth of enamel is accelerated [2]. The coordinated pH sensing and ion transportation of ameloblasts are critical for optimal enamel maturation [3,4].
Amelogenesis imperfecta (AI) is a heterogeneous collection of hereditary defects affecting tooth enamel. Depending on how the hereditary defect impacts the specific stage of enamel formation, AI is classified into hypoplastic, hypocalcified, and hypomatured types [5]. AI-affected individuals suffer from poor esthetics and masticatory discomfort including hypersensitivity to cold and hot stimuli and marked impact on the psychosocial wellbeing, such as social avoidance, distress, and low self-esteem [6,7].
Since the discovery of the amelogenins in the enamel matrix [8,9], many critical players in amelogenesis have been identified by biochemical analyses and genetic studies. To date, more than 20 genes are known to cause AI. Briefly, AMELX (amelogenin, OMIM *300391) [10], ENAM (enamelin, OMIM *606585) [11], AMBN (ameloblastin, OMIM *601259) [12], RELT (RELT TNF receptor, OMIM *611211) [13], and ACP4 (acid phosphatase 4, also known as acid phosphatase, testicular; OMIM *606362) [14] are related to hypoplastic AI; FAM83H (family with sequence similarity 83 member H, OMIM *611927) is involved in hypocalcified AI [15]; and ODAPH (odontogenesis associated phosphoprotein, also known as C4orf26; OMIM *614829) [16], MMP20 (matrix metalloproteinase 20, OMIM *604629) [17], KLK4 (kallikrein related peptidase 4, OMIM *603767) [18], WDR72 (WD repeat domain 72, OMIM *613214) [19], SLC24A4 (solute carrier family 24 member 4, OMIM *609840) [20], and GPR68 (G protein-coupled receptor 68, OMIM *601404) [21] are involved in hypomaturation AI.
In this study, we recruited three Turkish families with hypomaturation AI and performed mutational analyses with whole exome sequencing. The analyses revealed a novel frameshift mutation caused by an indel (insertion and deletion) in the GPR68 gene and a novel nonsense mutation and a previously reported missense mutation in the SLC24A4 gene. This report confirms the functional importance of GPR68 and SLC24A4 in maturation amelogenesis, contributes various enamel phenotypes, and expands the mutational spectrum to advance our understanding of enamel maturation.
2. Materials and Methods
2.1. Study Subject Enrollment
The study protocol was independently reviewed and approved by the institutional review boards of the Seoul National University Dental Hospital (CRI05003G and 10 December 2020), Istanbul University (No: 2008/931 and 20 September 2019), and the University of Michigan (H03-00001835-M1 and 6 May 2021). Informed consent was obtained from the participating family members. Clinical examinations were performed, and saliva samples were collected.
2.2. DNA Isolation and Whole Exome Sequencing
Genomic DNA was isolated from the saliva samples, and the quality and quantity were measured. The DNA samples of selected individuals from each family (Table S1) were submitted to the Center for Inherited Disease Research at the Johns Hopkins University (CIDR, Baltimore, Maryland). The exomes were captured with the Agilent SureSelect Human All Exon Enrichment System. The sequencing reads were generated using the Illumina HiSeq 2500 (Illumina, Inc., San Diego, CA, USA).
2.3. Bioinformatics
The obtained 125-bp paired-end sequence reads were aligned to the reference human genome assembly (hg37) with the Burrows-Wheeler Aligner [22]. Sequence variants were obtained after the application of a series of bioinformatics analysis programs including Samtools and Genome Analysis Tool Kit [23,24]. Annovar was used to annotate the sequence variants with dbSNP build 147. A minor allele frequency (MAF) of 0.01 was the cutoff value for variant filtering.
2.4. Sanger Sequencing
The identified mutations in the probands and the segregation among family members were interrogated by Sanger sequencing using the following primers: GPR68 exon 2 for family 1 (sense 5′-CCTTTCCTGCCTCTGACTTTC-3′ and antisense 5′-AGCACGTACTGCAGCCAGA-3′), SLC24A4 exon 7 for family 2 (sense 5′-GTGGCCTGGAGTTAGGAGGT-3′ and antisense 5′-AGTGCCAGGGGCAGAGAT-3′), and SLC24A4 exon 5 for family 3 (sense 5′-TTGGCTGTAGAGCGTCCAGT-3′ and antisense 5′-TGAGGCTCAGAGCTGACAAA-3′). Sanger sequencing was performed for all participating family members at Eurofins Genomics (Louisville, KY, USA).
3. Results
3.1. Family 1
Family 1 was a four-generation Turkish family with consanguineous marriage (Figure 1). There were several affected individuals in generation II and IV, suggesting an autosomal recessive inheritance pattern. The proband was an 11-year-old second child (IV:2) from a non-consanguineous marriage, and he had no remarkable past medical history, except for the generalized brown discoloration of his dentition. The discoloration was noticed in all teeth but was less severe in the cervical region of the anterior teeth. Panoramic radiograph did not show a clear contrast between the enamel and dentin, suggesting that the enamel was hypomineralized. His 5-year-old younger sister (VI:3) was also affected (Figure S1). The maternal grandmother (II:2) and a cousin (IV:4) from a consanguineous marriage were similarly affected.

Figure 1.
(a) Pedigree of family 1. The black arrow denotes the proband. Plus sign indicates participating individuals in this study. Number in the symbol indicates the number of siblings. Consanguineous marriage is shown by a double line. (b–d) Clinical photos of the proband (IV:2). Hypomatured enamel exhibits a brown discoloration, and the posterior teeth are treated with glass ionomer. (e) Panoramic radiograph shows hypomineralized enamel with a reduced radiodensity.
Mutational analysis of the whole exome sequencing data revealed a novel homozygous mutation in the GPR68 gene (Figure S2) which was previously reported to cause autosomal recessive hypomaturation AI without any other syndromic phenotype. The identified mutation was a deletion of six nucleotides (GGTCTA) at position 78-83 in the cDNA and an insertion of a nucleotide (C) (NM_003485.3:c.78_83delinsC), and this would shift the translational frame to result in a truncated protein (NP_003476.3:p.(Val27Cysfs*146)). This mutation was not listed in the Exome Aggregation Consortium (ExAC) database (https://exac.broadinstitute.org/ (accessed on 27 November 2021)) or Genome Aggregation Database (gnomAD: https://gnomad.broadinstitute.org/ (accessed on 27 November 2021)).
3.2. Family 2
The proband of family 2 was a 10-year-old first child (IV:1) from a consanguineous marriage in a four-generation Turkish family with hypomaturation AI (Figure 2). His dentition had a generalized brown discoloration, and the weak enamel on the first permanent molars had broken off. His younger sister (IV:2) was not affected, and they had no remarkable past medical histories. There were additional affected family members in generation II (II:2, II:5, and II:6), suggesting an autosomal recessive inheritance pattern.
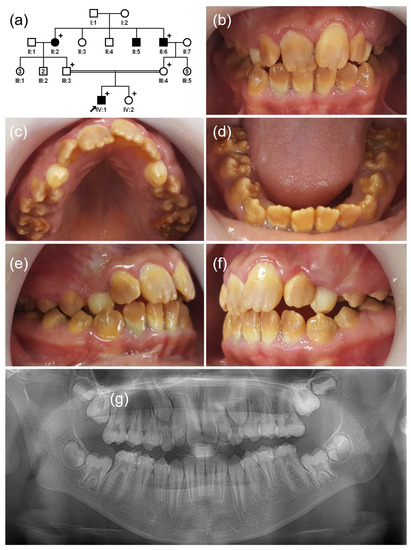
Figure 2.
(a) Pedigree of family 2. The black arrow denotes the proband. Plus sign indicates participating individuals in this study. Number in the symbol indicates the number of siblings. Consanguineous marriage is shown by a double line. (b–f) Clinical photos of the proband (IV:1). Hypomatured enamel exhibits a generalized brown discoloration. (g) Panoramic radiograph shows hypomineralized enamel with reduced radiodensity. Breakdown of weak enamel can be seen in the first molars.
Mutational analysis of the whole exome sequencing data with an autosomal recessive inheritance model revealed a novel homozygous mutation in SLC24A4 gene (Figure S3), which was previously proven to cause non-syndromic hypomaturation AI. The identified mutation was a nonsense mutation (NP_705932.2:p.(Arg205*)) caused by a transitional change of a cytosine to a thymine in exon 7 (NM_153646.4:c.613C>T). The mutation would have resulted in the decay of the mRNA by the nonsense-mediated surveillance system because the truncating mutation was in the early exon among the 17 coding exons [25,26]. The mutation was listed in the dbSNP database as rs141131742 and in gnomAD with an allele frequency of 2/251316.
3.3. Family 3
The proband of family 3 was a 10-year-old third child (V:3) from a consanguineous marriage in an extended five-generation Turkish family (Figure 3). The pregnancy and delivery of this child was uneventful, and there was no remarkable past medical history. Her teeth were brown, and the first molars were restored with composite resin due to the enamel breakdown probably resulting from masticatory force imposed on the weak, hypomineralized enamel.
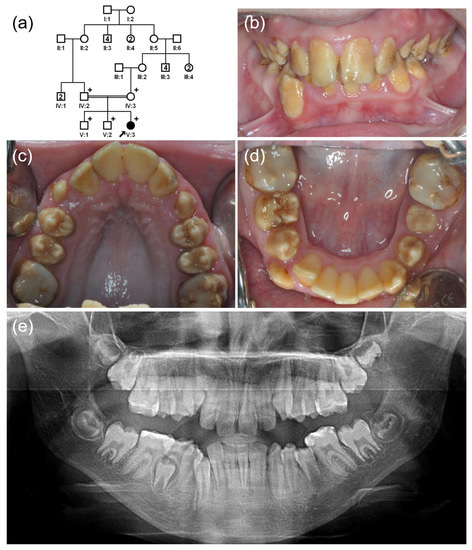
Figure 3.
(a) Pedigree of family 3. The black arrow denotes the proband. Plus sign indicates participating individuals in this study. Number in the symbol indicates the number of siblings. Consanguineous marriage is shown by a double line. (b–d) Clinical photos of the proband (V:3). Hypomatured enamel exhibits a brown discoloration, and the first molars are treated with a composite resin. (e) Panoramic radiograph shows hypomineralized enamel with a reduced radiodensity.
There were no other family members with similar enamel defect. Therefore, the mutational analysis was performed with the possibility of a spontaneous mutation and autosomal recessive inheritance model. A homozygous missense mutation (NP_705932.2:p.(Ala146Val)) in the SLC24A4 gene was identified (NM_153646.4:c.437C>T) (Figure S4). This mutation was listed in the dbSNP database as rs587777537 and was previously reported to cause non-syndromic autosomal recessive hypomaturation AI.
4. Discussion
GPR68 is one of the proton-sensing membrane receptors belonging to the G protein-coupled receptor (GPCR) superfamily [27]. There are more than 800 GPCRs in humans, and they are involved in diverse biological functions that sense and relay information from the extracellular to the intracellular environment [28]. Interestingly, they share similar structural features, having an extracellular N-terminus, intracellular C-terminus, and in-between seven transmembrane domains, with three extracellular and three intracellular loops [27]. GPR68 also has the same structure which interestingly is encoded by a single coding exon. There are 11 transcripts reported in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/ (accessed on 28 November 2021)), all with different noncoding exon(s). Even though the tissue-specific expression of each transcript remains to be further studied, the expression of the GPR68 protein in the ameloblast and underlying stellate reticulum was confirmed previously [21,29].
The mutation identified in this study was a delin frameshift mutation that occurred early in the transcript (NM_003485.3:c.78_83delinsC) (Table 1). This mutant transcript would introduce an early termination codon and produce a truncated protein with only 27 N-terminus plus 145 novel amino acids (p.(Val27Cysfs*146)) because it could escape from a nonsense-mediated mRNA decay system [25,26]. However, the mutant protein could not function like the wild-type protein (365 amino acids) due to the loss of most of its structural components.

Table 1.
Disease-causing mutations in GPR68 gene.
The clinical phenotype of GPR68 mutations presented in a previous study was slightly different from the typical hypomaturation AI; the brown discoloration was minimal in some affected siblings [21]. However, the affected enamel was apparently of full thickness but poorly mineralized and showed early loss of opaque enamel due to fracture. The clinical phenotype of the proband of family 1 in this study presented with typical brown discoloration, except the near-normal color in the cervical region of the permanent anterior teeth (especially in the maxillary central incisors). Other affected individuals exhibited typical clinical features of brown-discolored hypomaturation AI (Figure S1).
Families 2 and 3 had mutations in the SLC24A4 gene, which encodes a potassium-dependent sodium/calcium exchanger. Mutations in this gene have been proven to cause hypomaturation AI by disrupting the function of removing and supplying essential ions for the maturation of the enamel crystallites [20]. Additionally, it has been suggested to be associated with blond/brown hair and blue/green eyes [30]. The missense mutation identified in this study was previously reported [31]. Additionally, there were several previously reported loss of function mutations (Table 2). SLC24A4 is a transmembrane protein belonging to a sodium/potassium/calcium exchanger superfamily with a cytoplasmic N-terminus, 11 transmembrane domains, and an extracellular C-terminus [32]. The superfamily has highly conserved shared hydrophobic domains (α-1 and α-2 repeats), and the p.(Ala146Val) mutation is in the α-1 repeat [33].

Table 2.
Disease-causing mutations in SLC24A4 gene.
The clinical phenotypes of families 2 and 3 were similar to typical hypomaturation AI. There was a generalized dark brown discoloration in all teeth, and the poorly mineralized, relatively weak enamel tended to be prematurely lost to attrition or masticatory stress, especially in the posterior teeth.
In conclusion, we recruited three hypomaturation AI families, and mutational analyses revealed a GPR68 mutation in family 1 and SLC24A4 mutations in families 2 and 3. This is the second paper reporting a GPR68 mutation and suggests that the phenotype caused by the GPR68 mutation can be variable depending on the type of mutation and/or genetic background. Further functional studies with more mutation cases are needed to characterize the factors influencing the various clinical phenotype and to advance our understanding of the normal and pathologic enamel formation.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/jpm12010013/s1, Table S1: Statistics for exome sequencing; Figure S1: Clinical photos and panoramic radiograph of individuals IV:3 and IV:4 of family 1; Figure S2: Sequencing chromatograms of the participating individuals of family 1; Figure S3: Sequencing chromatograms of the participating individuals of family 2. Figure S4: Sequencing chromatograms of the participating individuals of family 3.
Author Contributions
H.Z., J.C.-C.H. and J.-W.K. contributed to performing the experiments, data analysis, interpretation, and drafting of the paper. F.S., Y.K., M.K., J.C.-C.H. and J.-W.K. contributed to diagnosis and recruitment of the family members and critically revised the paper. F.S., J.P.S., J.C.-C.H. and J.-W.K. contributed to designing and performing the research, interpretation and molecular diagnosis, drafting the paper, and critically revising the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (NRF-2018R1A5A2024418 and NRF-2020R1A2C2100543) and the National Institute of Dental and Craniofacial Research (DE015846).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the institutional review boards of the Seoul National University Dental Hospital, Istanbul University, and the University of Michigan.
Informed Consent Statement
Informed consent was obtained from all individual participants included in the study.
Data Availability Statement
The data presented in this study are openly available in ClinVar (http://www.ncbi.nlm.nih.gov/clinvar (accessed on 27 November 2021)), Submission ID: SUB10711085 and SUB10726703.
Acknowledgments
We are grateful to all family members who participated in this study.
Conflicts of Interest
All authors declare no conflict of interest.
References
- Simmer, J.P.; Papagerakis, P.; Smith, C.E.; Fisher, D.C.; Rountrey, A.; Zheng, L.; Hu, J.C.-C. Regulation of Dental Enamel Shape and Hardness. J. Dent. Res. 2010, 89, 1024–1038. [Google Scholar] [CrossRef]
- Smith, C. Cellular and Chemical Events during Enamel Maturation. Crit. Rev. Oral Biol. Med. 1998, 9, 128–161. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Nanci, A.; Kurtz, I.; Wright, J.T.; Paine, M.L. Regulation of pH During Amelogenesis. Calcif. Tissue Int. 2009, 86, 91–103. [Google Scholar] [CrossRef]
- Wright, J.T.; Carrion, I.A.; Morris, C. The Molecular Basis of Hereditary Enamel Defects in Humans. J. Dent. Res. 2015, 94, 52–61. [Google Scholar] [CrossRef]
- Witkop, C.J., Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: Problems in classification. J. Oral Pathol. Med. 1988, 17, 547–553. [Google Scholar] [CrossRef]
- Coffield, K.D.; Phillips, C.; Brady, M.; Roberts, M.W.; Strauss, R.P.; Wright, J.T. The psychosocial impact of developmental dental defects in people with hereditary amelogenesis imperfecta. J. Am. Dent. Assoc. 2005, 136, 620–630. [Google Scholar] [CrossRef]
- Sujak, S.L.; Kadir, R.A.; Dom, T.N.M. Esthetic perception and psychosocial impact of developmental enamel defects among Malaysian adolescents. J. Oral Sci. 2004, 46, 221–226. [Google Scholar] [CrossRef]
- Fincham, A.G.; Belcourt, A.B.; Termine, J.D.; Butler, W.T.; Cothran, W.C. Dental enamel matrix: Sequences of two amelogenin polypeptides. Biosci. Rep. 1981, 1, 771–778. [Google Scholar] [CrossRef]
- Fincham, A.G.; Simmer, J.P. Amelogenin proteins of developing dental enamel. In Ciba Foundation Symposium 205-Dental Enamel: Dental Enamel: Ciba Foundation Symposium 205; Wiley & Sons, Ltd.: Chichester, UK, 2007; Volume 205, pp. 118–134. [Google Scholar] [CrossRef]
- Lagerström, M.; Dahl, N.; Nakahori, Y.; Nakagome, Y.; Bäckman, B.; Landegren, U.; Pettersson, U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1). Genomics 1991, 10, 971–975. [Google Scholar] [CrossRef]
- Rajpar, M.H.; Harley, K.; Laing, C.; Davies, R.M.; Dixon, M.J. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum. Mol. Genet. 2001, 10, 1673–1677. [Google Scholar] [CrossRef]
- Poulter, J.A.; Murillo, G.; Brookes, S.J.; Smith, C.E.L.; Parry, D.A.; Silva, S.; Kirkham, J.; Inglehearn, C.F.; Mighell, A.J. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum. Mol. Genet. 2014, 23, 5317–5324. [Google Scholar] [CrossRef]
- Kim, J.W.; Zhang, H.; Seymen, F.; Koruyucu, M.; Hu, Y.; Kang, J.; Kim, Y.J.; Ikeda, A.; Kasimoglu, Y.; Bayram, M.; et al. Mutations in RELT cause autosomal recessive amelogenesis imperfecta. Clin. Genet. 2019, 95, 375–383. [Google Scholar] [CrossRef]
- Seymen, F.; Kim, Y.J.; Lee, Y.J.; Kang, J.; Kim, T.-H.; Choi, H.; Koruyucu, M.; Kasimoglu, Y.; Tuna, E.B.; Gencay, K.; et al. Recessive Mutations in ACPT, Encoding Testicular Acid Phosphatase, Cause Hypoplastic Amelogenesis Imperfecta. Am. J. Hum. Genet. 2016, 99, 1199–1205. [Google Scholar] [CrossRef][Green Version]
- Kim, J.-W.; Lee, S.-K.; Lee, Z.H.; Park, J.-C.; Lee, K.-E.; Lee, M.-H.; Park, J.-T.; Seo, B.-M.; Hu, J.C.-C.; Simmer, J.P. FAM83H Mutations in Families with Autosomal-Dominant Hypocalcified Amelogenesis Imperfecta. Am. J. Hum. Genet. 2008, 82, 489–494. [Google Scholar] [CrossRef]
- Parry, D.; Brookes, S.; Logan, C.; Poulter, J.; El-Sayed, W.; Al-Bahlani, S.; Al Harasi, S.; Sayed, J.; Raïf, E.M.; Shore, R.C.; et al. Mutations in C4orf26, Encoding a Peptide with In Vitro Hydroxyapatite Crystal Nucleation and Growth Activity, Cause Amelogenesis Imperfecta. Am. J. Hum. Genet. 2012, 91, 565–571. [Google Scholar] [CrossRef]
- Kim, J.-W.; Simmer, J.P.; Hart, T.C.; Hart, P.S.; Ramaswami, M.D.; Bartlett, J.D.; Hu, J.C.-C. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J. Med. Genet. 2005, 42, 271–275. [Google Scholar] [CrossRef]
- Hart, P.S.; Hart, T.C.; Michalec, M.D.; Ryu, O.H.; Simmons, D.; Hong, S.; Wright, J.T. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J. Med. Genet. 2004, 41, 545–549. [Google Scholar] [CrossRef]
- El-Sayed, W.; Parry, D.A.; Shore, R.C.; Ahmed, M.; Jafri, H.; Rashid, Y.; Al-Bahlani, S.; Al Harasi, S.; Kirkham, J.; Inglehearn, C.F.; et al. Mutations in the Beta Propeller WDR72 Cause Autosomal-Recessive Hypomaturation Amelogenesis Imperfecta. Am. J. Hum. Genet. 2009, 85, 699–705. [Google Scholar] [CrossRef]
- Parry, D.; Poulter, J.; Logan, C.; Brookes, S.J.; Jafri, H.; Ferguson, C.H.; Anwari, B.M.; Rashid, Y.; Zhao, H.; Johnson, C.A.; et al. Identification of Mutations in SLC24A4, Encoding a Potassium-Dependent Sodium/Calcium Exchanger, as a Cause of Amelogenesis Imperfecta. Am. J. Hum. Genet. 2013, 92, 307–312. [Google Scholar] [CrossRef]
- Parry, D.A.; Smith, C.E.; El-Sayed, W.; Poulter, J.A.; Shore, R.C.; Logan, C.V.; Mogi, C.; Sato, K.; Okajima, F.; Harada, A.; et al. Mutations in the pH-Sensing G-protein-Coupled Receptor GPR68 Cause Amelogenesis Imperfecta. Am. J. Hum. Genet. 2016, 99, 984–990. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Van Der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Miller, J.; Pearce, D.A. Nonsense-mediated decay in genetic disease: Friend or foe? Mutat. Res. Mutat. Res. 2014, 762, 52–64. [Google Scholar] [CrossRef]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Sisignano, M.; Fischer, M.; Geisslinger, G. Proton-Sensing GPCRs in Health and Disease. Cells 2021, 10, 2050. [Google Scholar] [CrossRef]
- Rowe, J.B.; Kapolka, N.J.; Taghon, G.J.; Morgan, W.M.; Isom, D.G. The evolution and mechanism of GPCR proton sensing. J. Biol. Chem. 2021, 296, 100167. [Google Scholar] [CrossRef]
- Sato, K.; Mogi, C.; Mighell, A.J.; Okajima, F. A missense mutation of Leu74Pro of OGR1 found in familial amelogenesis imperfecta actually causes the loss of the pH-sensing mechanism. Biochem. Biophys. Res. Commun. 2020, 526, 920–926. [Google Scholar] [CrossRef]
- Sulem, P.; Gudbjartsson, D.; Stacey, S.N.; Helgason, A.; Rafnar, T.; Magnusson, K.P.; Manolescu, A.; Karason, A.; Palsson, A.; Thorleifsson, G.; et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007, 39, 1443–1452. [Google Scholar] [CrossRef]
- Wang, S.-K.; Choi, M.; Richardson, A.; Reid, B.; Seymen, F.; Yildirim, M.; Tuna, E.; Gençay, K.; Simmer, J.; Hu, J. STIM1 and SLC24A4 Are Critical for Enamel Maturation. J. Dent. Res. 2014, 93, 94S–100S. [Google Scholar] [CrossRef]
- Li, X.-F.; Kraev, A.S.; Lytton, J. Molecular Cloning of a Fourth Member of the Potassium-dependent Sodium-Calcium Exchanger Gene Family, NCKX4. J. Biol. Chem. 2002, 277, 48410–48417. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, T.; Uehara, A.; Imanaga, I.; Shigekawa, M. The Na+/Ca2+ Exchanger NCX1 Has Oppositely Oriented Reentrant Loop Domains That Contain Conserved Aspartic Acids Whose Mutation Alters Its Apparent Ca2+Affinity. J. Biol. Chem. 2000, 275, 38571–38580. [Google Scholar] [CrossRef] [PubMed]
- Alam Khan, S.; Khan, M.A.; Muhammad, N.; Bashir, H.; Khan, N.; Muhammad, N.; Yilmaz, R.; Khan, S.; Wasif, N. A novel nonsense variant in SLC24A4 causing a rare form of amelogenesis imperfecta in a Pakistani family. BMC Med. Genet. 2020, 21, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.R.; Reid, B.M.; Seymen, F.; Koruyucu, M.; Tuna, E.B.; Simmer, J.P.; Hu, J.C.-C. Hypomaturation amelogenesis imperfecta caused by a novel SLC24A4 mutation. Oral Surgery Oral Med. Oral Pathol. Oral Radiol. 2014, 119, e77–e81. [Google Scholar] [CrossRef] [PubMed]
- Seymen, F.; Gencay, K.; Lee, K.-E.; Le, C.T.; Lee, Z.; Kim, J.-W.; Yildirim, M. Exonal Deletion of SLC24A4 Causes Hypomaturation Amelogenesis Imperfecta. J. Dent. Res. 2014, 93, 366–370. [Google Scholar] [CrossRef]
- Lepperdinger, U.; Maurer, E.; Witsch-Baumgartner, M.; Stigler, R.; Zschocke, J.; Lussi, A.; Kapferer-Seebacher, I. Expanding the phenotype of hypomaturation amelogenesis imperfecta due to a novel SLC24A4 variant. Clin. Oral Investig. 2020, 24, 3519–3525. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).