The Association of IgE Levels with ADAM33 Genetic Polymorphisms among Asthmatic Patients
Abstract
1. Introduction
2. Materials and Methods
2.1. Populations
2.2. Total Immunoglobulin E Level Measurement
2.3. DNA Extraction
2.4. Genotyping of ADAM33 Gene by PCR-RFLP
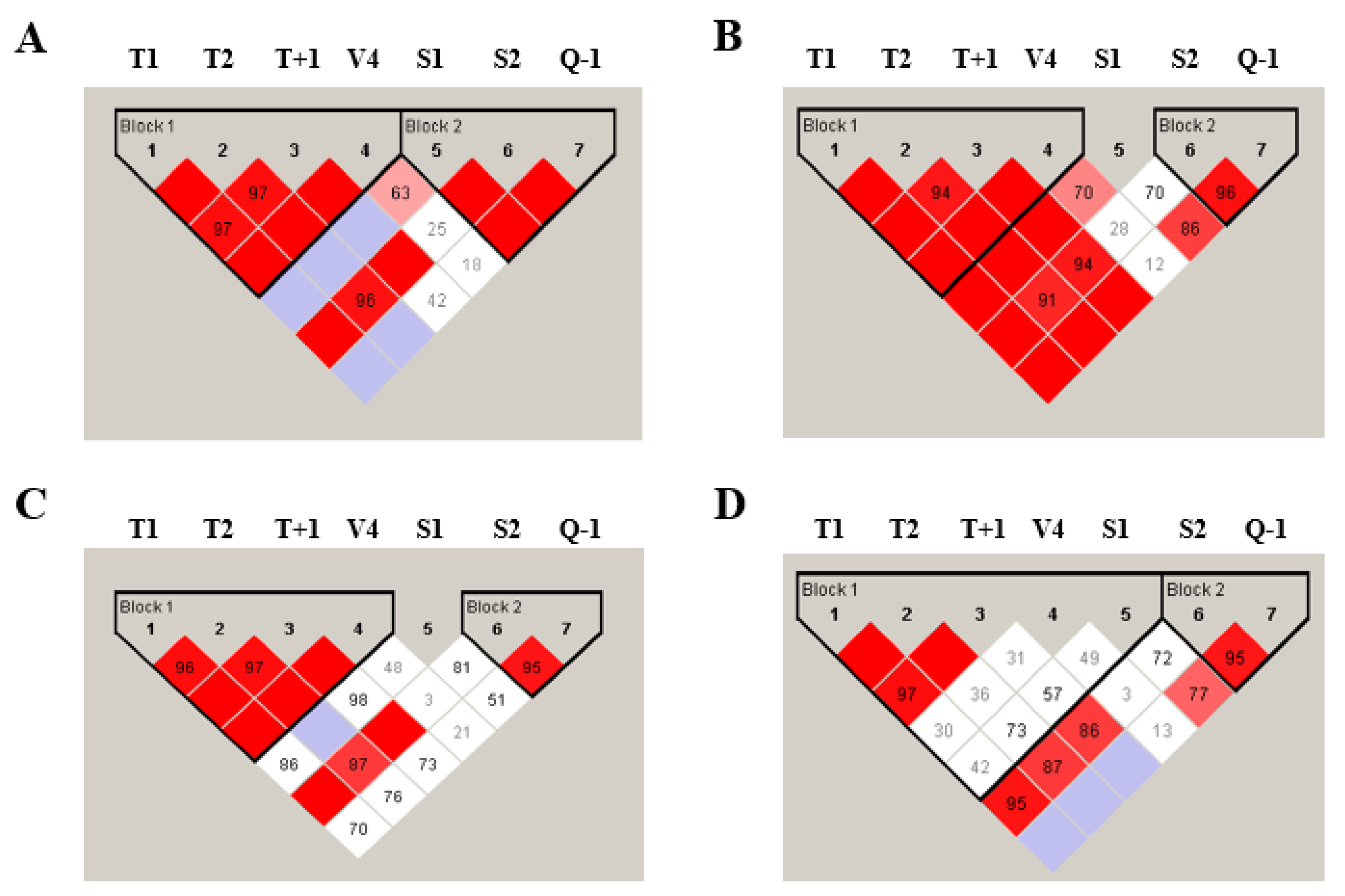
2.5. Linkage Disequilibrium and Haplotype Distribution
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berenguer, A.G.; Fernandes, A.T.; Oliveira, S.; Rodrigues, M.; Ornelas, P.; Romeira, D.; Serrao, T.; Rosa, A.; Camara, R. Genetic polymorphisms and asthma: Findings from a case-control study in the Madeira island population. Biol. Res. 2014, 47, 40. [Google Scholar] [CrossRef]
- Weinberg, E.G. Urbanization and childhood asthma: An African perspective. J. Allergy Clin. Immunol. 2000, 105, 224–231. [Google Scholar] [CrossRef]
- Li, H.F.; Yan, L.P.; Wang, K.; Li, X.T.; Liu, H.X.; Tan, W. Association between ADAM33 polymorphisms and asthma risk: A systematic review and meta-analysis. Respir. Res. 2019, 20, 38. [Google Scholar] [CrossRef]
- Bijanzadeh, M.; Mahesh, P.A.; Ramachandra, N.B. An understanding of the genetic basis of asthma. Indian J. Med. Res. 2011, 134, 149–161. [Google Scholar] [PubMed]
- Liang, S.; Wei, X.; Gong, C.; Wei, J.; Chen, Z.; Deng, J. A disintegrin and metalloprotease 33 (ADAM33) gene polymorphisms and the risk of asthma: A meta-analysis. Hum. Immunol. 2013, 74, 648–657. [Google Scholar] [CrossRef]
- Van Eerdewegh, P.; Little, R.D.; Dupuis, J.; Del Mastro, R.G.; Falls, K.; Simon, J.; Torrey, D.; Pandit, S.; McKenny, J.; Braunschweiger, K.; et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 2002, 418, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Awasthi, S.; Gao, P. ADAM metallopeptidase domain 33 (ADAM33): A promising target for asthma. Mediat. Inflamm. 2014, 2014, 572025. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Lin, R.; Wang, C.C.; Rei, J.; Sun, Y.; Yang, Y.L.; Lin, Y.Y. ADAM33 gene polymorphisms identified to be associated with asthma in a Chinese Li population. Biomed. Rep. 2017, 6, 323–328. [Google Scholar] [CrossRef][Green Version]
- AL-Zubaidy, B.S.; AL-Mahdawi, Z.M.M.; AL-Azaawie, A.F. Detection of IgE, Some Antioxidant Parameter Levels and their Association with Polymorphism of ADAM33 Gene in Asthma Patients. Res. J. Pharm. Technol. 2019, 12, 516–522. [Google Scholar] [CrossRef]
- Sharma, S.; Kathuria, P.C.; Gupta, C.K.; Nordling, K.; Ghosh, B.; Singh, A.B. Total serum immunoglobulin E levels in a case-control study in asthmatic/allergic patients, their family members, and healthy subjects from India. Clin. Exp. Allergy 2006, 36, 1019–1027. [Google Scholar] [CrossRef]
- Palomares, O.; Sanchez-Ramon, S.; Davila, I.; Prieto, L.; Perez de Llano, L.; Lleonart, M.; Domingo, C.; Nieto, A. dIvergEnt: How IgE Axis Contributes to the Continuum of Allergic Asthma and Anti-IgE Therapies. Int. J. Mol. Sci. 2017, 18, 1328. [Google Scholar] [CrossRef]
- Yang, K.D.; Chang, J.C.; Chuang, H.; Liang, H.M.; Kuo, H.C.; Lee, Y.S.; Hsu, T.Y.; Ou, C.Y. Gene-gene and gene-environment interactions on IgE production in prenatal stage. Allergy 2010, 65, 731–739. [Google Scholar] [CrossRef]
- Raby, B.A.; Silverman, E.K.; Kwiatkowski, D.J.; Lange, C.; Lazarus, R.; Weiss, S.T. ADAM33 polymorphisms and phenotype associations in childhood asthma. J. Allergy Clin. Immunol. 2004, 113, 1071–1078. [Google Scholar] [CrossRef]
- Howard, T.D.; Postma, D.S.; Jongepier, H.; Moore, W.C.; Koppelman, G.H.; Zheng, S.L.; Xu, J.; Bleecker, E.R.; Meyers, D.A. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J. Allergy Clin. Immunol. 2003, 112, 717–722. [Google Scholar] [CrossRef]
- Lind, D.L.; Choudhry, S.; Ung, N.; Ziv, E.; Avila, P.C.; Salari, K.; Ha, C.; Lovins, E.G.; Coyle, N.E.; Nazario, S.; et al. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am. J. Respir. Crit. Care Med. 2003, 168, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Godava, M.; Kopriva, F.; Bohmova, J.; Vodicka, R.; Dusek, L.; Cvanova, M.; Muzik, J.; Markova, M.; Schneiderova, E.; Vrtel, R. Association of STAT6 and ADAM33 single nucleotide polymorphisms with asthma bronchiale and IgE level and its possible epigenetic background. Biomed. Pap. Med. Fac. Univ. Palacky. Olomouc Czech Repub. 2012, 156, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Townshend, J.; Hails, S.; McKean, M. Diagnosis of asthma in children. BMJ 2007, 335, 198–202. [Google Scholar] [CrossRef] [PubMed]
- Unal, D.; Gelincik, A.; Elitok, A.; Demir, S.; Olgac, M.; Coskun, R.; Kocaaga, M.; Colakoglu, B.; Buyukozturk, S. Impact of high serum Immunoglobulin E levels on the risk of atherosclerosis in humans. Asia Pac. Allergy 2017, 7, 74–81. [Google Scholar] [CrossRef]
- Zihlif, M.; Mahafza, T.; Obeidat, N.M.; Froukh, T.; Shaban, M.; Al-Akhras, F.M.; Zihlif, N.; Naffa, R. Frequency of genetic polymorphisms of ADAM33 and their association with allergic rhinitis among Jordanians. Gene 2013, 531, 462–466. [Google Scholar] [CrossRef]
- Vergara, C.I.; Acevedo, N.; Jimenez, S.; Martinez, B.; Mercado, D.; Gusmao, L.; Barnes, K.C.; Caraballo, L. A Six-SNP haplotype of ADAM33 is associated with asthma in a population of Cartagena, Colombia. Int. Arch. Allergy Immunol. 2010, 152, 32–40. [Google Scholar] [CrossRef]
- El-Falaki, M.M.; Wilson, M.M.; Ezzat, G.M.; Mokhtar, D.A.; El Baz, M.S.; Hamed, D.H. A disintegrin and metalloproteinase 33 (ADAM33) gene polymorphism association with asthma in Egyptian children. Egypt. J. Med Human Genet. 2013, 14, 55–62. [Google Scholar] [CrossRef][Green Version]
- Lee, J.H.; Park, H.S.; Park, S.W.; Jang, A.S.; Uh, S.T.; Rhim, T.; Park, C.S.; Hong, S.J.; Holgate, S.T.; Holloway, J.W.; et al. ADAM33 polymorphism: Association with bronchial hyper-responsiveness in Korean asthmatics. Clin. Exp. Allergy 2004, 34, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Su, D.; Zhang, X.; Sui, H.; Lu, F.; Jin, L.; Zhang, J. Association of ADAM33 gene polymorphisms with adult allergic asthma and rhinitis in a Chinese Han population. BMC Med. Genet. 2008, 9, 82. [Google Scholar] [CrossRef]
- Zhang, X.; Su, D.; Zhang, X.; Sui, H.; Jin, L.; Lü, F.; Zhang, J. Association of ADAM33 gene polymorphisms with adult concomitant allergic rhinitis and asthma in Chinese Han population. Mol. Biol. Rep. 2009, 36, 1505–1509. [Google Scholar] [CrossRef]
- Karaca, S.; Civelek, E.; Karaca, M. Allergy-specific Phenome-Wide Association Study for Immunogenes in Turkish Children. Sci. Rep. 2016, 6, 33152. [Google Scholar] [CrossRef] [PubMed]
- Werner, M.; Herbon, N.; Gohlke, H.; Altmuller, J.; Knapp, M.; Heinrich, J.; Wjst, M. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin. Exp. Allergy 2004, 34, 26–31. [Google Scholar] [CrossRef]
- Wiester, L.M.; Giachelli, C.M. Expression and function of the integrin alpha9beta1 in bovine aortic valve interstitial cells. J. Heart Valve Dis. 2003, 12, 605–616. [Google Scholar] [PubMed]
| Phenotype | Sample Size | % Female | % Male | Av. Age ± SD |
|---|---|---|---|---|
| Adult asthma | 151 | 0.71 | 0.29 | 44.73 ± 15.41 |
| Adult control | 110 | 0.47 | 0.53 | 38.74 ± 14.85 |
| Children asthma | 99 | 0.31 | 0.69 | 5.96 ± 4.63 |
| Children control | 49 | 0.38 | 0.62 | 7.53 ± 4.83 |
| SNP Name | Nucleotide Change | Total Frequency | Frequency in Control | Frequency in Asthma | HWE Exact p |
|---|---|---|---|---|---|
| T1 | A/G | 0.2 | 0.18 | 0.22 | 0.45 |
| T2 | G/A | 0.21 | 0.19 | 0.23 | 0.36 |
| T + 1 | G/A | 0.21 | 0.18 | 0.23 | 0.72 |
| V4 | C/G | 0.2 | 0.19 | 0.2 | 0.68 |
| S1 | G/A | 0.06 | 0.05 | 0.06 | 0.6 |
| S2 | C/G | 0.37 | 0.32 | 0.41 | 0.9 |
| Q-1 | G/A | 0.16 | 0.12 | 0.19 | 0.073 |
| SNP Name | Nucleotide Change | Total Frequency | Frequency in Control | Frequency in Asthma | HWE Exact p |
|---|---|---|---|---|---|
| T1 | A/G | 0.19 | 0.19 | 0.2 | 0.81 |
| T2 | G/A | 0.22 | 0.22 | 0.22 | 0.85 |
| T + 1 | G/A | 0.2 | 0.2 | 0.21 | 0.22 |
| V4 | C/G | 0.25 | 0.2 | 0.31 | 0.049 |
| S1 | G/A | 0.08 | 0.07 | 0.09 | 0.37 |
| S2 | C/G | 0.41 | 0.41 | 0.41 | 1.0 |
| Q-1 | G/A | 0.2 | 0.2 | 0.21 | 0.84 |
| Model | Genotype | Adult (n = 261) IgE Levels: IU/mL (n) | Genotype Comparison (p-Value) | Children (n = 148) IgE Levels: IU/mL (n) | Genotype Comparison (p-Value) | |
|---|---|---|---|---|---|---|
| T1 | Codominant | AA | 71.1 ± 3.6 (168) | AA vs. AG (0.0084) | 93.0 ± 9.1 (100) | AA vs. AG (0.5109) |
| AG | 93.1 ± 2.2 (81) | AA vs. GG (0.0235) | 51.0 ± 4.8 (41) | AA vs. GG (0.1383) | ||
| GG | 127.2 ± 7.8 (12) | AG vs. GG (0.6344) | 157.5 ± 2.1 (7) | AG vs. GG (0.3067) | ||
| Dominant | AA | (0.0047) | (0.0806) | |||
| AG + GG | ||||||
| Recessive | AA + AG | (0.1447) | (0.1028) | |||
| GG | ||||||
| T2 | Codominant | GG | 77.1 ± 6.6 (165) | GG vs. GA (0.0111) | 99.6 ± 1.9 (92) | GG vs. GA (0.5906) |
| GA | 93.3 ± 2.4 (83) | GG vs. AA (0.0064) | 49.3 ± 2.1 (49) | GG vs. AA (0.1497) | ||
| AA | 139.4 ± 2.8 (13) | GA vs. AA (0.3914) | 157.1 ± 1.3 (7) | GA vs. AA (0.2734) | ||
| Dominant | GG | (0.0038) | (0.0729) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.0467) | (0.0071) | |||
| AA | ||||||
| T + 1 | Codominant | GG | 70.6 ± 3.5 (164) | GG vs. GA (0.0044) | 98.5 ± 9.47 (98) | GG vs. GA (0.6622) |
| GA | 92.4 ± 2.62 (84) | GG vs. AA (0.0230) | 55.3 ± 3.9 (42) | GG vs. AA (0.1745) | ||
| AA | 117.4 ± 2.2 (13) | GA vs. AA (0.7278) | 139.2 ± 4.5 (8) | GA vs. AA (0.2734) | ||
| Dominant | GG | (0.0029) | (0.0291) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.1888) | (0.0093) | |||
| AA | ||||||
| V4 | Codominant | CC | 85.3 ± 6.4 (169) | CC vs. CG (0.6329) | 86.5 ± 4.1 (93) | CC vs. CG (0.5830) |
| CG | 93.7 ± 4.7 (80) | CC vs. GG (0.0001) | 49.1 ± 7.5 (48) | CC vs. GG (0.0144) | ||
| GG | 40.0 ± 1.2 (12) | CG vs. GG (0.0002) | 23.2 ± 5.3 (7) | CG vs. GG (0.0920) | ||
| Dominant | CC | (0.0768) | (0.0345) | |||
| CG + GG | ||||||
| Recessive | CC + CG | (0.0001) | (0.0001) | |||
| GG | ||||||
| S1 | Codominant | GG | 90.1 ± 4.6 (230) | GG vs. GA (0.0001) | 85.4 ± 3.2 (124) | GG vs. GA (0.1529) |
| GA | 26.5 ± 2.5 (28) | GG vs. AA (0.0001) | 87.2 ± 4.7 (24) | |||
| AA | 0.8 ± 0.05 (3) | GA vs. AA (0.2503) | None | |||
| Dominant | GG | (0.0001) | N/A | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.0001) | N/A | |||
| AA | ||||||
| S2 | Codominant | CC | 98.5 ± 5.5 (103) | CC vs. CG (0.2055) | 124.4 ± 7.5 (56) | CC vs. CG (0.9958) |
| CG | 81.6 ± 6.1 (122) | CC vs. GG (0.0404) | 60.7 ± 7.1 (69) | CC vs. GG (0.0923) | ||
| GG | 65.1 ± 1.8 (36) | CG vs. GG (0.2936) | 43.9 ± 2.9 (23) | CG vs. GG (0.0582) | ||
| Dominant | CC | (0.0963) | (0.2211) | |||
| CG + GG | ||||||
| Recessive | CC + CG | (0.1061) | (0.0152) | |||
| GG | ||||||
| Q-1 | Codominant | TT | 80.9 ± 3.1 (189) | TT vs. TC (0.0009) | 98.0 ± 8.7 (97) | TT vs. TC (0.5724) |
| TC | 45.5 ± 3.6 (62) | TT vs. CC (0.3344) | 67.8 ± 3.3 (36) | |||
| CC | 6.9 ± 3.8 (10) | TC vs. CC (0.5633) | 10.8 ± 1.8 (5) | |||
| Dominant | TT | (0.0013) | (0.0875) | |||
| CT + CC | ||||||
| Recessive | TT + CT | (0.6842) | (0.0001) | |||
| CC |
| Model | Genotype | Control (n = 110) IgE Levels: IU/mL (n) | Genotype Comparison (p-Value) | Asthma (n = 151) IgE Levels: IU/mL (n) | Genotype Comparison (p-Value) | |
|---|---|---|---|---|---|---|
| T1 | Codominant | AA | 14.9 ± 2.1 (75) | AA vs. AG (0.2664) | 116.4 ± 3.6 (93) | AA vs. AG (0.0367) |
| AG | 25.1 ± 4.8 (31) | AA vs. GG (0.0402) | 135.4 ± 4.7 (50) | AA vs. GG (0.0420) | ||
| GG | 3.4 ± 1.2 (4) | AG vs. GG (0.1963) | 189.1 ± 12.5 (8) | AG vs. GG (0.6116) | ||
| Dominant | AA | (0.0795) | (0.0240) | |||
| AG + GG | ||||||
| Recessive | AA + AG | (0.0188) | (0.0407) | |||
| GG | ||||||
| T2 | Codominant | GG | 15.3 ± 2.1 (73) | GG vs. GA (0.4522) | 126.3 ± 8.9 (92) | GG vs. GA (0.0353) |
| GA | 23.6 ± 4.6 (33) | GG vs. AA (0.0454) | 139.4 ± 4.0 (50) | GG vs. AA (0.0248) | ||
| AA | 3.4 ± 1.2 (4) | GA vs. AA (0.1607) | 199.9 ± 5.4 (9) | GA vs. AA (0.5096) | ||
| Dominant | GG | (0.1570) | (0.0175) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.0188) | (0.0150) | |||
| AA | ||||||
| T + 1 | Codominant | GG | 15.9 ± 2.2 (74) | GG vs. GA (0.7063) | 115.7 ± 2.1 (90) | GG vs. GA (0.0344) |
| GA | 22.5 ± 4.6 (32) | GG vs. AA (0.0597) | 135.5 ± 3.3 (52) | GG vs. AA (0.0779) | ||
| AA | 3.4 ± 1.2 (4) | GA vs. AA (0.1359) | 168.1 ± 1.4 (9) | GA vs. AA (0.7501) | ||
| Dominant | GG | (0.3069) | (0.0270) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.0188) | (0.0765) | |||
| AA | ||||||
| V4 | Codominant | CC | 17.2 ± 2.5 (75) | CC vs. CG (0.2281) | 139.7 ± 8.1 (94) | CC vs. CG (0.9058) |
| CG | 16.5 ± 3.8 (28) | CC vs. GG (0.0647) | 135.4 ± 6.8 (52) | CC vs. GG (0.0339) | ||
| GG | 21.3 ± 6.9 (7) | CG vs. GG (0.4108) | 66.2 ± 7.76 (5) | CG vs. GG (0.0650) | ||
| Dominant | CC | (0.1150) | (0.8174) | |||
| CG + GG | ||||||
| Recessive | CC + CG | (0.0494) | (0.0236) | |||
| GG | ||||||
| S1 | Codominant | GG | 17.9 ± 2.2 (99) | GG vs. GA (0.3436) | 144.8 ± 4.3 (131) | GG vs. GA (0.001) |
| GA | 12.0 ± 4.1 (11) | 72.8 ± 8.4 (17) | GG vs. AA (0.001) | |||
| AA | None | 0.80 ± 0.05 (3) | GA vs. AA (0.6143) | |||
| Dominant | GG | N/A | (0.0001) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.0001) | ||||
| AA | ||||||
| S2 | Codominant | CC | 16.2 ± 2.7 (52) | CC vs. CG (0.8768) | 182.5 ± 9.09 (51) | CC vs. CG (0.6011) |
| CG | 16.8 ± 3.0 (45) | CC vs. GG (0.7129) | 119.5 ± 4.36 (77) | CC vs. GG (0.7842) | ||
| GG | 23.9 ± 4.1 (13) | CG vs. GG (0.4698) | 88.5 ± 2.28 (23) | CG vs. GG (0.5157) | ||
| Dominant | CC | (0.5982) | (0.1281) | |||
| CG + GG | ||||||
| Recessive | CC + CG | (0.2437) | (0.3601) | |||
| GG | ||||||
| Q-1 | Codominant | TT | 17.3 ± 2.3 (84) | TT vs. TC (0.5607) | 131.8 ± 4.0 (105) | TT vs. TC (0.0224) |
| TC | 18.0 ± 4.6 (25) | TT vs. CC (0.2417) | 64.1 ± 3.3 (37) | TT vs. CC (0.8103) | ||
| CC | 3.7 ± 0.1 (3) | TC vs. CC (0.3069) | 8.4 ± 2.0 (7) | TC vs. CC (0.4156) | ||
| Dominant | TT | (0.3717) | (0.1123) | |||
| CT + CC | ||||||
| Recessive | TT + CT | (0.2721) | (0.1187) | |||
| CC |
| Model | Genotype | Control (n = 99) IgE Levels: IU/mL (n) | Genotype Comparison (p-Value) | Asthma (n = 49) IgE Levels: IU/mL (n) | Genotype Comparison (p-Value) | |
|---|---|---|---|---|---|---|
| T1 | Codominant | AA | 19.7 ± 2.7 (68) | AA vs. AG (0.5680) | 248.9 ± 2.5 (32) | AA vs. AG (0.5109) |
| AG | 22.7 ± 4.1 (27) | AA vs. GG (0.9403) | 105.5 ± 7.3 (14) | AA vs. GG (0.1383) | ||
| GG | 20.6 ± 6.2 (4) | AG vs. GG (0.8742) | 340.0 ± 2.3 (3) | AG vs. GG (0.3067) | ||
| Dominant | AA | (0.0179) | (0.2772) | |||
| AG + GG | ||||||
| Recessive | AA + AG | (0.4438) | (0.1447) | |||
| GG | ||||||
| T2 | Codominant | GG | 20.5 ± 2.9 (61) | GG vs. GA (0.8897) | 255.5 ± 5.0 (31) | GG vs. GA (0.5906) |
| GA | 26.6 ± 5.1 (34) | GG vs. AA (0.3014) | 101.6 ± 4.9 (15) | GG vs. AA (0.1497) | ||
| AA | 19.9 ± 6.5 (4) | GA vs. AA (0.3881) | 340.0 ± 2.3 (3) | GA vs. AA (0.2734) | ||
| Dominant | GG | (0.6909) | (0.3373) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.2947) | (0.1461) | |||
| AA | ||||||
| T + 1 | Codominant | GG | 19.9 ± 2.6 (67) | GG vs. GA (0.4440) | 268.4 ± 0.47 (31) | GG vs. GA (0.6622) |
| GA | 29.6 ± 6.2 (27) | GG vs. AA (0.3902) | 101.6 ± 4.9 (15) | GG vs. AA (0.1745) | ||
| AA | 18.83 ± 5.1 (5) | GA vs. AA (0.7032) | 340.0 ± 2.3 (3) | GA vs. AA (0.2734) | ||
| Dominant | GG | (0.3517) | (0.3975) | |||
| GA + AA | ||||||
| Recessive | GG + GA | (0.4544) | (0.1632) | |||
| AA | ||||||
| V4 | Codominant | CC | 25.6 ± 3.5 (62) | CC vs. CG (0.1723) | 208.4 ± 1.7 (31) | CC vs. CG (0.5830) |
| CG | 16.6 ± 3.1 (34) | CC vs. GG (0.0164) | 128.1 ± 5.6 (14) | CC vs. GG (0.0144) | ||
| GG | 22.6 ± 8.5 (3) | CG vs. GG (0.1809) | 23.7 ± 7.9 (4) | CG vs. GG (0.0920) | ||
| Dominant | CC | (0.0839) | (0.1709) | |||
| CG + GG | ||||||
| Recessive | CC + CG | (0.0430) | (0.0168) | |||
| GG | ||||||
| S1 | Codominant | GG | 23.5 ± 2.8 (85) | GG vs. GA (0.2337) | 220.5 ± 9.3 (39) | GG vs. GA (0.1529) |
| GA | 16.3 ± 4.7 (14) | 186.5 ± 3.9 (10) | ||||
| AA | None | None | ||||
| Dominant | GG | N/A | N/A | |||
| GA + AA | ||||||
| Recessive | GG + GA | N/A | N/A | |||
| AA | ||||||
| S2 | Codominant | CC | 23.6 ± 4.0 (35) | CC vs. CG (0.2847) | 292.4 ± 3.5 (21) | CC vs. CG (0.9958) |
| CG | 22.2 ± 4.2 (47) | CC vs. GG (0.3025) | 143.1 ± 9.2 (22) | CC vs. GG (0.0923) | ||
| GG | 20.9 ± 3.8 (17) | CG vs. GG (0.0630) | 109.1 ± 9.2 (6) | CG vs. GG (0.0582) | ||
| Dominant | CC | (0.7217) | (0.5259) | |||
| CG + GG | ||||||
| Recessive | CC + CG | (0.0893) | (0.0444) | |||
| GG | ||||||
| Q-1 | Codominant | TT | 25.1 ± 3.4 (64) | TT vs. TC (0.0339) | 239.4 ± 9.3 (33) | TT vs. TC (0.5724) |
| TC | 18.7 ± 3.6 (30) | TT vs. CC (0.0176) | 160.1 ± 1.4 (16) | |||
| CC | 10.8 ± 1.8 (5) | TC vs. CC (0.4316) | ||||
| Dominant | TT | (0.0127) | N/A | |||
| CT + CC | ||||||
| Recessive | TT + CT | (0.0829) | N/A | |||
| CC |
| ADAM33 Haplotype | Frequency | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T1 | T2 | T + 1 | V4 | S1 | S2 | Q-1 | Healthy Adults | Asthma Adults | Healthy Children | Asthma Children |
| A | G | G | C | C | G | G | 0.53 | 0.43 * | 0.49 | 0.42 # |
| G | A | A | C | C | C | G | 0.18 | 0.22 | 0.17 | 0.16 |
| A | G | G | G | C | G | G | 0.14 | 0.15 | 0.10 | 0.15 |
| A | G | G | C | C | C | A | 0.07 | 0.12 * | 0.09 | 0.09 |
| A | G | G | G | T | C | A | 0.04 | 0.05 | 0.03 | 0.04 |
| A | G | G | C | C | C | A | 0.02 | 0 | 0.01 | 0 |
| A | G | G | C | T | C | A | 0.02 | 0 | 0.02 | 0.03 |
| A | G | G | G | C | C | A | 0 | 0.02 | 0.05 | 0.04 |
| A | G | G | G | T | C | G | 0 | 0 | 0.01 | 0 |
| G | A | A | C | C | C | A | 0 | 0 | 0.02 | 0 |
| G | A | A | G | C | G | A | 0 | 0 | 0 | 0.04 # |
| A | G | G | G | T | G | G | 0 | 0 | 0 | 0.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zihlif, M.; Imraish, A.; Al-Rawashdeh, B.; Qteish, A.; Husami, R.; Husami, R.; Tahboub, F.; Jarrar, Y.; Lee, S.-J. The Association of IgE Levels with ADAM33 Genetic Polymorphisms among Asthmatic Patients. J. Pers. Med. 2021, 11, 329. https://doi.org/10.3390/jpm11050329
Zihlif M, Imraish A, Al-Rawashdeh B, Qteish A, Husami R, Husami R, Tahboub F, Jarrar Y, Lee S-J. The Association of IgE Levels with ADAM33 Genetic Polymorphisms among Asthmatic Patients. Journal of Personalized Medicine. 2021; 11(5):329. https://doi.org/10.3390/jpm11050329
Chicago/Turabian StyleZihlif, Malek, Amer Imraish, Baeth Al-Rawashdeh, Aya Qteish, Raihan Husami, Rawand Husami, Farah Tahboub, Yazun Jarrar, and Su-Jun Lee. 2021. "The Association of IgE Levels with ADAM33 Genetic Polymorphisms among Asthmatic Patients" Journal of Personalized Medicine 11, no. 5: 329. https://doi.org/10.3390/jpm11050329
APA StyleZihlif, M., Imraish, A., Al-Rawashdeh, B., Qteish, A., Husami, R., Husami, R., Tahboub, F., Jarrar, Y., & Lee, S.-J. (2021). The Association of IgE Levels with ADAM33 Genetic Polymorphisms among Asthmatic Patients. Journal of Personalized Medicine, 11(5), 329. https://doi.org/10.3390/jpm11050329







