Analysis of miRNA Expression in Patients with Rheumatoid Arthritis during Olokizumab Treatment
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Procedures
2.3. RNA Extraction and Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Assay
2.4. Prediction of miRNA Targets
2.5. Statistical Analysis
3. Results
3.1. Clinical Features and General Characteristics of the Patients
3.2. MiRNA Expression Is Associated with Olokizumab TREATMENT
3.3. MiRNA Expression Is Associated with Some Patient Characteristics
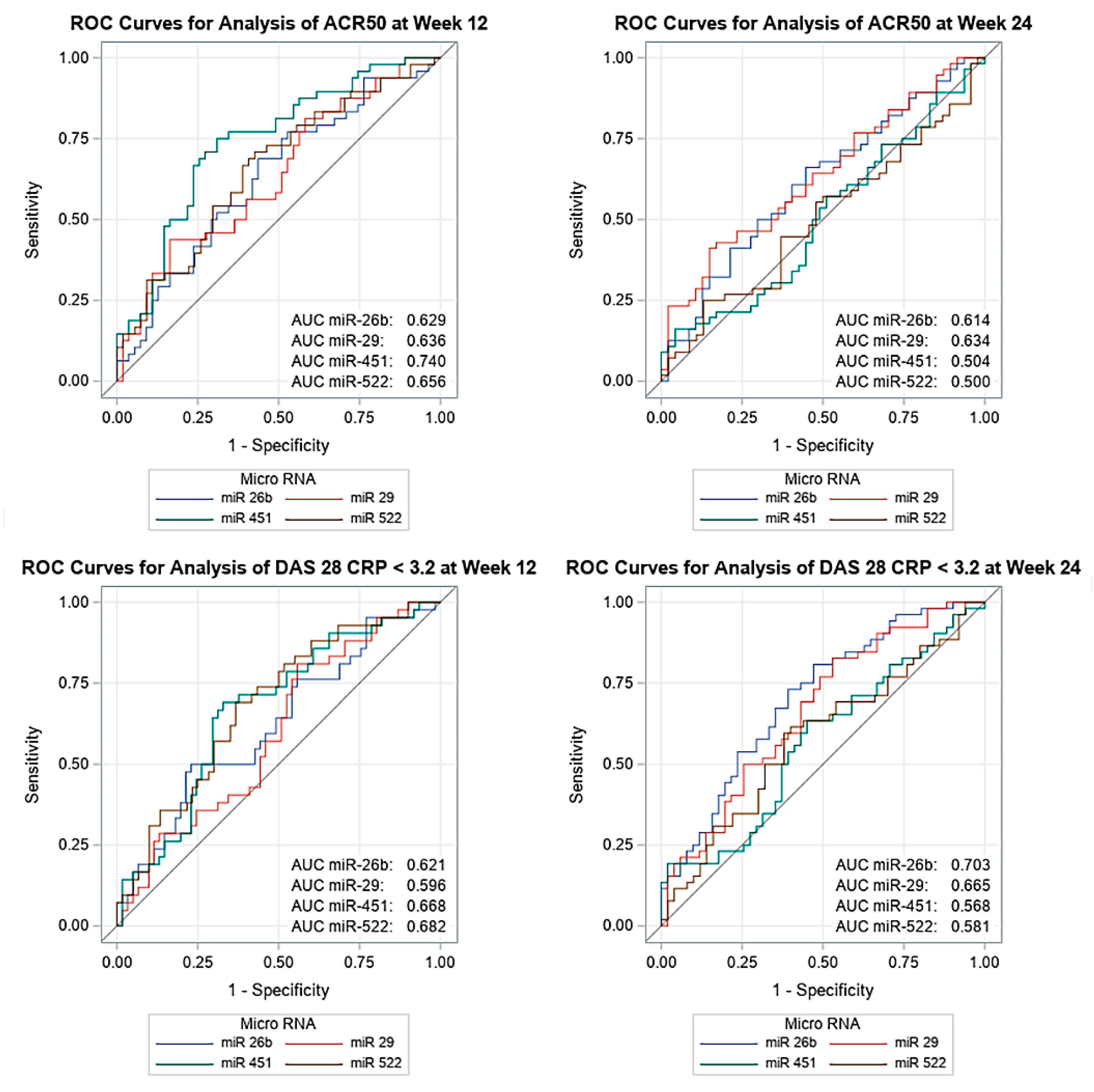
3.4. MiRNA Expression Is Associated with Olokizumab Therapy Efficiency
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smolen, J.S.; Aletaha, D.; McInnes, I.B. Rheumatoid arthritis. Lancet 2016, 388, 2023–2038. [Google Scholar] [CrossRef]
- Wang, J.; Yan, S.; Yang, J.; Lu, H.; Xu, D.; Wang, Z. Non-coding RNAs in Rheumatoid Arthritis: From Bench to Bedside. Front. Immunol. 2020, 10, 3129. [Google Scholar] [CrossRef]
- Gaffo, A.; Saag, K.G.; Curtis, J.R. Treatment of rheumatoid arthritis. Am. J. Heal. Pharm. 2006, 63, 2451–2465. [Google Scholar] [CrossRef]
- Kim, G.W.; Lee, N.R.; Pi, R.H.; Lim, Y.S.; Lee, Y.M.; Lee, J.M.; Jeong, H.S.; Chung, S.H. IL-6 inhibitors for treatment of rheumatoid arthritis: Past, present, and future. Arch. Pharmacal Res. 2015, 38, 575–584. [Google Scholar] [CrossRef] [PubMed]
- Srirangan, S.; Choy, E.H. The role of Interleukin 6 in the pathophysiology of rheumatoid arthritis. Ther. Adv. Musculoskelet. Dis. 2010, 2, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Genovese, M.C.; Fleischmann, R.; Furst, D.; Janssen, N.; Carter, J.; Dasgupta, B.; Bryson, J.; Duncan, B.; Zhu, W.; Pitzalis, C.; et al. Efficacy and safety of olokizumab in patients with rheumatoid arthritis with an inadequate response to TNF inhibitor therapy: Outcomes of a randomised Phase IIb study. Ann. Rheum. Dis. 2014, 73, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Tavasolian, F.; Abdollahi, E.; Rezaei, R.; Momtazi-Borojeni, A.A.; Henrotin, Y.; Sahebkar, A. Altered Expression of MicroRNAs in Rheumatoid Arthritis. J. Cell. Biochem. 2017, 119, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Nemtsova, M.; Zaletaev, D.V.; Bure, I.V.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Alekseeva, E.A.; Beloukhova, M.I.; Deviatkin, A.A.; Lukashev, A.N.; Zamyatnin, A.A.J. Epigenetic Changes in the Pathogenesis of Rheumatoid Arthritis. Front. Genet. 2019, 10, 570. [Google Scholar] [CrossRef]
- Baulina, N.; Kulakova, O.G.; Favorova, O.O. MicroRNAs: The Role in Autoimmune Inflammation. Acta Nat. 2016, 8, 21–33. [Google Scholar] [CrossRef]
- Tokar, T.; Pastrello, C.; Rossos, A.E.M.; Abovsky, M.; Hauschild, A.-C.; Tsay, M.; Lu, R.; Jurisica, I. mirDIP 4.1—Integrative database of human microRNA target predictions. Nucleic Acids Res. 2017, 46, D360–D370. [Google Scholar] [CrossRef]
- Brown, K.R.; Otasek, D.; Ali, M.; McGuffin, M.J.; Xie, W.; Devani, B.; Van Toch, I.L.; Jurisica, I. NAViGaTOR: Network Analysis, Visualization and Graphing Toronto. Bioinformatics 2009, 25, 3327–3329. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, M.J.; Jamshidi, A.; Chopra, A.; Aslani, S.; Akhlaghi, M.; Mahmoudi, M. Implications of the noncoding RNAs in rheumatoid arthritis pathogenesis. J. Cell. Physiol. 2018, 234, 335–347. [Google Scholar] [CrossRef]
- Li, X.; Tian, F.; Wang, F. Rheumatoid Arthritis-Associated MicroRNA-155 Targets SOCS1 and Upregulates TNF-α and IL-1β in PBMCs. Int. J. Mol. Sci. 2013, 14, 23910–23921. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Yu, P.; Liu, Y.; Wang, S.; Li, R.; Shi, J.; Zhang, X.; Li, Y.; Sun, X.; Zhou, B.; et al. Upregulated MicroRNA-155 Expression in Peripheral Blood Mononuclear Cells and Fibroblast-Like Synoviocytes in Rheumatoid Arthritis. Clin. Dev. Immunol. 2013, 2013, 296139. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Si, X.; Sun, J.; Yue, L.; Wang, J.; Yu, Z. miR-522 Modulated the Expression of Proinflammatory Cytokines and Matrix Metalloproteinases Partly via Targeting Suppressor of Cytokine Signaling 3 in Rheumatoid Arthritis Synovial Fibroblasts. DNA Cell Biol. 2018, 37, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.-J.; Li, X.-W.; Lu, J.-L.; Long, Z.-X.; Liang, J.-Q.; Wei, S.-B.; Lu, C.-X.; Lu, W.-Z. MiR-20a regulates fibroblast-like synoviocyte proliferation and apoptosis in rheumatoid arthritis. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 3886–3893. [Google Scholar] [PubMed]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef]
- Wang, Z.-C.; Lü, H.; Zhou, Q.; Yu, S.-M.; Mao, Y.-L.; Zhang, H.-J.; Zhang, P.-C.; Yan, W.-J. MiR-451 inhibits synovial fibroblasts proliferation and inflammatory cytokines secretion in rheumatoid arthritis through mediating p38MAPK signaling pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 14562–14567. [Google Scholar]
- Li, S.; Jin, Z.; Lu, X. MicroRNA-192 suppresses cell proliferation and induces apoptosis in human rheumatoid arthritis fibroblast-like synoviocytes by downregulating caveolin 1. Mol. Cell. Biochem. 2017, 432, 123–130. [Google Scholar] [CrossRef]
- Migita, K.; Iwanaga, N.; Izumi, Y.; Kawahara, C.; Kumagai, K.; Nakamura, T.; Koga, T.; Kawakami, A. TNF-α-induced miR-155 regulates IL-6 signaling in rheumatoid synovial fibroblasts. BMC Res. Notes 2017, 10, 1–6. [Google Scholar] [CrossRef]
- Abdul-Maksoud, R.S.; Sediq, A.M.; Kattaia, A.; Elsayed, W.; Ezzeldin, N.; Galil, S.A.; Ibrahem, R.; Kattaia, A. Serum miR-210 and miR-155 expression levels as novel biomarkers for rheumatoid arthritis diagnosis. Br. J. Biomed. Sci. 2017, 74, 209–213. [Google Scholar] [CrossRef]
- Stanczyk, J.; Pedrioli, D.M.L.; Brentano, F.; Sanchez-Pernaute, O.; Kolling, C.; Gay, R.E.; Detmar, M.; Gay, S.; Kyburz, D. Altered expression of MicroRNA in synovial fibroblasts and synovial tissue in rheumatoid arthritis. Arthritis Rheum. 2008, 58, 1001–1009. [Google Scholar] [CrossRef]
- Kurowska-Stolarska, M.; Alivernini, S.; Ballantine, L.E.; Asquith, D.L.; Millar, N.L.; Gilchrist, D.S.; Reilly, J.; Ierna, M.; Fraser, A.R.; Stolarski, B.; et al. MicroRNA-155 as a proinflammatory regulator in clinical and experimental arthritis. Proc. Natl. Acad. Sci. USA 2011, 108, 11193–11198. [Google Scholar] [CrossRef]
- Singh, A.; Patro, P.S.; Aggarwal, A. MicroRNA-132, miR-146a, and miR-155 as potential biomarkers of methotrexate response in patients with rheumatoid arthritis. Clin. Rheumatol. 2018, 38, 877–884. [Google Scholar] [CrossRef]
- Castro-Villegas, C.; Pérez-Sánchez, C.; Escudero, A.; Filipescu, I.; Verdu, M.; Ruiz-Limón, P.; Aguirre, M.A.; Jiménez-Gomez, Y.; Font, P.; Rodriguez-Ariza, A.; et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFα. Arthritis Res. Ther. 2015, 17, 1–15. [Google Scholar] [CrossRef]
- Filková, M.; Aradi, B.; Senolt, L.; Ospelt, C.; Vettori, S.; Mann, H.; Filer, A.; Raza, K.; Buckley, C.D.; Snow, M.; et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann. Rheum. Dis. 2014, 73, 1898–1904. [Google Scholar] [CrossRef]
- Dunaeva, M.; Blom, J.; Thurlings, R.; Pruijn, G.J. Circulating serum miR-223-3p and miR-16-5p as possible biomarkers of early rheumatoid arthritis. Clin. Exp. Immunol. 2018, 193, 376–385. [Google Scholar] [CrossRef]
- Ephilippe, L.; Alsaleh, G.; Pichot, A.; Ostermann, E.; Zuber, G.; Frisch, B.; Sibilia, J.; Pfeffer, S.; Bahram, S.; Wachsmann, D.; et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 2012, 72, 1071–1079. [Google Scholar] [CrossRef]
- Li, X.-F.; Shen, W.-W.; Sun, Y.-Y.; Li, W.-X.; Sun, Z.-H.; Liu, Y.; Zhang, L.; Huang, C.; Meng, X.; Li, J. MicroRNA-20a negatively regulates expression of NLRP3-inflammasome by targeting TXNIP in adjuvant-induced arthritis fibroblast-like synoviocytes. Jt. Bone Spine 2016, 83, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Gaur, N.; Karouzakis, E.; Glück, S.; Bagdonas, E.; Jüngel, A.; Michel, B.A.; Gay, R.E.; Gay, S.; Frank-Bertoncelj, M.; Neidhart, M. MicroRNAs interfere with DNA methylation in rheumatoid arthritis synovial fibroblasts. RMD Open 2016, 2, e000299. [Google Scholar] [CrossRef][Green Version]
- Liu, Y.; Han, Y.; Qu, H.; Fang, J.; Ye, M.; Yin, W. Correlation of microRNA expression profile with clinical response to tumor necrosis factor inhibitor in treating rheumatoid arthritis patients: A prospective cohort study. J. Clin. Lab. Anal. 2019, 33, e22953. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, F.; Guo, J. miR-137 decreases proliferation, migration and invasion in rheumatoid arthritis fibroblast-like synoviocytes. Mol. Med. Rep. 2017, 17, 3312–3317. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Yan, P.; Chen, Y.; Chen, Y.; Yang, J.; Xu, G.; Mao, H.; Qiu, Y. MicroRNA-26b inhibits cell proliferation and cytokine secretion in human RASF cells via the Wnt/GSK-3β/β-catenin pathway. Diagn. Pathol. 2015, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zou, Q.; Song, H.; Song, F.; Wang, L.; Zhang, G.; Shen, X. A study of miRNAs targets prediction and experimental validation. Protein Cell 2010, 1, 979–986. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, A.; Xiang, J.; Lv, Y.; Zhang, X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol. Rep. 2016, 36, 1385–1392. [Google Scholar] [CrossRef]
- Martínez-Hernández, R.; De La Fuente, H.; Lamana, A.; Sampedro-Núñez, M.; Ramos-Levi, A.; Serrano-Somavilla, A.; García-Vicuña, R.; Ortiz, A.M.; Daudén, E.; Llamas-Velasco, M.; et al. Utility of circulating serum miRNA profiles to evaluate the potential risk and severity of immune-mediated inflammatory disorders. J. Autoimmun. 2020, 111, 102472. [Google Scholar] [CrossRef]
- Anaparti, V.; Smolik, I.; Meng, X.; Spicer, V.; Mookherjee, N.; El-Gabalawy, H. Whole blood microRNA expression pattern differentiates patients with rheumatoid arthritis, their seropositive first-degree relatives, and healthy unrelated control subjects. Arthritis Res. 2017, 19, 1–11. [Google Scholar] [CrossRef]
- Mallinson, D.J.; Dunbar, D.R.; Ridha, S.; Sutton, E.R.; De La Rosa, O.; Dalemans, W.; Lombardo, E. Identification of Potential Plasma microRNA Stratification Biomarkers for Response to Allogeneic Adipose-Derived Mesenchymal Stem Cells in Rheumatoid Arthritis. Stem Cells Transl. Med. 2017, 6, 1202–1206. [Google Scholar] [CrossRef]
- Jinnin, M. microRNA in autoimmune disorders. Jpn. J. Clin. Immunol. 2011, 34, 439–446. [Google Scholar] [CrossRef][Green Version]
- Churov, A.V.; Oleinik, E.K.; Knip, M. MicroRNAs in rheumatoid arthritis: Altered expression and diagnostic potential. Autoimmun. Rev. 2015, 14, 1029–1037. [Google Scholar] [CrossRef]
- Murata, K.; Yoshitomi, H.; Furu, M.; Ishikawa, M.; Shibuya, H.; Ito, H.; Matsuda, S. MicroRNA-451 Down-Regulates Neutrophil Chemotaxis via p38 MAPK. Arthritis Rheumatol. 2014, 66, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Shaw, S.; Bourne, T.; Meier, C.; Carrington, B.; Gelinas, R.; Henry, A.; Popplewell, A.; Adams, R.; Baker, T.; Rapecki, S.; et al. Discovery and characterization of olokizumab. mAbs 2014, 6, 773–781. [Google Scholar] [CrossRef] [PubMed]

| miRNA | Sequence |
|---|---|
| miR-29 | 5′-GACTGATTTCTTTTGGTGTTCAAAAA-3′ |
| miR-16 | 5′-TAGCAGCACGTAAATATTGGCA-3′ |
| miR-155 | 5′-TTAATGCTAATCGTGATAGGGGTAAAA-3′ |
| mir-20a | 5′-TAAAGTGCTTATAGTGCAGGTAAAA-3′ |
| miR-451 | 5′-AAACCGTTACCATTACTGAGTAAAA-3′ |
| mir-26b | 5′-GTTCAAGTAATTCAGGATAGGAAAA-3′ |
| mir-522 | 5′-CTCTAGAGGGAAGCGCTTTCT-3′ |
| miR-192 | 5′-CTGACCTATGAATTGACAGCAAA-3′ |
| miR-137 | 5′-ACGGGTATTCTTGGGTGGATAA-3′ |
| Variables | Statistical Values | Patients (N = 103) |
|---|---|---|
| Age (years) | n mean ± SD min, max | 103 51.8 ± 12.17 27, 82 |
| Gender | n | 103 |
| Female Male | n (%) n (%) | 90 (87.4%) 13 (12.6%) |
| Weight (kg) | n mean ± SD min, max | 103 74.14 ± 15.515 41.0, 121.0 |
| Disease duration (years) | n Median Q1, Q3 | 103 6.29 3.37, 11.20 |
| Disease severity | n | 101 |
| Moderate (DAS28-CRP > 3.2 to ≤5.1) High (DAS28-CRP >5.1) | n (%) n (%) | 7 (6.8%) 94 (91.3%) |
| DAS28-CRP | n mean ± SD min, max | 101 6.03 ± 0.676 4.5, 8.1 |
| CDAI | n mean ± SD min, max | 101 40.47 ± 9.064 24.8, 69.3 |
| HAQ-DI | n mean ± SD min, max | 101 1.7339 ± 0.4882 0.500, 2.875 |
| CRP (mg/mL) | n mean ± SD min, max | 10,323.5 ± 22.85 1, 120 |
| Anti-CCP (IU/mL) | n mean ± SD min, max | 101 531.35 ± 671.132 0.4, 3408.4 |
| RF (IU/mL) | n mean ± SD min, max | 103 193.1 ± 224.15 7, 1250 |
| Anti-CCP >10 IU/mL ≤10 IU/mL | n n (%) n (%) | 101 86 (83.5%) 15 (14.6%) |
| RF ≥15 IU/mL <15 IU/mL | n n (%) n (%) | 101 90 (87.4%) 13 (12.6%) |
| Basal Anti-CCP and RF Low/Low Medium High/High | n n (%) n (%) n (%) | 101 15 (14.6%) 72 (69.9%) 14 (13.6%) |
| Variables Weeks | Olokizumab (N = 103) | |
|---|---|---|
| Response based on DAS28 (<3.2) | ||
| Week 12 yes no Week 24 yes no | n (%) n (%) n (%) n (%) | 103 42 (40.8%) 61 (59.2%) 103 52 (50.5%) 51 (49.5%) |
| Response based on ARC20 | ||
| Week 12 yes no Week 24 Yes no | n (%) n (%) n (%) n (%) | 103 71 (68.9%) 32 (31.1%) 103 83 (80.6%) 20 (19.4%) |
| Response based on ARC50 | ||
| Week 12 yes no Week 24 yes no | n (%) n (%) n (%) n (%) | 103 48 (46.6%) 55 (53.4%) 103 56 (54.4%) 47 (45.6%) |
| MiRNA | Olokizumab (N = 103) | ||||
|---|---|---|---|---|---|
| Baseline Value * | Week 12 * | p-Value ** | Week 24 * | p-Value ** | |
| miR-137 | n = 101 −14.0901 −15.4189, −13.3029 | n = 94 −13.8384 −14.7926, −12.9781 | 0.1741 | n = 92 −14.1488 −15.3060, −12.8427 | 0.9689 |
| miR-155 | n = 103 −7.7179 −9.6218, −6.0038 | n = 97 −6.6529 −8.7346, −5.3672 | 0.0123 | n = 83 −6.8945 −8.9418, −5.6004 | 0.1211 |
| miR-16 | n = 103 −10.2718 −11.8904, −8.8278 | n = 97 −9.3376 −10.9355, −8.2970 | 0.0001 | n = 83 −9.6046 −11.3095, −8.3791 | 0.1147 |
| miR-192 | n = 102 −9.4246 −11.0052, −8.4285 | n = 96 −8.8499 −10.5248, −8.0931 | 0.0151 | n = 83 −9.0553 −10.8217, −7.8205 | 0.1326 |
| miR-20a | n = 103 −12.5434 −13.5346, −11.2284 | n = 97 −12.0229 −13.2547, −10.8907 | 0.0089 | n = 83 −11.9538 −13.6044, −11.1414 | 0.9821 |
| miR-26b | n = 103 −13.9983 −14.8531, −13.1991 | n = 97 −14.0670 −14.7470, −13.1972 | 0.8457 | n = 83 −14.3008 −15.2316, −13.4501 | 0.0291 |
| miR-29 | n = 103 −12.0607 −12.8295, −11.2436 | n = 97 −11.8202 −12.7251, −11.1674 | 0.0204 | n = 83 −12.0506 −12.8377, −11.2525 | 0.8998 |
| miR-451 | n = 103 −12.0970 −13.8915, −10.5405 | n = 97 −11.3203 −12.9140, −9.2073 | 0.0018 | n = 83 −11.1599 −13.1725, −9.5124 | 0.0234 |
| miR-522 | n = 102 −17.8232 −19.0515, −16.1847 | n = 95 −17.9271 −18.9235, −16.6540 | 0.2977 | n = 82 −18.3636 *** −19.3113, −17.2911 | <0.0001 |
| Olokizumab (N = 103) | p-value | |
|---|---|---|
| Disease severity miR-26b | 0.0486 | |
| Moderate (DAS28-CRP >3.2 to ≤5.1) | ||
| n | 7 | |
| Mean ± SD | −12.7001 ± 1.9300 | |
| Median | −13.4802 | |
| Q1, Q3 | −13.9373, −12.2775 | |
| Min, max | −14.2987, −8.6716 | |
| High (DAS28-CRP > 5.1) | ||
| n | 94 | |
| Mean ± SD | −14.0451 ± 1.2415 | |
| Median | −14.0730 | |
| Q1, Q3 | −14.9105, −13.2551 | |
| Min, max | −16.6515, −10.7522 | |
| Disease severity miR-29 | 0.0141 | |
| Moderate (DAS28-CRP >3.2 to ≤5.1) | ||
| n | 7 | |
| Mean ± SD | −10.8471 ± 1.5037 | |
| Median | −11.2337 | |
| Q1, Q3 | −11.2997, −11.1783 | |
| Min, max | −12.2331, −7.5453 | |
| High (DAS28-CRP > 5.1) | ||
| n | 94 | |
| Mean ± SD | −12.0729 ± 1.2746 | |
| Median | −12.1499 | |
| Q1, Q3 | −12.8570, −11.4248 | |
| Min, max | −14.9079, −8.2635 | |
| Anti-CCP miR-522 | 0.0447 | |
| Positive | ||
| n | 85 | |
| Mean ± SD | −17.3993 ± 2.1323 | |
| Median | −17.6365 | |
| Q1, Q3 | −18.9118, −15.6329 | |
| Min, max | −23.1875, −9.9698 | |
| Negative | ||
| n | 15 | |
| Mean ± SD | −18.5592 ± 1.3619 | |
| Median | −18.1645 | |
| Q1, Q3 | −19.5899, −17.4640 | |
| Min, max | −21.4611, −16.3436 |
| miRNAs | Expression in RA | Localization | Target Genes | Potential Role in RA | Reference |
|---|---|---|---|---|---|
| miR-155 | increased | peripheral blood mononuclear cell (PBMC) | SOCS1 | inflammation and joint damage | [13] |
| increased | synovial fibroblasts (SF) | RIP1JKK | NF-kB signaling pathway regulation | [20] | |
| increased | SF | IKBKE | decrease MMP3 expression, proliferation and FLS, inflammatory processes regulation | [14] | |
| increased | serum | - | inflammatory processes regulation | [21] | |
| increased | SF | MMP-3, MMP-1 | blocks cytokine induction of MMP-1 and MMP-3 | [22] | |
| increased | synovial macrophages, monocytes | SHIP-1 | increase secretion of proinflammatory cytokines IL-6 and TNF-a | [23] | |
| decreased | whole blood | - | potential biomarker of methotrexate therapy response | [24] | |
| miR-16 | decreased | synovial fluid | - | NF-kB signaling pathway regulation | [25] |
| decreased | serum | - | potential biomarker for RA diagnostic | [26,27] | |
| increased | SF | Bcl-2 | apoptosis regulation | [17] | |
| miR-20a | decreased | SF | ASK1 | IL-6, TNF and IL-1b regulation, anti-inflammatory effect | [28] |
| decreased | SF | STAT3 | anti-inflammatory effect; suppress proliferation and induce apoptosis | [16] | |
| decreased | SF | TXNIP | regulate secretion of proinflammatory cytokines | [29] | |
| miR-451 | decreased | SF | - | regulate FLS proliferation and secretion of proinflammatory cytokines | [18] |
| miR-29 | decreased | SF | DNMT3a | DNA methylation regulation | [30] |
| miR-192 | decreased | SF | CAV-1 | regulate proliferation and apoptosis | [19] |
| decreased | SF | - | biomarker of therapy with TNF inhibitors response | [31] | |
| miR-137 | decreased | SF | CXCL12 | regulate proliferation, migration and invasion | [32] |
| miR-26b | decreased | SF | GSK-3P | decrease proliferation and cytokine secretion, affecting signaling pathway Wnt/GSK-3b/b-katenin | [33] |
| miR-522 | increased | SF | SOCS3 | regulate expression of MMPs and proinflammatory cytokines | [15] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bure, I.V.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Alekseeva, E.A.; Bondareva, K.I.; Kalinkin, A.I.; Lukashev, A.N.; Tarasov, V.V.; Zamyatnin, A.A., Jr.; Nemtsova, M.V. Analysis of miRNA Expression in Patients with Rheumatoid Arthritis during Olokizumab Treatment. J. Pers. Med. 2020, 10, 205. https://doi.org/10.3390/jpm10040205
Bure IV, Mikhaylenko DS, Kuznetsova EB, Alekseeva EA, Bondareva KI, Kalinkin AI, Lukashev AN, Tarasov VV, Zamyatnin AA Jr., Nemtsova MV. Analysis of miRNA Expression in Patients with Rheumatoid Arthritis during Olokizumab Treatment. Journal of Personalized Medicine. 2020; 10(4):205. https://doi.org/10.3390/jpm10040205
Chicago/Turabian StyleBure, Irina V., Dmitry S. Mikhaylenko, Ekaterina B. Kuznetsova, Ekaterina A. Alekseeva, Kristina I. Bondareva, Alexey I. Kalinkin, Alexander N. Lukashev, Vadim V. Tarasov, Andrey A. Zamyatnin, Jr., and Marina V. Nemtsova. 2020. "Analysis of miRNA Expression in Patients with Rheumatoid Arthritis during Olokizumab Treatment" Journal of Personalized Medicine 10, no. 4: 205. https://doi.org/10.3390/jpm10040205
APA StyleBure, I. V., Mikhaylenko, D. S., Kuznetsova, E. B., Alekseeva, E. A., Bondareva, K. I., Kalinkin, A. I., Lukashev, A. N., Tarasov, V. V., Zamyatnin, A. A., Jr., & Nemtsova, M. V. (2020). Analysis of miRNA Expression in Patients with Rheumatoid Arthritis during Olokizumab Treatment. Journal of Personalized Medicine, 10(4), 205. https://doi.org/10.3390/jpm10040205






