Multivariate Analysis of Fecal Metabolites from Children with Autism Spectrum Disorder and Gastrointestinal Symptoms before and after Microbiota Transfer Therapy
Abstract
:1. Introduction
2. Materials and Methods
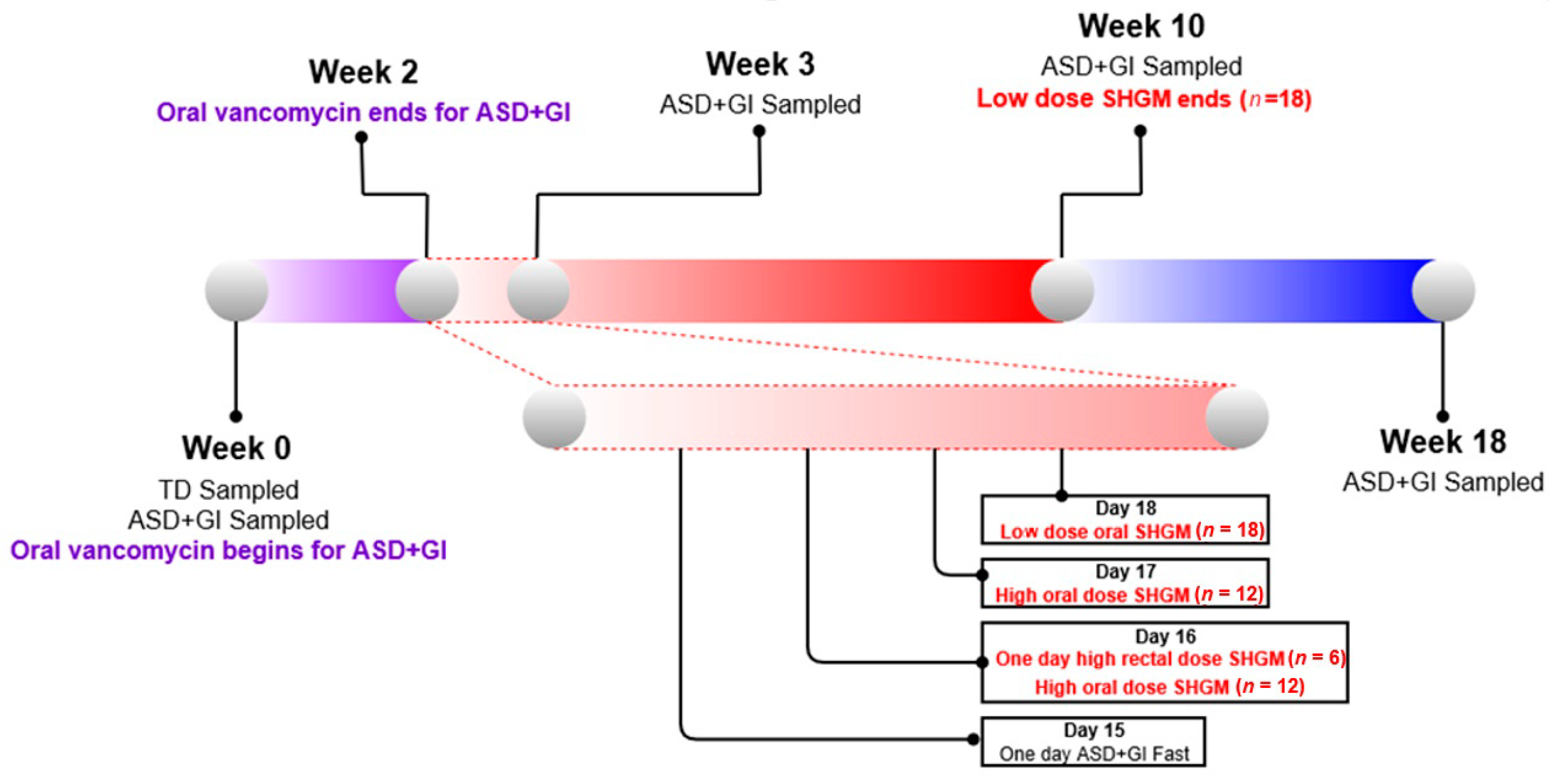
2.1. Study Design
2.2. Metabolite Measurements
2.3. Statistical Analysis
2.3.1. Preprocessing
2.3.2. Univariate Analysis
2.3.3. Multivariate Analysis
2.4. Cross-Validation
2.5. Model Evaluation
3. Results
3.1. Univariate Analysis
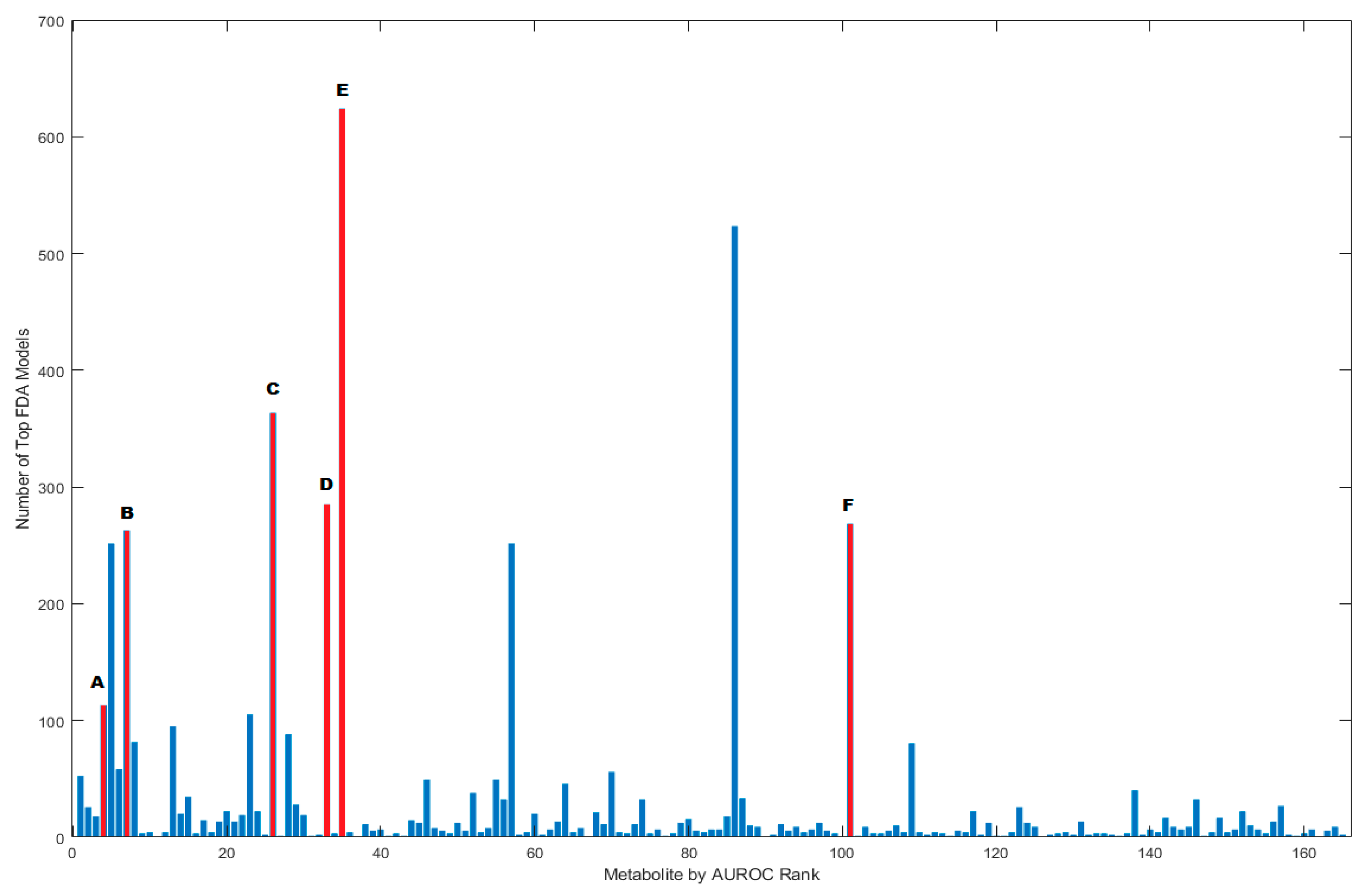
3.2. FDA Models
3.3. Correlation Analysis
3.4. Assessing Effects of MTT
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Metabolite | Week 0 AUROC | Week 18 AUROC | TD Mean Week 0 | ASD Mean Week 0 | Week 0 vs. Week 18 ASD p-Value | Week 0 ASD vs. TD Un-Adjusted p-Value | Leave-1-Out | Leave 2-Out | Leave 3-Out |
|---|---|---|---|---|---|---|---|---|---|
| carnitine | 0.77 | 0.68 | 9.81 × 106 | 3.35 × 107 | 7.64 × 10−1 | 4.78 × 10−3 | 0.00 | 0.00 | 0.00 |
| sphingosine | 0.75 | 0.58 | 7.18 × 106 | 2.13 × 107 | 1.10 × 10−1 | 8.15 × 10−3 | 0.00 | 0.00 | 0.00 |
| 2′-deoxyadenosine | 0.75 | 0.59 | 8.08 × 105 | 2.32 × 105 | 1.83 × 10−2 | 9.59 × 10−3 | 0.00 | 0.00 | 0.00 |
| indole | 0.74 | 0.72 | 4.74 × 105 | 1.05 × 106 | 5.80 × 10−1 | 1.04 × 10−2 | 0.00 | 0.00 | 0.00 |
| adenine | 0.74 | 0.81 | 6.70 × 106 | 2.91 × 106 | 5.17 × 10−1 | 1.35 × 10−2 | 0.00 | 0.00 | 0.03 |
| N-stearoyl-sphingosine | 0.73 | 0.61 | 1.01 × 106 | 4.05 × 106 | 1.03 × 10−1 | 1.40 × 10−2 | 0.00 | 0.00 | 0.04 |
| imidazole | 0.71 | 0.64 | 1.02 × 107 | 3.95 × 107 | 1.50 × 10−1 | 1.83 × 10−2 | 0.00 | 0.00 | 0.13 |
| 10-nonadecenoate | 0.71 | 0.59 | 2.42 × 106 | 5.49 × 106 | 1.19 × 10−2 | 2.18 × 10−2 | 0.00 | 0.01 | 0.25 |
| p-cresol | 0.71 | 0.53 | 1.55 × 106 | 4.00 × 106 | 6.42 × 10−2 | 2.53 × 10−2 | 0.00 | 0.16 | 0.27 |
| cystathionine | 0.71 | 0.65 | 6.07 × 104 | 9.12 × 104 | 5.98 × 10−1 | 2.53 × 10−2 | 0.00 | 0.17 | 0.25 |
| 5alpha-androstan-3beta,17alpha-diol | 0.71 | 0.63 | 9.86 × 104 | 6.65 × 104 | 4.50 × 10−3 | 2.55 × 10−2 | 0.00 | 0.31 | 0.32 |
| 3-(3-hydroxyphenyl)propionate | 0.71 | 0.66 | 2.24 × 107 | 2.67 × 107 | 8.62 × 10−1 | 2.73 × 10−2 | 0.00 | 0.24 | 0.30 |
| 1-(1-enyl-oleoyl)-GPE | 0.71 | 0.87 | 3.31 × 105 | 2.57 × 105 | 2.47 × 10−1 | 2.81 × 10−2 | 0.00 | 0.24 | 0.29 |
| gamma-glutamyl-histidine | 0.71 | 0.58 | 1.90 × 105 | 1.95 × 105 | 1.83 × 10−1 | 2.94 × 10−2 | 0.00 | 0.25 | 0.38 |
| Deoxy-carnitine | 0.71 | 0.68 | 2.82 × 108 | 1.09 × 109 | 6.46 × 10−1 | 2.94 × 10−2 | 0.00 | 0.26 | 0.42 |
| diaminopimelate | 0.70 | 0.62 | 8.57 × 105 | 1.14 × 106 | 7.16 × 10−1 | 3.11 × 10−2 | 0.00 | 0.32 | 0.40 |
| tyramine | 0.70 | 0.70 | 2.17 × 107 | 8.33 × 106 | 7.86 × 10−1 | 3.17 × 10−2 | 0.21 | 0.27 | 0.36 |
| gulonate | 0.70 | 0.53 | 2.43 × 105 | 1.52 × 106 | 9.12 × 10−2 | 3.40 × 10−2 | 0.11 | 0.42 | 0.46 |
| gamma-tocotrienol | 0.70 | 0.62 | 5.04 × 106 | 3.24 × 106 | 3.51 × 10−1 | 3.49 × 10−2 | 0.26 | 0.42 | 0.46 |
| 4-hydroxyphenylacetate | 0.70 | 0.56 | 9.97 × 105 | 2.75 × 106 | 2.75 × 10−1 | 3.66 × 10−2 | 0.32 | 0.42 | 0.44 |
| delta-tocopherol | 0.70 | 0.53 | 2.50 × 106 | 1.62 × 106 | 7.38 × 10−2 | 3.85 × 10−2 | 0.42 | 0.49 | 0.48 |
| phenethylamine | 0.69 | 0.55 | 4.35 × 105 | 9.09 × 105 | 6.42 × 10−2 | 3.93 × 10−2 | 0.39 | 0.45 | 0.49 |
| Propionyl-glycine | 0.69 | 0.61 | 1.56 × 105 | 3.92 × 105 | 6.09 × 10−1 | 3.93 × 10−2 | 0.47 | 0.51 | 0.53 |
| N-acetyl-sphingosine | 0.69 | 0.63 | 9.84 × 104 | 2.63 × 105 | 3.33 × 10−1 | 4.22 × 10−2 | 0.45 | 0.51 | 0.47 |
| betaine | 0.69 | 0.60 | 3.39 × 106 | 5.05 × 106 | 5.37 × 10−1 | 4.55 × 10−2 | 0.63 | 0.50 | 0.47 |
| adenosine | 0.69 | 0.65 | 5.81 × 105 | 3.41 × 105 | 5.58 × 10−1 | 4.84 × 10−2 | 0.50 | 0.59 | 0.60 |
| ornithine | 0.68 | 0.51 | 2.05 × 107 | 2.74 × 107 | 6.89 × 10−2 | 5.19 × 10−2 | 0.71 | 0.61 | 0.65 |
| N-palmitoyl-sphingosine | 0.68 | 0.54 | 3.01 × 106 | 1.04 × 107 | 2.57 × 10−2 | 5.33 × 10−2 | 1.00 | 1.00 | 0.67 |
| galactonate | 0.68 | 0.67 | 4.63 × 105 | 2.05 × 106 | 7.63 × 10−1 | 5.37 × 10−2 | 0.66 | 0.62 | 0.67 |
| N1-Methyl-2-pyridone-5-carboxamide | 0.68 | 0.74 | 1.63 × 105 | 4.37 × 105 | 7.62 × 10−1 | 5.44 × 10−2 | 1.00 | 1.00 | 0.69 |
| 1-palmitoylglycerol | 0.68 | 0.54 | 1.08 × 107 | 1.55 × 107 | 1.69 × 10−1 | 5.72 × 10−2 | 1.00 | 1.00 | 0.71 |
| phosphocholine | 0.67 | 0.59 | 3.01 × 105 | 1.25 × 107 | 5.36 × 10−2 | 5.93 × 10−2 | 0.79 | 0.72 | 0.72 |
| theobromine | 0.67 | 0.62 | 2.60 × 106 | 5.81 × 106 | 1.56 × 10−2 | 5.93 × 10−2 | 0.76 | 0.70 | 0.72 |
| hydroxyproline | 0.67 | 0.60 | 2.96 × 106 | 9.08 × 106 | 6.92 × 10−1 | 6.10 × 10−2 | 0.79 | 0.73 | 0.74 |
| l-urobilin | 0.67 | 0.56 | 4.11 × 107 | 3.08 × 107 | 1.55 × 10−2 | 6.35 × 10−2 | 0.71 | 0.80 | 0.73 |
| 3,5-dihydroxybenzoic | 0.67 | 0.60 | 1.61 × 105 | 9.06 × 104 | 3.29 × 10−1 | 6.52 × 10−2 | 1.00 | 1.00 | 0.95 |
| carboxyethyl-GABA | 0.67 | 0.55 | 1.52 × 107 | 1.01 × 107 | 2.00 × 10−1 | 6.52 × 10−2 | 1.00 | 1.00 | 0.95 |
| oxalate | 0.67 | 0.53 | 1.03 × 106 | 2.11 × 106 | 7.91 × 10−2 | 6.58 × 10−2 | 0.74 | 0.80 | 0.78 |
| Palmitoyl-carnitine | 0.67 | 0.53 | 4.93 × 105 | 4.68 × 106 | 5.18 × 10−2 | 6.77 × 10−2 | 0.79 | 0.79 | 0.79 |
| Copro-stanol | 0.67 | 0.60 | 2.21 × 106 | 1.31 × 106 | 3.17 × 10−1 | 6.86 × 10−2 | 0.87 | 0.79 | 0.79 |
| 5-hydroxylysine | 0.66 | 0.57 | 1.03 × 106 | 2.53 × 106 | 3.35 × 10−1 | 6.99 × 10−2 | 0.82 | 0.82 | 0.80 |
| Saccharopine | 0.66 | 0.53 | 1.50 × 106 | 4.10 × 105 | 9.49 × 10−2 | 7.22 × 10−2 | 0.89 | 0.82 | 0.79 |
| 3-(4-hydroxyphenyl)lactate | 0.66 | 0.58 | 1.56 × 106 | 1.88 × 106 | 2.37 × 10−2 | 7.22 × 10−2 | 0.82 | 0.82 | 0.81 |
| Stearoyl-carnitine | 0.66 | 0.60 | 1.65 × 106 | 4.06 × 106 | 1.03 × 10−1 | 7.69 × 10−2 | 0.92 | 0.85 | 0.82 |
| biliverdin | 0.66 | 0.59 | 1.29 × 105 | 2.88 × 105 | 3.67 × 10−1 | 7.69 × 10−2 | 0.89 | 0.85 | 0.82 |
| carnosine | 0.66 | 0.57 | 8.48 × 104 | 8.23 × 104 | 3.44 × 10−1 | 8.31 × 10−2 | 0.87 | 0.91 | 0.85 |
| pentadecanoate | 0.66 | 0.64 | 2.40 × 108 | 1.97 × 108 | 8.62 × 10−1 | 8.44 × 10−2 | 0.87 | 0.87 | 0.87 |
| hexadecanedioate | 0.66 | 0.55 | 4.54 × 105 | 4.28 × 105 | 1.25 × 10−1 | 8.72 × 10−2 | 0.89 | 0.90 | 0.87 |
| 10-hydroxystearate | 0.66 | 0.60 | 1.16 × 109 | 1.93 × 109 | 8.25 × 10−3 | 9.27 × 10−2 | 0.95 | 0.92 | 0.88 |
| Sphinganine | 0.66 | 0.54 | 1.67 × 107 | 2.34 × 107 | 3.19 × 10−1 | 9.28 × 10−2 | 0.89 | 0.91 | 0.89 |
| trigonelline | 0.66 | 0.81 | 3.08 × 106 | 6.92 × 106 | 1.41 × 10−1 | 9.28 × 10−2 | 0.92 | 0.92 | 0.88 |
| Indole-lactate | 0.66 | 0.64 | 4.74 × 105 | 1.80 × 106 | 8.80 × 10−3 | 9.46 × 10−2 | 1.00 | 1.00 | 0.87 |
| Dihomo-linoleate | 0.65 | 0.58 | 1.11 × 107 | 2.92 × 107 | 7.38 × 10−2 | 9.76 × 10−2 | 1.00 | 1.00 | 1.00 |
| Phyto-sphingosine | 0.65 | 0.60 | 8.22 × 105 | 1.25 × 106 | 5.58 × 10−1 | 9.77 × 10−2 | 1.00 | 0.95 | 0.90 |
| gamma-tocopherol/beta-tocopherol | 0.65 | 0.51 | 5.43 × 107 | 4.05 × 107 | 1.03 × 10−1 | 9.86 × 10−2 | 0.92 | 0.91 | 0.89 |
| acesulfame | 0.65 | 0.59 | 3.59 × 104 | 7.24 × 105 | 6.27 × 10−1 | 9.86 × 10−2 | 0.92 | 0.92 | 0.90 |
| N-methyl-pipecolate | 0.65 | 0.63 | 4.88 × 106 | 9.71 × 106 | 7.16 × 10−1 | 9.86 × 10−2 | 0.97 | 0.94 | 0.88 |
| 2-methylserine | 0.65 | 0.66 | 9.22 × 105 | 7.05 × 105 | 7.16 × 10−1 | 9.86 × 10−2 | 0.97 | 0.93 | 0.90 |
| 2-aminobutyrate | 0.65 | 0.61 | 1.27 × 107 | 2.65 × 107 | 4.02 × 10−1 | 1.02 × 10−1 | 0.97 | 0.94 | 0.92 |
| N-palmitoyl-sphinganine | 0.65 | 0.64 | 1.99 × 106 | 4.43 × 106 | 1.55 × 10−2 | 1.02 × 10−1 | 0.95 | 0.94 | 0.91 |
| caffeate | 0.65 | 0.60 | 4.23 × 105 | 4.23 × 105 | 5.19 × 10−1 | 1.06 × 10−1 | 0.97 | 0.95 | 0.94 |
| piperine | 0.65 | 0.59 | 2.41 × 107 | 1.59 × 107 | 1.96 × 10−2 | 1.11 × 10−1 | 1.00 | 0.96 | 0.93 |
| N-propionyl-methionine | 0.65 | 0.65 | 1.55 × 105 | 3.58 × 105 | 8.87 × 10−1 | 1.11 × 10−1 | 1.00 | 0.96 | 0.93 |
| alpha-CEHC | 0.65 | 0.50 | 1.02 × 105 | 5.68 × 105 | 6.53 × 10−2 | 1.11 × 10−1 | 0.97 | 0.96 | 0.93 |
| 5-aminovalerate | 0.65 | 0.55 | 3.29 × 107 | 1.16 × 108 | 4.57 × 10−1 | 1.11 × 10−1 | 1.00 | 1.00 | 0.94 |
| 2-aminophenol | 0.65 | 0.56 | 7.90 × 105 | 6.54 × 105 | 4.02 × 10−1 | 1.17 × 10−1 | 1.00 | 1.00 | 0.93 |
| O-sulfo-l-tyrosine | 0.65 | 0.50 | 9.04 × 104 | 3.24 × 105 | 1.26 × 10−1 | 1.17 × 10−1 | 0.97 | 0.95 | 0.93 |
| N-acetyl-valine | 0.64 | 0.69 | 1.99 × 105 | 9.36 × 105 | 9.50 × 10−1 | 1.18 × 10−1 | 1.00 | 0.96 | 0.94 |
| norvaline | 0.64 | 0.52 | 4.20 × 106 | 1.65 × 107 | 2.05 × 10−1 | 1.18 × 10−1 | 1.00 | 0.96 | 0.94 |
| tryptamine | 0.64 | 0.93 | 3.40 × 105 | 1.63 × 106 | 1.19 × 10−2 | 1.18 × 10−1 | 1.00 | 0.96 | 0.94 |
| myristate | 0.64 | 0.73 | 3.65 × 108 | 3.05 × 108 | 3.04 × 10−1 | 1.18 × 10−1 | 1.00 | 1.00 | 0.94 |
| Eicosenoate | 0.64 | 0.59 | 5.55 × 107 | 1.30 × 108 | 7.91 × 10−2 | 1.19 × 10−1 | 1.00 | 1.00 | 0.95 |
| cholesterol | 0.64 | 0.56 | 3.70 × 106 | 1.35 × 107 | 3.67 × 10−1 | 1.20 × 10−1 | 1.00 | 0.98 | 0.95 |
| 3-ureidopropionate | 0.64 | 0.58 | 6.14 × 105 | 3.26 × 105 | 6.92 × 10−1 | 1.24 × 10−1 | 1.00 | 0.95 | 0.93 |
| diglycerol | 0.64 | 0.60 | 3.13 × 106 | 1.50 × 106 | 5.17 × 10−1 | 1.25 × 10−1 | 1.00 | 0.97 | 0.95 |
| N-acetylneuraminate | 0.64 | 0.52 | 4.66 × 106 | 9.65 × 106 | 1.25 × 10−1 | 1.25 × 10−1 | 1.00 | 0.96 | 0.94 |
| Succinyl-carnitine | 0.64 | 0.65 | 6.00 × 105 | 3.56 × 106 | 8.87 × 10−1 | 1.29 × 10−1 | 1.00 | 0.97 | 0.97 |
| 2′-deoxyinosine | 0.64 | 0.76 | 4.88 × 106 | 7.84 × 106 | 1.69 × 10−1 | 1.30 × 10−1 | 1.00 | 0.99 | 0.97 |
| 3-aminoisobutyrate | 0.64 | 0.54 | 2.82 × 105 | 3.10 × 106 | 3.41 × 10−1 | 1.32 × 10−1 | 1.00 | 0.99 | 0.96 |
| d-urobilin | 0.64 | 0.54 | 7.74 × 105 | 2.68 × 106 | 6.42 × 10−2 | 1.36 × 10−1 | 1.00 | 0.99 | 0.97 |
| 1-methylnicotinamide | 0.64 | 0.68 | 1.02 × 105 | 2.95 × 105 | 5.58 × 10−1 | 1.36 × 10−1 | 1.00 | 0.98 | 0.97 |
| N-acetyl-alanine | 0.64 | 0.58 | 3.06 × 106 | 3.98 × 106 | 3.84 × 10−1 | 1.38 × 10−1 | 1.00 | 1.00 | 0.98 |
| aspartate | 0.64 | 0.54 | 1.90 × 108 | 2.44 × 108 | 1.03 × 10−1 | 1.40 × 10−1 | 1.00 | 0.98 | 0.96 |
| trans-urocanate | 0.64 | 0.50 | 4.71 × 107 | 3.96 × 107 | 1.17 × 10−1 | 1.40 × 10−1 | 1.00 | 0.98 | 0.97 |
| 3-carboxyadipate | 0.64 | 0.66 | 8.99 × 106 | 1.47 × 107 | 5.58 × 10−1 | 1.40 × 10−1 | 1.00 | 0.98 | 0.96 |
| 1,7-dimethylurate | 0.64 | 0.60 | 8.91 × 105 | 2.31 × 106 | 2.87 × 10−2 | 1.40 × 10−1 | 1.00 | 0.98 | 0.97 |
| 2-piperidinone | 0.64 | 0.64 | 2.17 × 107 | 3.82 × 107 | 9.37 × 10−1 | 1.40 × 10−1 | 1.00 | 1.00 | 0.97 |
| pheophorbide | 0.64 | 0.61 | 4.12 × 106 | 4.78 × 105 | 6.94 × 10−1 | 1.41 × 10−1 | 1.00 | 1.00 | 1.00 |
| acisoga | 0.63 | 0.63 | 3.52 × 105 | 6.39 × 105 | 6.92 × 10−1 | 1.41 × 10−1 | 1.00 | 1.00 | 1.00 |
| sulfate | 0.63 | 0.55 | 6.68 × 106 | 1.65 × 107 | 1.10 × 10−1 | 1.41 × 10−1 | 1.00 | 1.00 | 1.00 |
| 1-(1-enyl-palmitoyl)-GPE | 0.63 | 0.79 | 8.53 × 105 | 7.57 × 105 | 5.11 × 10−2 | 1.41 × 10−1 | 0.97 | 0.96 | 0.95 |
| histidine | 0.63 | 0.55 | 9.65 × 107 | 1.10 × 108 | 2.00 × 10−1 | 1.43 × 10−1 | 1.00 | 1.00 | 0.97 |
| Maltotetraose | 0.63 | 0.53 | 5.10 × 105 | 6.33 × 105 | 2.66 × 10−1 | 1.48 × 10−1 | 1.00 | 1.00 | 0.98 |
| maltose | 0.63 | 0.63 | 7.98 × 106 | 5.18 × 106 | 9.62 × 10−1 | 1.48 × 10−1 | 1.00 | 1.00 | 0.98 |
| 2-methylcitrate/homocitrate | 0.63 | 0.66 | 6.24 × 105 | 9.91 × 105 | 9.87 × 10−1 | 1.48 × 10−1 | 1.00 | 0.98 | 0.97 |
| trimethylamine | 0.63 | 0.55 | 3.98 × 105 | 1.53 × 106 | 3.35 × 10−1 | 1.48 × 10−1 | 1.00 | 0.99 | 0.97 |
| linoleoyl-linolenoyl-glycerol | 0.63 | 0.53 | 8.05 × 106 | 2.50 × 106 | 1.33 × 10−1 | 1.48 × 10−1 | 1.00 | 0.99 | 0.97 |
| thymidine | 0.63 | 0.66 | 5.10 × 106 | 7.32 × 106 | 9.87 × 10−1 | 1.52 × 10−1 | 1.00 | 0.98 | 0.98 |
| Pyri-doxate | 0.63 | 0.52 | 1.40 × 107 | 1.76 × 107 | 1.69 × 10−1 | 1.52 × 10−1 | 1.00 | 0.98 | 0.98 |
| sarcosine | 0.63 | 0.66 | 2.59 × 105 | 6.59 × 105 | 6.12 × 10−1 | 1.56 × 10−1 | 0.97 | 0.95 | 0.94 |
| 2-hydroxy-3-methylvalerate | 0.63 | 0.54 | 1.27 × 106 | 1.74 × 106 | 5.06 × 10−1 | 1.56 × 10−1 | 1.00 | 0.99 | 0.98 |
| gamma-glutamyl-phenylalanine | 0.63 | 0.61 | 2.27 × 105 | 2.94 × 105 | 9.50 × 10−1 | 1.56 × 10−1 | 1.00 | 0.99 | 0.98 |
| Linolenate | 0.63 | 0.59 | 1.98 × 108 | 9.80 × 107 | 9.87 × 10−1 | 1.56 × 10−1 | 1.00 | 0.98 | 0.97 |
| 3-hydroxy-3-methylglutarate | 0.63 | 0.59 | 1.40 × 107 | 1.99 × 107 | 5.17 × 10−1 | 1.56 × 10−1 | 1.00 | 0.98 | 0.97 |
| 2′-deoxyuridine | 0.63 | 0.69 | 2.81 × 106 | 3.79 × 106 | 6.46 × 10−1 | 1.56 × 10−1 | 1.00 | 1.00 | 0.98 |
| Quino-linate | 0.63 | 0.52 | 4.89 × 104 | 1.15 × 105 | 9.77 × 10−2 | 1.57 × 10−1 | 1.00 | 1.00 | 0.98 |
| I-urobilinogen | 0.63 | 0.60 | 1.77 × 106 | 1.02 × 107 | 3.91 × 10−2 | 1.65 × 10−1 | 1.00 | 1.00 | 0.98 |
| N-acetyl-cadaverine | 0.63 | 0.61 | 4.95 × 107 | 1.14 × 108 | 7.64 × 10−1 | 1.65 × 10−1 | 1.00 | 1.00 | 0.99 |
| 7-ketolithocholate | 0.63 | 0.51 | 4.53 × 106 | 1.32 × 106 | 1.92 × 10−1 | 1.69 × 10−1 | 1.00 | 1.00 | 0.98 |
| Carboxy-ibuprofen | 0.63 | 0.53 | 1.47 × 106 | 1.75 × 104 | 1.48 × 10−1 | 1.74 × 10−1 | 1.00 | 0.99 | 0.98 |
| Phenyl-lactate | 0.62 | 0.57 | 1.83 × 106 | 1.83 × 106 | 4.13 × 10−2 | 1.74 × 10−1 | 1.00 | 0.99 | 0.98 |
| kynurenate | 0.62 | 0.59 | 1.30 × 106 | 9.08 × 105 | 9.05 × 10−2 | 1.74 × 10−1 | 1.00 | 0.99 | 0.98 |
| citrate | 0.62 | 0.58 | 1.29 × 106 | 2.64 × 106 | 5.91 × 10−1 | 1.74 × 10−1 | 1.00 | 1.00 | 0.99 |
| 5alpha-androstan-3beta,17beta-diol | 0.62 | 0.53 | 1.85 × 105 | 7.81 × 104 | 1.06 × 10−1 | 1.74 × 10−1 | 1.00 | 1.00 | 0.99 |
| Octadecane-dioate | 0.62 | 0.66 | 1.10 × 106 | 8.50 × 105 | 2.67 × 10−2 | 1.74 × 10−1 | 1.00 | 0.99 | 0.98 |
| Oleoyl-carnitine | 0.62 | 0.54 | 4.89 × 105 | 4.10 × 106 | 1.35 × 10−1 | 1.74 × 10−1 | 1.00 | 0.99 | 0.98 |
| 4-androsten-3alpha,17alpha-diol | 0.62 | 0.50 | 2.30 × 105 | 7.59 × 104 | 2.02 × 10−1 | 1.83 × 10−1 | 1.00 | 1.00 | 0.99 |
| 8-hydroxyguanine | 0.62 | 0.55 | 1.15 × 105 | 1.50 × 105 | 1.58 × 10−1 | 1.83 × 10−1 | 1.00 | 1.00 | 0.98 |
| skatol | 0.62 | 0.50 | 5.08 × 105 | 3.96 × 105 | 1.71 × 10−1 | 1.83 × 10−1 | 1.00 | 1.00 | 0.99 |
| Ethyl-malonate | 0.62 | 0.66 | 5.50 × 106 | 6.23 × 106 | 4.20 × 10−1 | 1.83 × 10−1 | 1.00 | 1.00 | 0.99 |
| lactate | 0.62 | 0.52 | 2.39 × 106 | 3.29 × 106 | 1.41 × 10−1 | 1.83 × 10−1 | 1.00 | 1.00 | 0.99 |
| N6-carboxymethyllysine | 0.62 | 0.60 | 2.90 × 106 | 3.69 × 106 | 5.37 × 10−1 | 1.86 × 10−1 | 1.00 | 1.00 | 1.00 |
| Nervonate | 0.62 | 0.53 | 6.63 × 106 | 1.61 × 107 | 3.67 × 10−1 | 1.86 × 10−1 | 1.00 | 1.00 | 1.00 |
| inosine | 0.62 | 0.57 | 3.45 × 106 | 6.39 × 106 | 7.16 × 10−1 | 1.86 × 10−1 | 1.00 | 1.00 | 0.99 |
| Docosapentaenoate | 0.61 | 0.55 | 3.98 × 106 | 1.35 × 107 | 7.90 × 10−2 | 1.89 × 10−1 | 1.00 | 1.00 | 0.99 |
| Acetyl-carnitine | 0.61 | 0.54 | 7.50 × 105 | 8.06 × 106 | 4.77 × 10−1 | 1.89 × 10−1 | 1.00 | 1.00 | 0.98 |
| N-methyl-GABA | 0.61 | 0.59 | 5.35 × 106 | 6.36 × 106 | 9.35 × 10−1 | 1.93 × 10−1 | 1.00 | 1.00 | 0.99 |
| 2-aminoadipate | 0.61 | 0.66 | 1.11 × 106 | 1.27 × 106 | 7.40 × 10−1 | 1.95 × 10−1 | 1.00 | 1.00 | 0.99 |
| N-methyl-phenylalanine | 0.61 | 0.57 | 1.65 × 106 | 1.25 × 106 | 7.38 × 10−2 | 1.95 × 10−1 | 1.00 | 1.00 | 0.99 |
| cystine | 0.61 | 0.59 | 8.29 × 104 | 1.14 × 105 | 9.49 × 10−3 | 1.98 × 10−1 | 1.00 | 1.00 | 1.00 |
| 3-hydroxystearate | 0.61 | 0.54 | 6.21 × 106 | 4.61 × 106 | 7.88 × 10−1 | 2.00 × 10−1 | 1.00 | 1.00 | 0.99 |
| gluconate | 0.61 | 0.51 | 1.08 × 106 | 3.21 × 106 | 6.90 × 10−1 | 2.02 × 10−1 | 1.00 | 1.00 | 1.00 |
| diacylglycerol | 0.61 | 0.51 | 6.93 × 105 | 2.55 × 105 | 3.02 × 10−1 | 2.03 × 10−1 | 1.00 | 1.00 | 0.99 |
| dipicolinate | 0.61 | 0.51 | 4.99 × 105 | 4.94 × 105 | 2.46 × 10−1 | 2.03 × 10−1 | 1.00 | 1.00 | 0.99 |
| quinate | 0.61 | 0.61 | 9.58 × 106 | 2.75 × 107 | 9.37 × 10−1 | 2.03 × 10−1 | 1.00 | 1.00 | 0.99 |
| O-acetyl-homoserine | 0.61 | 0.52 | 4.56 × 105 | 3.96 × 105 | 4.20 × 10−1 | 2.14 × 10−1 | 1.00 | 1.00 | 0.99 |
| glutamate, | 0.61 | 0.51 | 2.42 × 106 | 2.03 × 106 | 2.75 × 10−1 | 2.14 × 10−1 | 1.00 | 1.00 | 0.99 |
| 6-hydroxynicotinate | 0.61 | 0.58 | 1.82 × 106 | 8.38 × 105 | 3.67 × 10−1 | 2.18 × 10−1 | 1.00 | 1.00 | 1.00 |
| pyridoxine | 0.61 | 0.61 | 1.57 × 106 | 2.91 × 106 | 5.47 × 10−2 | 2.18 × 10−1 | 1.00 | 1.00 | 1.00 |
| 2,4,6-trihydroxybenzoate | 0.61 | 0.55 | 3.58 × 104 | 5.36 × 105 | 6.48 × 10−1 | 2.20 × 10−1 | 1.00 | 1.00 | 1.00 |
| theophylline | 0.61 | 0.57 | 1.46 × 105 | 8.09 × 104 | 6.37 × 10−1 | 2.21 × 10−1 | 1.00 | 1.00 | 1.00 |
| 1-methylimidazoleacetate | 0.61 | 0.53 | 2.51 × 106 | 2.47 × 106 | 1.17 × 10−1 | 2.21 × 10−1 | 1.00 | 1.00 | 1.00 |
| 1-methylhistamine | 0.61 | 0.55 | 1.63 × 105 | 5.62 × 105 | 4.69 × 10−1 | 2.25 × 10−1 | 1.00 | 1.00 | 0.99 |
| phenylacetate | 0.61 | 0.58 | 1.69 × 107 | 2.48 × 107 | 7.16 × 10−1 | 2.25 × 10−1 | 1.00 | 1.00 | 0.99 |
| 4-hydroxyphenylpyruvate | 0.61 | 0.57 | 5.26 × 105 | 5.38 × 105 | 6.69 × 10−1 | 2.25 × 10−1 | 1.00 | 1.00 | 1.00 |
| cysteine | 0.61 | 0.61 | 3.35 × 106 | 4.04 × 106 | 8.62 × 10−1 | 2.25 × 10−1 | 1.00 | 1.00 | 1.00 |
| N-acetylcysteine | 0.61 | 0.51 | 3.09 × 105 | 3.95 × 105 | 2.23 × 10−1 | 2.25 × 10−1 | 1.00 | 1.00 | 0.99 |
| 13-methylmyristate | 0.61 | 0.72 | 3.09 × 105 | 3.95 × 105 | 2.89 × 10−1 | 2.27 × 10−1 | 1.00 | 1.00 | 1.00 |
| AMP | 0.61 | 0.52 | 3.56 × 108 | 3.17 × 108 | 2.44 × 10−1 | 2.29 × 10−1 | 1.00 | 1.00 | 1.00 |
| 2′-deoxyguanosine | 0.61 | 0.75 | 1.20 × 105 | 5.63 × 104 | 1.79 × 10−1 | 2.36 × 10−1 | 1.00 | 1.00 | 1.00 |
| 5-methyluridine | 0.61 | 0.64 | 2.32 × 106 | 3.59 × 106 | 8.62 × 10−1 | 2.36 × 10−1 | 1.00 | 1.00 | 1.00 |
| 1,3-dimethylurate | 0.61 | 0.58 | 7.67 × 105 | 9.16 × 105 | 5.20 × 10−2 | 2.36 × 10−1 | 1.00 | 1.00 | 1.00 |
| adrenate | 0.60 | 0.64 | 3.54 × 104 | 2.94 × 104 | 9.61 × 10−1 | 2.45 × 10−1 | 1.00 | 1.00 | 0.99 |
| tryptophan | 0.60 | 0.51 | 1.44 × 107 | 1.86 × 107 | 3.67 × 10−1 | 2.47 × 10−1 | 1.00 | 1.00 | 1.00 |
| dimethylarginine | 0.60 | 0.52 | 2.18 × 106 | 6.82 × 105 | 4.77 × 10−1 | 2.47 × 10−1 | 1.00 | 1.00 | 1.00 |
| 4-acetamidobutanoate | 0.60 | 0.50 | 7.64 × 107 | 8.86 × 107 | 3.34 × 10−1 | 2.47 × 10−1 | 1.00 | 1.00 | 1.00 |
| gamma-glutamyl-glutamine | 0.60 | 0.52 | 3.61 × 106 | 5.50 × 106 | 3.51 × 10−1 | 2.48 × 10−1 | 1.00 | 1.00 | 1.00 |
| caproate | 0.60 | 0.72 | 1.31 × 106 | 1.72 × 106 | 1.83 × 10−3 | 2.48 × 10−1 | 1.00 | 1.00 | 1.00 |
| 4-methylcatechol | 0.60 | 0.54 | 1.41 × 106 | 1.44 × 106 | 5.61 × 10−1 | 2.48 × 10−1 | 1.00 | 1.00 | 1.00 |
| nicotiana-amine | 0.60 | 0.55 | 3.44 × 107 | 3.48 × 107 | 9.67 × 10−2 | 2.54 × 10−1 | 1.00 | 1.00 | 0.99 |
| 1,3-propanediol | 0.60 | 0.56 | 3.19 × 104 | 8.27 × 104 | 5.73 × 10−1 | 2.58 × 10−1 | 1.00 | 1.00 | 1.00 |
| N-acetylserine | 0.60 | 0.59 | 1.87 × 106 | 1.07 × 106 | 6.42 × 10−2 | 2.59 × 10−1 | 1.00 | 1.00 | 1.00 |
| erythronate | 0.60 | 0.64 | 9.27 × 104 | 4.29 × 105 | 4.38 × 10−1 | 2.60 × 10−1 | 1.00 | 1.00 | 1.00 |
| 1-palmitoyl-GPE | 0.60 | 0.64 | 2.07 × 106 | 3.41 × 106 | 6.92 × 10−1 | 2.60 × 10−1 | 1.00 | 1.00 | 1.00 |
| 4-androsten-3beta,17beta-diol | 0.60 | 0.61 | 3.81 × 106 | 5.96 × 106 | 6.02 × 10−1 | 2.60 × 10−1 | 1.00 | 1.00 | 1.00 |
References
- Maenner, M.J.; Shaw, K.A.; Baio, J.; Washington, A.; Patrick, M.; DiRienzo, M.; Christensen, D.L.; Wiggins, L.D.; Pettygrove, S.; Andrews, J.G. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years—Autism and Developmental Disabilities Monitoring Network—Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2016. MMWR. Morb. Mortal. Wkly. Rep. Surveill. Summ. 2020, 69, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gaugler, T.L.; Klei, L.; Sanders, S.J.; Bodea, C.A.; Goldberg, A.P.; Lee, A.B.; Mahajan, M.; Manaa, D.; Pawitan, Y.; Reichert, J.; et al. Most genetic risk for autism resides with common variation. Nat. Genet. 2014, 46, 881–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandy, W.; Lai, M.-C. Annual Research Review: The role of the environment in the developmental psychopathology of autism spectrum condition. J. Child Psychol. Psychiatry 2016, 57, 271–292. [Google Scholar] [CrossRef] [PubMed]
- Meltzer, A.; Van De Water, J. The Role of the Immune System in Autism Spectrum Disorder. Neuropsychopharmacology 2016, 42, 284–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, E.Y. Gastrointestinal Issues in Autism Spectrum Disorder. Harv. Rev. Psych. 2014, 22, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Frye, R.E.; Rose, S.; Slattery, J.; Macfabe, D.F. Gastrointestinal dysfunction in autism spectrum disorder: The role of the mitochondria and the enteric microbiome. Microb. Ecol. Heal. Dis. 2015, 26, 27458. [Google Scholar] [CrossRef]
- Gorkiewicz, G.; Moschen, A.R. Gut microbiome: A new player in gastrointestinal disease. Virchows Arch. 2017, 472, 159–172. [Google Scholar] [CrossRef] [Green Version]
- Hills, J.R.D.; Pontefract, B.A.; Mishcon, H.R.; Black, C.A.; Sutton, S.C.; Theberge, C.R. Gut Microbiome: Profound Implications for Diet and Disease. Nutrition 2019, 11, 1613. [Google Scholar] [CrossRef] [Green Version]
- Abreu, M.T.; Peek, R.M. Gastrointestinal Malignancy and the Microbiome. Gastroenterol. 2014, 146, 1534–1546.e3. [Google Scholar] [CrossRef] [Green Version]
- Hughes, H.K.; Rose, D.; Ashwood, P. The Gut Microbiota and Dysbiosis in Autism Spectrum Disorders. Curr. Neurol. Neurosci. Rep. 2018, 18, 81. [Google Scholar] [CrossRef]
- Vuong, H.E.; Hsiao, E. Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Boil. Psych. 2016, 81, 411–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.-W.; Park, J.G.; Ilhan, Z.E.; Wallstrom, G.; LaBaer, J.; Adams, J.B.; Krajmalnik-Brown, R. Reduced Incidence of Prevotella and Other Fermenters in Intestinal Microflora of Autistic Children. PLoS ONE 2013, 8, e68322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Han, Y.; Dy, A.B.C.; Hagerman, R.J. The Gut Microbiota and Autism Spectrum Disorders. Front. Cell. Neurosci. 2017, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Needham, B.D.; Adame, M.D.; Serena, G.; Rose, D.R.; Preston, G.M.; Conrad, M.C.; Campbell, A.S.; Donabedian, D.H.; Fasano, A.; Ashwood, P.; et al. Plasma and Fecal Metabolite Profiles in Autism Spectrum Disorder. In Plasma and Fecal Metabolite Profiles in Autism Spectrum Disorder. BioRxiv 2020; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 2020. [Google Scholar]
- Mohamadkhani, A. Gut Microbiota and Fecal Metabolome Perturbation in Children with Autism Spectrum Disorder. Midd. East J. Dig. Dis. 2018, 10, 205–212. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.-W.; Ilhan, Z.E.; Isern, N.G.; Hoyt, D.W.; Howsmon, D.P.; Shaffer, M.; Lozupone, C.; Hahn, J.; Adams, J.B.; Krajmalnik-Brown, R. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe 2018, 49, 121–131. [Google Scholar] [CrossRef]
- De Angelis, M.; Piccolo, M.; Vannini, L.; Siragusa, S.; De Giacomo, A.; Serrazzanetti, D.I.; Cristofori, F.; Guerzoni, M.E.; Gobbetti, M.; Francavilla, R. Fecal Microbiota and Metabolome of Children with Autism and Pervasive Developmental Disorder Not Otherwise Specified. PLoS ONE 2013, 8, e76993. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Liang, S.; Wang, M.; Gao, J.; Sun, C.; Wang, J.; Xia, W.; Wu, S.; Sumner, S.J.; Zhang, F.; et al. Potential serum biomarkers from a metabolomics study of autism. J. Psych. Neurosci. 2016, 41, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Berding, K.; Donovan, S.M. Diet Can Impact Microbiota Composition in Children with Autism Spectrum Disorder. Front. Neurosci. 2018, 12, 515. [Google Scholar] [CrossRef] [Green Version]
- Coretti, L.; Paparo, L.; Riccio, M.P.; Amato, F.; Cuomo, M.; Natale, A.; Borrelli, L.; Corrado, G.; De Caro, C.; Comegna, M.; et al. Gut Microbiota Features in Young Children With Autism Spectrum Disorders. Front. Microbiol. 2018, 9, 3146. [Google Scholar] [CrossRef] [Green Version]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.W.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism—Comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, J.; Chen, H.; Li, Y.; Amakye, W.K.; Liang, J.; Ma, B.; Chu, X.; Mao, L.; Zhang, Z. Fecal Short-Chain Fatty Acids Levels Were Not Associated With Autism Spectrum Disorders in Chinese Children: A Case–Control Study. Front. Neurosci. 2019, 13, 1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharon, G.; Cruz, N.J.; Kang, D.-W.; Gandal, M.J.; Wang, B.; Kim, Y.-M.; Zink, E.M.; Casey, C.P.; Taylor, B.C.; Lane, C.J.; et al. Human Gut Microbiota from Autism Spectrum Disorder Promote Behavioral Symptoms in Mice. Cell 2019, 177, 1600–1618.e17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.-W.; Adams, J.B.; Gregory, A.C.; Borody, T.J.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 10. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.-W.; Adams, J.B.; Coleman, D.M.; Pollard, E.L.; Maldonado, J.; McDonough-Means, S.; Caporaso, J.G.; Krajmalnik-Brown, R. Long-term benefit of Microbiota Transfer Therapy on autism symptoms and gut microbiota. Sci. Rep. 2019, 9, 5821. [Google Scholar] [CrossRef] [PubMed]
- Grimaldi, R.; Gibson, G.R.; Vulevic, J.; Giallourou, N.; Castro-Mejía, J.; Hansen, L.H.; Gibson, E.L.; Nielsen, D.S.; Costabile, A. A prebiotic intervention study in children with autism spectrum disorders (ASDs). Microbiome 2018, 6, 133. [Google Scholar] [CrossRef]
- Wang, Y.; Li, N.; Yang, J.-J.; Zhao, D.-M.; Chen, B.; Zhang, G.-Q.; Chen, S.; Cao, R.-F.; Yu, H.; Zhao, C.-Y.; et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol. Res. 2020, 157, 104784. [Google Scholar] [CrossRef]
- Adams, J.B.; Vargason, T.; Kang, D.-W.; Krajmalnik-Brown, R.; Hahn, J. Multivariate Analysis of Plasma Metabolites in Children with Autism Spectrum Disorder and Gastrointestinal Symptoms Before and After Microbiota Transfer Therapy. Processes 2019, 7, 806. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.; Adams, J.; Vargason, T.; Santiago, M.; Hahn, J.; Krajmalnik-Brown, R. Distinct fecal and plasma metabolites in children with Autism Spectrum Disorders and their modulation after microbiota transfer therapy. mShere 2020, in press. [Google Scholar]
- Long, T.; Hicks, M.A.; Yu, H.-C.; Biggs, W.H.; Kirkness, E.F.; Menni, C.; Zierer, J.; Small, K.S.; Mangino, M.; Messier, H.; et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 2017, 49, 568–578. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 7, 179–188. [Google Scholar] [CrossRef]
- Cawley, G.C.; Talbot, N.L. Efficient leave-one-out cross-validation of kernel fisher discriminant classifiers. Pattern Recognit. 2003, 36, 2585–2592. [Google Scholar] [CrossRef]
- Loftfield, E.; Vogtmann, E.; Sampson, J.N.; Moore, S.C.; Nelson, H.; Knight, R.; Chia, N.; Sinha, R. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidem. Biomark. Prev. 2016, 25, 1483–1490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linden, A. Measuring diagnostic and predictive accuracy in disease management: An introduction to receiver operating characteristic (ROC) analysis. J. Eval. Clin. Pract. 2006, 12, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Demarquoy, C.; Demarquoy, J. Autism and carnitine: A possible link. World J. Boil. Chem. 2019, 10, 7–16. [Google Scholar] [CrossRef]
- Bryan, P.-F.; Carvajal, K.; Alejandro, M.-T.E.; Elva, E.-P.S.; Gemma, F.; Luz, C. Sphingolipids as Mediators in the Crosstalk between Microbiota and Intestinal Cells: Implications for Inflammatory Bowel Disease. Mediat. Inflamm. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zierer, J.; Jackson, M.A.; Kastenmüller, G.; Mangino, M.; Long, T.; Telenti, A.; Mohney, R.P.; Small, K.S.; Bell, J.T.; Steves, C.J.; et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 2018, 50, 790–795. [Google Scholar] [CrossRef]
- Gu, L.; Gonzalez, F.J.; Kalow, W.; Tang, B.K. Biotransformation of caffeine, paraxanthine, theobromine and theophylline by cDNA-expressed human CYP1A2 and CYP2E1. Pharmacogenetics 1992, 2, 73–77. [Google Scholar] [CrossRef]
- James, S.J.; Cutler, P.; Melnyk, S.; Jernigan, S.; Janak, L.; Gaylor, D.W.; Neubrander, J.A. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am. J. Clin. Nutr. 2004, 80, 1611–1617. [Google Scholar] [CrossRef] [Green Version]
- Vargason, T.; Kruger, U.; McGuinness, D.L.; Adams, J.B.; Geis, E.; Gehn, E.; Coleman, D.; Hahn, J. Investigating plasma amino acids for differentiating individuals with autism spectrum disorder and typically developing peers. Res. Autism Spectr. Disord. 2018, 50, 60–72. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevi, F.; Zolla, L.; Gabriele, S.; Persico, A.M. Urinary metabolomics of young Italian autistic children supports abnormal tryptophan and purine metabolism. Mol. Autism 2016, 7, 47. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Wan, J.; Rong, H.; He, F.; Wang, H.; Zhou, J.; Cai, C.; Wang, Y.; Xu, R.; Yin, Z.; et al. Alterations in Gut Glutamate Metabolism Associated with Changes in Gut Microbiota Composition in Children with Autism Spectrum Disorder. mSystems 2019, 4, e00321-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I. Betaine is a positive regulator of mitochondrial respiration. Biochem. Biophys. Res. Commun. 2015, 456, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, L.; Papaleo, V.; Porcelli, V.; Scarcia, P.; Gaita, L.; Sacco, R.; Hager, J.; Rousseau, F.; Curatolo, P.; Manzi, B.; et al. Altered calcium homeostasis in autism-spectrum disorders: Evidence from biochemical and genetic studies of the mitochondrial aspartate/glutamate carrier AGC1. Mol. Psychiatry 2008, 15, 38–52. [Google Scholar] [CrossRef] [Green Version]
- Frye, R.E.; Melnyk, S.; Macfabe, D.F. Unique acyl-carnitine profiles are potential biomarkers for acquired mitochondrial disease in autism spectrum disorder. Transl. Psychiatry 2013, 3, e220. [Google Scholar] [CrossRef] [Green Version]
- Frye, R.E. Mitochondrial Dysfunction in Autism Spectrum Disorder: Unique Abnormalities and Targeted Treatments. Semin. Pediatr. Neurol. 2020, 35, 100829. [Google Scholar] [CrossRef]
- Frye, R.E.; Rossignol, D.A. Mitochondrial Dysfunction Can Connect the Diverse Medical Symptoms Associated with Autism Spectrum Disorders. Pediatr. Res. 2011, 69, 41–47. [Google Scholar] [CrossRef]
- Orozco, J.S.; Hertz-Picciotto, I.; Abbeduto, L.; Slupsky, C.M. Metabolomics analysis of children with autism, idiopathic-developmental delays, and Down syndrome. Transl. Psychiatry 2019, 9, 243. [Google Scholar] [CrossRef] [Green Version]
- Karu, N.; Deng, L.; Slae, M.; Guo, A.C.; Sajed, T.; Huynh, H.; Wine, E.; Wishart, D.S. A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Anal. Chim. Acta 2018, 1030, 1–24. [Google Scholar] [CrossRef]
- Ousley, O.; Cermak, T. Autism Spectrum Disorder: Defining Dimensions and Subgroups. Curr. Dev. Disord. Rep. 2013, 1, 20–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacco, R.; Lenti, C.; Saccani, M.; Curatolo, P.; Manzi, B.; Bravaccio, C.; Persico, A.M. Cluster Analysis of Autistic Patients Based on Principal Pathogenetic Components. Autism Res. 2012, 5, 137–147. [Google Scholar] [CrossRef] [PubMed]


| Metabolite Rank | Metabolite | AUROC Week 0 | AUROC Week 18 | Pre/Post MTT p-Value | Associated Sub-Pathway |
|---|---|---|---|---|---|
| 1 | Carnitine | 0.77 | 0.68 | Carnitine Metabolism | |
| 2 | Sphingosine | 0.75 | 0.58 | Sphingolipid Metabolism | |
| 3 | 2′-deoxyadenosine | 0.75 | 0.59 | 0.02 | Purine Metabolism, Adenine containing |
| 4 | Indole | 0.74 | 0.72 | Tryptophan Metabolism | |
| 5 | Adenine | 0.74 | 0.81 | Purine Metabolism, Adenine containing | |
| 6 | N-stearoyl-sphingosine (d18:1/18:0) | 0.73 | 0.61 | Sphingolipid Metabolism | |
| 7 | Imidazole Propionate | 0.71 | 0.64 | Histidine Metabolism | |
| 8 | 10-nonadecenoate (19:1n9) | 0.71 | 0.59 | 0.01 | Long Chain Fatty Acid |
| 9 | p-cresol sulfate | 0.71 | 0.53 | Phenylalanine and Tyrosine Metabolism | |
| 10 | Cystathionine | 0.71 | 0.65 | Methionine, Cysteine, SAM and Taurine Metabolism | |
| 11 | 5alpha-androstan-3beta,17alpha-diol monosulfate (1) | 0.71 | 0.63 | 0.01 | Steroid |
| 12 | 3-(3-hydroxyphenyl)propionate | 0.71 | 0.66 | Phenylalanine and Tyrosine Metabolism | |
| 13 | 1-(1-enyl-oleoyl)-GPE (P-18:1) | 0.71 | 0.87 | Lysoplasmalogen | |
| 14 | Deoxy-carnitine | 0.71 | 0.58 | Carnitine Metabolism | |
| 15 | Gamma-glutamyl-histidine | 0.71 | 0.68 | Gamma-glutamyl Amino Acid | |
| 16 | Diaminopimelate | 0.70 | 0.62 | Food Component/Plant | |
| 17 | Tyramine O-sulfate | 0.70 | 0.70 | Phenylalanine and Tyrosine Metabolism | |
| 18 | Gulonate | 0.70 | 0.53 | Ascorbate and Aldarate Metabolism | |
| 19 | gamma-tocotrienol | 0.70 | 0.62 | Tocopherol Metabolism | |
| 20 | 4-hydroxyphenylacetate | 0.70 | 0.56 | Phenylalanine and Tyrosine Metabolism | |
| 21 | Delta-tocopherol | 0.70 | 0.53 | Tocopherol Metabolism | |
| 22 | Phenethylamine | 0.69 | 0.55 | Phenylalanine and Tyrosine Metabolism | |
| 23 | Propionyl-glycine (C3) | 0.69 | 0.61 | Sphingolipid Metabolism | |
| 24 | N-acetyl-sphingosine | 0.69 | 0.63 | Sphingolipid Metabolism | |
| 25 | Betaine | 0.69 | 0.60 | Glycine, Serine and Threonine Metabolism | |
| 26 | Adenosine | 0.69 | 0.65 | Purine Metabolism, Adenine containing | |
| 27 | Ornithine | 0.68 | 0.51 | Urea cycle; Arginine and Proline Metabolism | |
| 28 | N-palmitoyl-sphingosine (d18:1/16:0) | 0.68 | 0.54 | 0.03 | Sphingolipid Metabolism |
| 29 | Galactonate | 0.68 | 0.67 | Fructose, Mannose and Galactose Metabolism | |
| 30 | N1-Methyl-2-pyridone-5-carboxamide | 0.68 | 0.74 | Nicotinate and Nicotinamide Metabolism | |
| 31 | 1-palmitoylglycerol (16:0) | 0.68 | 0.54 | Monoacylglycerol | |
| 32 | Phosphocholine | 0.67 | 0.59 | Phospholipid Metabolism | |
| 33 | Theobromine | 0.67 | 0.62 | 0.02 | Xanthine Metabolism |
| 34 | 3,5-dihydroxybenzoic acid | 0.67 | 0.60 | Food Component/Plant | |
| 35 | Hydroxyproline | 0.67 | 0.56 | Urea cycle; Arginine and Proline Metabolism | |
| 36 | l-urobilin | 0.67 | 0.60 | 0.02 | Hemoglobin and Porphyrin Metabolism |
| 37 | carboxyethyl-GABA | 0.67 | 0.55 | Glutamate Metabolism | |
| 38 | oxalate (ethane-dioate) | 0.67 | 0.53 | Ascorbate and Aldarate Metabolism | |
| 39 | Palmitoyl-carnitine (C16) | 0.67 | 0.53 | Fatty Acid Metabolism(Acyl Carnitine) | |
| 40 | Copro-stanol | 0.67 | 0.60 | Sterol | |
| 41 | Saccharopine | 0.66 | 0.57 | Lysine Metabolism | |
| 42 | 5-hydroxylysine | 0.66 | 0.53 | Lysine Metabolism | |
| 43 | Stearoyl-carnitine (C18) | 0.66 | 0.58 | Fatty Acid Metabolism(Acyl Carnitine) | |
| 44 | Biliverdin | 0.66 | 0.60 | Hemoglobin and Porphyrin Metabolism | |
| 45 | 3-(4-hydroxyphenyl)lactate (HPLA) | 0.66 | 0.59 | Phenylalanine and Tyrosine Metabolism | |
| 46 | Carnosine | 0.66 | 0.57 | Dipeptide Derivative | |
| 47 | 10-hydroxystearate | 0.66 | 0.64 | 0.01 | Fatty Acid, Monohydroxy |
| 48 | Pentadecanoate (15:0) | 0.66 | 0.55 | Long Chain Fatty Acid | |
| 49 | Hexadecanedioate (C16) | 0.66 | 0.60 | Fatty Acid, Dicarboxylate | |
| 50 | Sphinganine | 0.66 | 0.54 | Sphingolipid Metabolism |
| Number of Metabolites | Metabolite Combination | Fitted AUROC | Cross-Validated Results | ||
|---|---|---|---|---|---|
| β | TPR | TNR | |||
| 2 | Carnitine 2′deoxyadenosine | 0.88 | 0.01 | 1.00 | 0.20 |
| 0.05 | 1.00 | 0.35 | |||
| 0.10 | 1.00 | 0.50 | |||
| 0.20 | 0.89 | 0.75 | |||
| 3 | Adenosine theobromine hydroxyproline | 0.93 | 0.01 | 1.00 | 0.30 |
| 0.05 | 0.94 | 0.55 | |||
| 0.10 | 0.94 | 0.75 | |||
| 0.20 | 0.72 | 0.85 | |||
| 4 | indole | 0.98 | 0.01 | 0.94 | 0.35 |
| 1-(1-enyl-oleoyl)-GPE (P-18:1) | 0.05 | 0.89 | 0.60 | ||
| Hydroxyproline | 0.10 | 0.83 | 0.60 | ||
| Carnosine | 0.20 | 0.83 | 0.80 | ||
| 5 | Imidazole Propionate | 1.00 | 0.01 | 1.00 | 0.85 |
| Hydroxyproline | 0.05 | 0.94 | 0.95 | ||
| Theobromine | 0.10 | 0.89 | 0.95 | ||
| 2-hydroxy-3-methylvalerate Adenosine | 0.20 | 0.78 | 0.95 | ||
| 5 | Imidazole Propionate | 1.00 | 0.01 | 1.00 | 0.85 |
| Hydroxyproline | 0.05 | 0.94 | 0.95 | ||
| Theobromine | 0.10 | 0.89 | 0.95 | ||
| 2-hydroxy-3-methylvalerate | 0.20 | 0.78 | 0.95 | ||
| Indole | |||||
| Metabolite | Correlation Coefficient | p-Value |
|---|---|---|
| Indole | ||
| Carnitine | 0.67 | <0.001 |
| Indole-lactate | 0.56 | <0.001 |
| Saccharopine | 0.42 | 0.007 |
| Stearoyl-carnitine | 0.39 | 0.015 |
| 3-(3-hydroxyphenyl)propionate | 0.33 | 0.043 |
| Oxalate | 0.33 | 0.043 |
| Imidazole Propionate | ||
| Galactonate | 0.78 | <0.001 |
| Gulonate | 0.76 | <0.001 |
| Palmitoyl-carnitine | 0.72 | <0.001 |
| Saccharopine | 0.7 | <0.001 |
| Phosphocholine | 0.69 | <0.001 |
| Cystathionine | 0.62 | <0.001 |
| Phenethylamine | 0.61 | <0.001 |
| Betaine | 0.61 | <0.001 |
| 3-(4-hydroxyphenyl)lactate | 0.6 | <0.001 |
| N-propionyl-methionine | 0.58 | <0.001 |
| N-palmitoyl-sphingosine | 0.41 | 0.011 |
| 3,5-dihydroxybenzoic | 0.4 | 0.014 |
| 3-(3-hydroxyphenyl)propionate | 0.39 | 0.017 |
| Stearoyl-carnitine | 0.38 | 0.018 |
| 1-palmitoylglycerol | 0.37 | 0.023 |
| Gamma-glutamyl-histidine | 0.36 | 0.027 |
| Biliverdin | 0.34 | 0.037 |
| Carnitine | 0.32 | 0.048 |
| Adenosine | ||
| Adenine | 0.74 | <0.001 |
| 2′-deoxyadenosine | 0.54 | <0.001 |
| 5-hydroxylysine | 0.36 | 0.0254 |
| Hydroxyproline | 0.36 | 0.0256 |
| 5-hydroxylysine | 0.36 | 0.0254 |
| 1-(1-enyl-oleoyl)-GPE | 0.34 | 0.0366 |
| Theobromine ** | ||
| None | ||
| Hydroxyproline | ||
| 2-hydroxy-3-methylvalerate *** | 0.61 | <0.001 |
| delta-tocopherol | 0.41 | 0.011 |
| 2′-deoxyadenosine | 0.38 | 0.017 |
| Adenosine | 0.36 | 0.026 |
| Copro-stanol | 0.36 | 0.026 |
| 5alpha-androstan-3beta,17alpha-diol | 0.35 | 0.030 |
| p-cresol | −0.32 | 0.050 |
| Betaine | −0.33 | 0.043 |
| Oxalate | −0.35 | 0.031 |
| N-palmitoyl-sphingosine | −0.37 | 0.023 |
| 2-hydroxy-3-methylvalerate *** | ||
| Gulonate | 0.81 | <0.001 |
| Imidazole propionate | 0.79 | <0.001 |
| Galactonate | 0.78 | <0.001 |
| Phosphocholine | 0.75 | <0.001 |
| 5-hydroxylysine | 0.72 | <0.001 |
| Hydroxyproline | 0.61 | <0.001 |
| Betaine | 0.6 | <0.001 |
| Phenethylamine | 0.59 | <0.001 |
| 1-(1-enyl-oleoyl)-GPE (P-18:1) | 0.55 | <0.001 |
| Cystathionine | 0.53 | <0.001 |
| 1-palmitoylglycerol (16:0) | 0.51 | <0.001 |
| Biliverdin | 0.51 | 0.001 |
| Propionyl-glycine (C3) | 0.45 | 0.005 |
| 3-(4-hydroxyphenyl)lactate (HPLA) | 0.43 | 0.008 |
| 3-(3-hydroxyphenyl)propionate | 0.34 | 0.039 |
| Delta-tocopherol | −0.33 | 0.042 |
| Copro-stanol | −0.35 | 0.033 |
| ASD Week 0 | ASD Week 3 | ASD Week 10 | ASD Week 18 | TD Week 0 | |
|---|---|---|---|---|---|
| Imidazole Propionate (25th/75th percentile) | 0.37 (0.19, 3.38) | 0.55 (0.08, 8.57) | 0.29 (0.08, 0.80) | 0.14 (0.03, 0.46) | 0.12 (0.09, 0.29) |
| Hydroxyproline (25th/75th percentile) | 0.96 (0.72, 4.83) | 1.06 (0.42, 3.67) | 1.27 (0.24, 3.34) | 0.80 (0.54, 3.60) | 0.60 (0.29, 1.20) |
| Theobromine (25th/75th percentile) | 0.89 (0.47, 2.38) | 0.47 (0.47, 1.68) | 0.47 (0.16, 0.47) | 0.47 (0.43, 0.47) | 0.46 (0.34, 0.64) |
| 2-hydroxy-3-methylvalerate (25th/75th percentile) | 0.53 (0.43, 0.75) | 0.43 (0.18, 0.56) | 0.34 (0.06, 0.50) | 0.52 (0.21, 0.63) | 0.44 (0.21, 0.61) |
| Indole (25th/75th percentile) | 1.13 (0.25, 1.83) | 0.66 (0.32, 1.75) | 0.85 (0.18, 1.86) | 0.86 (0.26, 1.52) | 0.39 (0.15, 0.59) |
| Adenosine (25th/75th percentile) | 0.67 (0.36, 0.88) | 0.77 (0.50, 1.01) | 0.73 (0.47, 0.90) | 0.57 (0.26, 0.86) | 0.40 (0.18, 0.86) |
| OFM-I Median discriminant score (25th/75th percentile) | 3.90 (2.33, 5.72) | 1.90 (0.72, 9.52) | 1.84 (0.90, 3.71) | 1.73 (0.71, 2.62) | 0.46 (0.21, 1.35) |
| Type II error | 5% | 53% | 50% | 56% | - |
| OFM-A Median discriminant score (25th/75th percentile) | 3.51 (2.28, 5.73) | 2.87 (1.13, 9.43) | 2.18 (1.07, 4.18) | 1.36 (0.54, 2.44) | 0.62 (0.35, 1.05) |
| Type II error | 5% | 53% | 39% | 56% | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qureshi, F.; Adams, J.; Hanagan, K.; Kang, D.-W.; Krajmalnik-Brown, R.; Hahn, J. Multivariate Analysis of Fecal Metabolites from Children with Autism Spectrum Disorder and Gastrointestinal Symptoms before and after Microbiota Transfer Therapy. J. Pers. Med. 2020, 10, 152. https://doi.org/10.3390/jpm10040152
Qureshi F, Adams J, Hanagan K, Kang D-W, Krajmalnik-Brown R, Hahn J. Multivariate Analysis of Fecal Metabolites from Children with Autism Spectrum Disorder and Gastrointestinal Symptoms before and after Microbiota Transfer Therapy. Journal of Personalized Medicine. 2020; 10(4):152. https://doi.org/10.3390/jpm10040152
Chicago/Turabian StyleQureshi, Fatir, James Adams, Kathryn Hanagan, Dae-Wook Kang, Rosa Krajmalnik-Brown, and Juergen Hahn. 2020. "Multivariate Analysis of Fecal Metabolites from Children with Autism Spectrum Disorder and Gastrointestinal Symptoms before and after Microbiota Transfer Therapy" Journal of Personalized Medicine 10, no. 4: 152. https://doi.org/10.3390/jpm10040152
APA StyleQureshi, F., Adams, J., Hanagan, K., Kang, D.-W., Krajmalnik-Brown, R., & Hahn, J. (2020). Multivariate Analysis of Fecal Metabolites from Children with Autism Spectrum Disorder and Gastrointestinal Symptoms before and after Microbiota Transfer Therapy. Journal of Personalized Medicine, 10(4), 152. https://doi.org/10.3390/jpm10040152






