Identification of Rare Mutations of Two Presynaptic Cytomatrix Genes BSN and PCLO in Schizophrenia and Bipolar Disorder
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Next Generation Sequencing Analysis
2.3. Sanger Sequencing
2.4. Bioinformatics Analysis
3. Results
3.1. Identification of the R1087Q Mutation of BSN
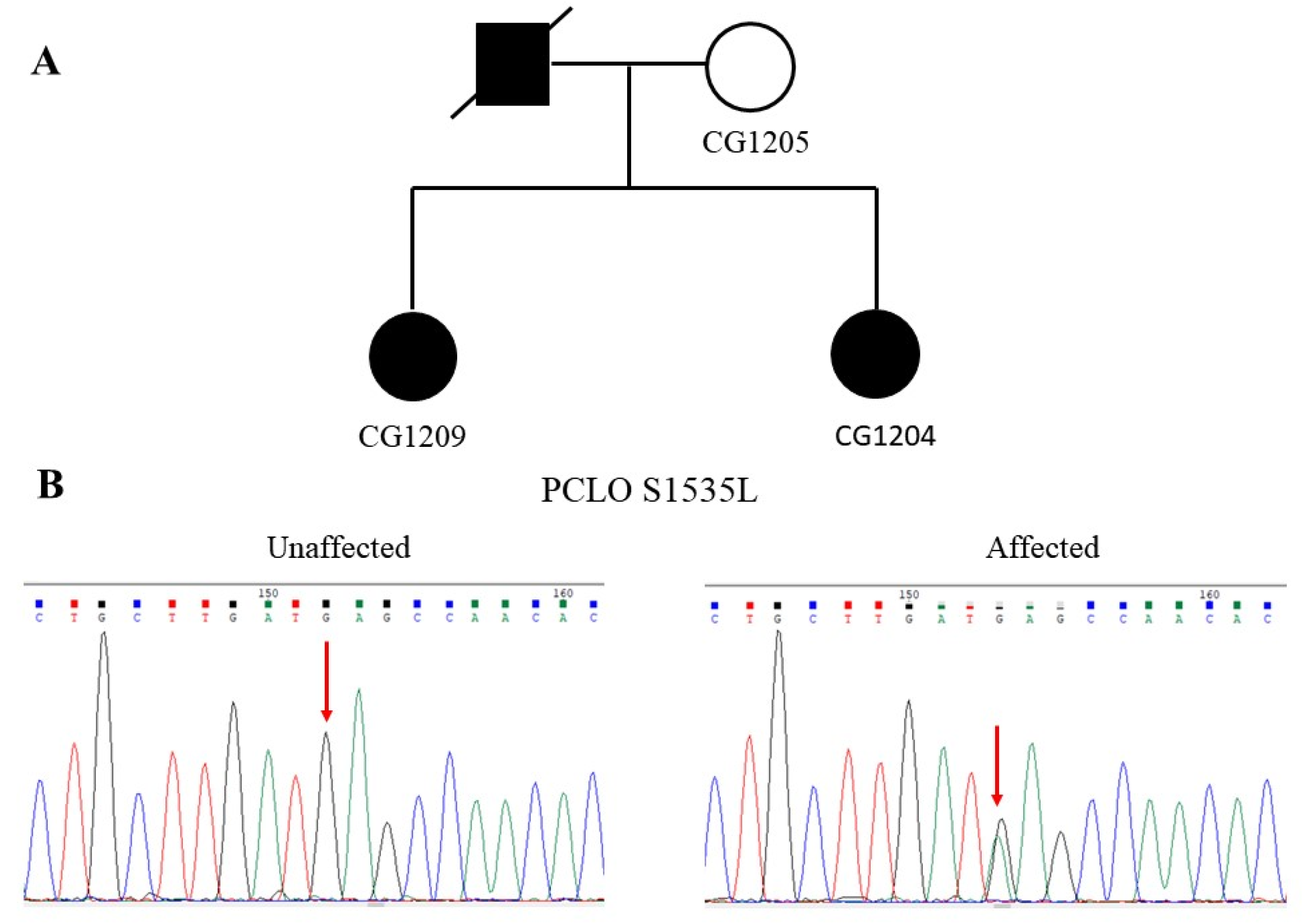
3.2. Identification of the S1535L Mutation of PCLO
3.3. Identification of the H5142R Mutation of PCLO
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Trifu, S.C.; Kohn, B.; Vlasie, A.; Patrichi, B.E. Genetics of schizophrenia (Review). Exp. Ther. Med. 2020, 20, 3462–3468. [Google Scholar] [CrossRef]
- Gordovez, F.J.A.; McMahon, F.J. The genetics of bipolar disorder. Mol. Psychiatry 2020, 25, 544–559. [Google Scholar] [CrossRef]
- O’Donovan, M.C.; Craddock, N.J.; Owen, M.J. Genetics of psychosis; insights from views across the genome. Hum. Genet. 2009, 126, 3–12. [Google Scholar] [CrossRef]
- Stahl, E.A.; Breen, G.; Forstner, A.J.; McQuillin, A.; Ripke, S.; Trubetskoy, V.; Mattheisen, M.; Wang, Y.; Coleman, J.R.I.; Gaspar, H.A.; et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat. Genet. 2019, 51, 793–803. [Google Scholar] [CrossRef]
- Cross-Disorder Group of the Psychiatric Genomics. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 2013, 381, 1371–1379. [Google Scholar] [CrossRef]
- Schoch, S.; Gundelfinger, E.D. Molecular organization of the presynaptic active zone. Cell Tissue Res. 2006, 326, 379–391. [Google Scholar] [CrossRef]
- Dresbach, T.; Qualmann, B.; Kessels, M.M.; Garner, C.C.; Gundelfinger, E.D. The presynaptic cytomatrix of brain synapses. Cell Mol. Life Sci. 2001, 58, 94–116. [Google Scholar] [CrossRef]
- Gundelfinger, E.D.; Fejtova, A. Molecular organization and plasticity of the cytomatrix at the active zone. Curr. Opin. Neurobiol. 2012, 22, 423–430. [Google Scholar] [CrossRef]
- Fejtova, A.; Gundelfinger, E.D. Molecular organization and assembly of the presynaptic active zone of neurotransmitter release. Results Probl. Cell Differ. 2006, 43, 49–68. [Google Scholar] [CrossRef]
- Hamada, S.; Ohtsuka, T. CAST: Its molecular structure and phosphorylation-dependent regulation of presynaptic plasticity. Neurosci. Res. 2018, 127, 25–32. [Google Scholar] [CrossRef]
- Torres, V.I.; Inestrosa, N.C. Vertebrate presynaptic active zone assembly: A role accomplished by diverse molecular and cellular mechanisms. Mol. Neurobiol. 2018, 55, 4513–4528. [Google Scholar] [CrossRef]
- Gundelfinger, E.D.; Reissner, C.; Garner, C.C. Role of Bassoon and Piccolo in assembly and molecular organization of the active zone. Front. Synaptic Neurosci. 2015, 7, 19. [Google Scholar] [CrossRef] [PubMed]
- Kittel, R.J.; Heckmann, M. Synaptic vesicle proteins and active zone plasticity. Front. Synaptic Neurosci. 2016, 8, 8. [Google Scholar] [CrossRef]
- Hashida, H.; Goto, J.; Zhao, N.; Takahashi, N.; Hirai, M.; Kanazawa, I.; Sakaki, Y. Cloning and mapping of ZNF231, a novel brain-specific gene encoding neuronal double zinc finger protein whose expression is enhanced in a neurodegenerative disorder, multiple system atrophy (MSA). Genomics 1998, 54, 50–58. [Google Scholar] [CrossRef]
- Tom Dieck, S.; Sanmarti-Vila, L.; Langnaese, K.; Richter, K.; Kindler, S.; Soyke, A.; Wex, H.; Smalla, K.H.; Kampf, U.; Franzer, J.T.; et al. Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J. Cell Biol. 1998, 142, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Winter, C.; tom Dieck, S.; Boeckers, T.M.; Bockmann, J.; Kampf, U.; Sanmarti-Vila, L.; Langnaese, K.; Altrock, W.; Stumm, M.; Soyke, A.; et al. The presynaptic cytomatrix protein Bassoon: Sequence and chromosomal localization of the human BSN gene. Genomics 1999, 57, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Fenster, S.D.; Garner, C.C. Gene structure and genetic localization of the PCLO gene encoding the presynaptic active zone protein Piccolo. Int. J. Dev. Neurosci. 2002, 20, 161–171. [Google Scholar] [CrossRef]
- Ackermann, F.; Schink, K.O.; Bruns, C.; Izsvak, Z.; Hamra, F.K.; Rosenmund, C.; Garner, C.C. Critical role for Piccolo in synaptic vesicle retrieval. eLife 2019, 8, e46629. [Google Scholar] [CrossRef]
- Ivanova, D.; Dirks, A.; Fejtova, A. Bassoon and piccolo regulate ubiquitination and link presynaptic molecular dynamics with activity-regulated gene expression. J. Physiol. 2016, 594, 5441–5448. [Google Scholar] [CrossRef]
- Kononenko, N.; Pechstein, A.; Haucke, V. Synaptic requiem: A duet for Piccolo and Bassoon. EMBO J. 2013, 32, 920–922. [Google Scholar] [CrossRef][Green Version]
- Yabe, I.; Yaguchi, H.; Kato, Y.; Miki, Y.; Takahashi, H.; Tanikawa, S.; Shirai, S.; Takahashi, I.; Kimura, M.; Hama, Y.; et al. Mutations in bassoon in individuals with familial and sporadic progressive supranuclear palsy-like syndrome. Sci. Rep. 2018, 8, 819. [Google Scholar] [CrossRef]
- Eto, K.; Sakai, N.; Shimada, S.; Shioda, M.; Ishigaki, K.; Hamada, Y.; Shinpo, M.; Azuma, J.; Tominaga, K.; Shimojima, K.; et al. Microdeletions of 3p21.31 characterized by developmental delay, distinctive features, elevated serum creatine kinase levels, and white matter involvement. Am. J. Med. Genet. A 2013, 161A, 3049–3056. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, P.F.; de Geus, E.J.; Willemsen, G.; James, M.R.; Smit, J.H.; Zandbelt, T.; Arolt, V.; Baune, B.T.; Blackwood, D.; Cichon, S.; et al. Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol. Psychiatry 2009, 14, 359–375. [Google Scholar] [CrossRef] [PubMed]
- Bochdanovits, Z.; Verhage, M.; Smit, A.B.; de Geus, E.J.; Posthuma, D.; Boomsma, D.I.; Penninx, B.W.; Hoogendijk, W.J.; Heutink, P. Joint reanalysis of 29 correlated SNPs supports the role of PCLO/Piccolo as a causal risk factor for major depressive disorder. Mol. Psychiatry 2009, 14, 650–652. [Google Scholar] [CrossRef]
- Mbarek, H.; Milaneschi, Y.; Hottenga, J.J.; Ligthart, L.; de Geus, E.J.C.; Ehli, E.A.; Willemsen, G.; Davies, G.E.; Smit, J.H.; Boomsma, D.I.; et al. Genome-wide significance for PCLO as a gene for najor depressive disorder. Twin Res. Hum. Genet. 2017, 20, 267–270. [Google Scholar] [CrossRef]
- Minelli, A.; Scassellati, C.; Cloninger, C.R.; Tessari, E.; Bortolomasi, M.; Bonvicini, C.; Giacopuzzi, M.; Frisoni, G.B.; Gennarelli, M. PCLO gene: Its role in vulnerability to major depressive disorder. J. Affect. Disord 2012, 139, 250–255. [Google Scholar] [CrossRef]
- Choi, K.H.; Higgs, B.W.; Wendland, J.R.; Song, J.; McMahon, F.J.; Webster, M.J. Gene expression and genetic variation data implicate PCLO in bipolar disorder. Biol. Psychiatry 2011, 69, 353–359. [Google Scholar] [CrossRef]
- Uno, K.; Nishizawa, D.; Seo, S.; Takayama, K.; Matsumura, S.; Sakai, N.; Ohi, K.; Nabeshima, T.; Hashimoto, R.; Ozaki, N.; et al. The piccolo intronic single nucleotidepolymorphism rs13438494 regulates dopamine and serotonin uptake and shows associations with dependence-like behavior in genomic association study. Curr. Mol. Med. 2015, 15, 265–274. [Google Scholar] [CrossRef]
- Ahmed, M.Y.; Chioza, B.A.; Rajab, A.; Schmitz-Abe, K.; Al-Khayat, A.; Al-Turki, S.; Baple, E.L.; Patton, M.A.; Al-Memar, A.Y.; Hurles, M.E.; et al. Loss of PCLO function underlies pontocerebellar hypoplasia type III. Neurology 2015, 84, 1745–1750. [Google Scholar] [CrossRef]
- Siddique, A.; Willoughby, J.; Study, D.D.D.; McNeill, A. A 7q21.11 microdeletion presenting with apparent intellectual disability without epilepsy. Am. J. Med. Genet. A 2017, 173, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Weidenhofer, J.; Bowden, N.A.; Scott, R.J.; Tooney, P.A. Altered gene expression in the amygdala in schizophrenia: Up-regulation of genes located in the cytomatrix active zone. Mol. Cell Neurosci. 2006, 31, 243–250. [Google Scholar] [CrossRef]
- Coyle, J.T. NMDA receptor and schizophrenia: A brief history. Schizophr. Bull. 2012, 38, 920–926. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Fukushima, T.; Shimizu, E.; Komatsu, N.; Watanabe, H.; Shinoda, N.; Nakazato, M.; Kumakiri, C.; Okada, S.; Hasegawa, H.; et al. Decreased serum levels of D-serine in patients with schizophrenia: Evidence in support of the N-methyl-D-aspartate receptor hypofunction hypothesis of schizophrenia. Arch. Gen. Psychiatry 2003, 60, 572–576. [Google Scholar] [CrossRef]
- Cho, S.E.; Na, K.S.; Cho, S.J.; Kang, S.G. Low d-serine levels in schizophrenia: A systematic review and meta-analysis. Neurosci. Lett. 2016, 634, 42–51. [Google Scholar] [CrossRef] [PubMed]
- El-Tallawy, H.N.; Saleem, T.H.; El-Ebidi, A.M.; Hassan, M.H.; Gabra, R.H.; Farghaly, W.M.; Abo El-Maali, N.; Sherkawy, H.S. Clinical and biochemical study of d-serine metabolism among schizophrenia patients. Neuropsychiatr. Dis. Treat. 2017, 13, 1057–1063. [Google Scholar] [CrossRef] [PubMed]
- Bendikov, I.; Nadri, C.; Amar, S.; Panizzutti, R.; De Miranda, J.; Wolosker, H.; Agam, G. A CSF and postmortem brain study of D-serine metabolic parameters in schizophrenia. Schizophr. Res. 2007, 90, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Madeira, C.; Freitas, M.E.; Vargas-Lopes, C.; Wolosker, H.; Panizzutti, R. Increased brain D-amino acid oxidase (DAAO) activity in schizophrenia. Schizophr. Res. 2008, 101, 76–83. [Google Scholar] [CrossRef]
- Burnet, P.W.; Eastwood, S.L.; Bristow, G.C.; Godlewska, B.R.; Sikka, P.; Walker, M.; Harrison, P.J. D-amino acid oxidase activity and expression are increased in schizophrenia. Mol. Psychiatry 2008, 13, 658–660. [Google Scholar] [CrossRef]
- Popiolek, M.; Ross, J.F.; Charych, E.; Chanda, P.; Gundelfinger, E.D.; Moss, S.J.; Brandon, N.J.; Pausch, M.H. D-amino acid oxidase activity is inhibited by an interaction with bassoon protein at the presynaptic active zone. J. Biol. Chem. 2011, 286, 28867–28875. [Google Scholar] [CrossRef]
- Nitta, A.; Izuo, N.; Hamatani, K.; Inagaki, R.; Kusui, Y.; Fu, K.; Asano, T.; Torii, Y.; Habuchi, C.; Sekiguchi, H.; et al. Schizophrenia-like behavioral impairments in mice with suppressed expression of piccolo in the medial prefrontal cortex. J. Pers. Med. 2021, 11, 607. [Google Scholar] [CrossRef] [PubMed]


| Mutation | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Ta (°C) | Size (bp) |
|---|---|---|---|---|
| BSN R1087Q | TGCGGGAGGAAGAGGAGCTGCTT | GGAGCGGTGTAGCTCCTCCATCT | 60 | 227 |
| PCLO S1535L | CTTCTGGCTCTCAGTACTGC | CCTTCCAGCAAGGACCATAA | 60 | 318 |
| PCLO H5142R | CCCACTCTTATGTTTGCCTCTC | GACCTTGATTGGTGAAGCCTG | 60 | 214 |
| Gene Name and dbSNP Number | Location | Allele Frequency | Functional Prediction | |||||
|---|---|---|---|---|---|---|---|---|
| Taiwan Biobank | ALFA | TOPMED | 1000 Genomes | PROVEAN (Score) | PolyPhen-2 (Score) | SIFT (Score) | ||
| BSN rs200987266 | NC_000003.11:g.49690249G > A NM_003458.3:c.3260G > A NM_003458.3:p.Arg1087Gln | 0 | 0 | 0.000008 | 0.0002 | Deleterious (−3.16) | Probably damaging (1.000) | Damaging (0.000) |
| PCLO rs757482765 | NC_000007.13:g.82585665G > A NM_033026.5:c.4604C > T NM_033026.5:p.Ser1535Leu | 0 | 0 | 0.000032 | 0.000056 | Deleterious (−2.67) | Probably damaging (0.999) | Damaging (0.002) |
| PCLO rs141283244 | NC_000007.13:g.82387895T > C NM_033026.5:c.15425A > G NM_033026.5:p.His5142Arg | 0.00824 | 0 | 0.000191 | 0.000229 | Deleterious (−3.27) | Probably damaging (0.996) | Damaging (0.002) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, C.-H.; Huang, Y.-S.; Liao, D.-L.; Huang, C.-Y.; Lin, C.-H.; Fang, T.-H. Identification of Rare Mutations of Two Presynaptic Cytomatrix Genes BSN and PCLO in Schizophrenia and Bipolar Disorder. J. Pers. Med. 2021, 11, 1057. https://doi.org/10.3390/jpm11111057
Chen C-H, Huang Y-S, Liao D-L, Huang C-Y, Lin C-H, Fang T-H. Identification of Rare Mutations of Two Presynaptic Cytomatrix Genes BSN and PCLO in Schizophrenia and Bipolar Disorder. Journal of Personalized Medicine. 2021; 11(11):1057. https://doi.org/10.3390/jpm11111057
Chicago/Turabian StyleChen, Chia-Hsiang, Yu-Shu Huang, Ding-Lieh Liao, Cheng-Yi Huang, Chia-Heng Lin, and Ting-Hsuan Fang. 2021. "Identification of Rare Mutations of Two Presynaptic Cytomatrix Genes BSN and PCLO in Schizophrenia and Bipolar Disorder" Journal of Personalized Medicine 11, no. 11: 1057. https://doi.org/10.3390/jpm11111057
APA StyleChen, C.-H., Huang, Y.-S., Liao, D.-L., Huang, C.-Y., Lin, C.-H., & Fang, T.-H. (2021). Identification of Rare Mutations of Two Presynaptic Cytomatrix Genes BSN and PCLO in Schizophrenia and Bipolar Disorder. Journal of Personalized Medicine, 11(11), 1057. https://doi.org/10.3390/jpm11111057








