Visual Interpretation of Convolutional Neural Network Predictions in Classifying Medical Image Modalities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
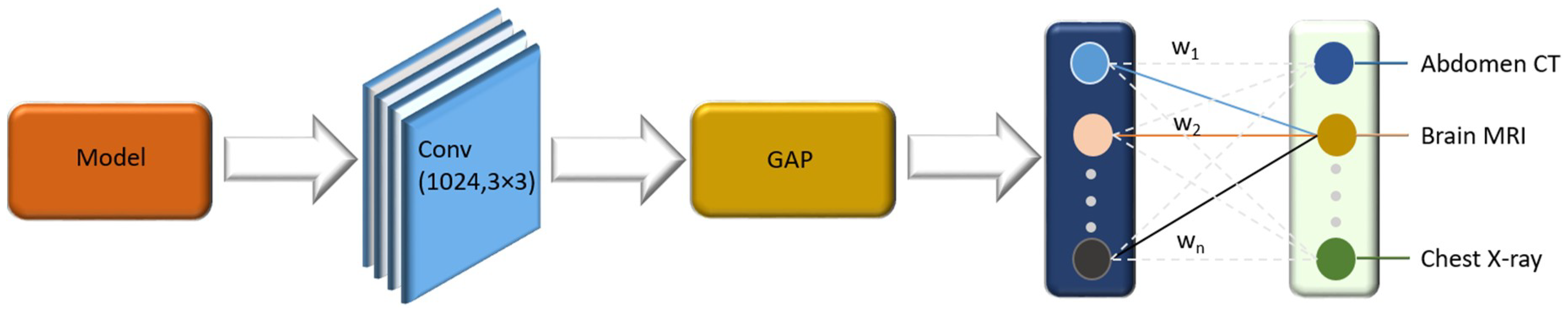
2.2. Convolutional Neural Network (CNN) Configuration
3. Class-Selective Relevance Map (CRM)
3.1. Class Activation Map (CAM)
3.2. Gradient-Weighted Class Activation Map (Grad-CAM)
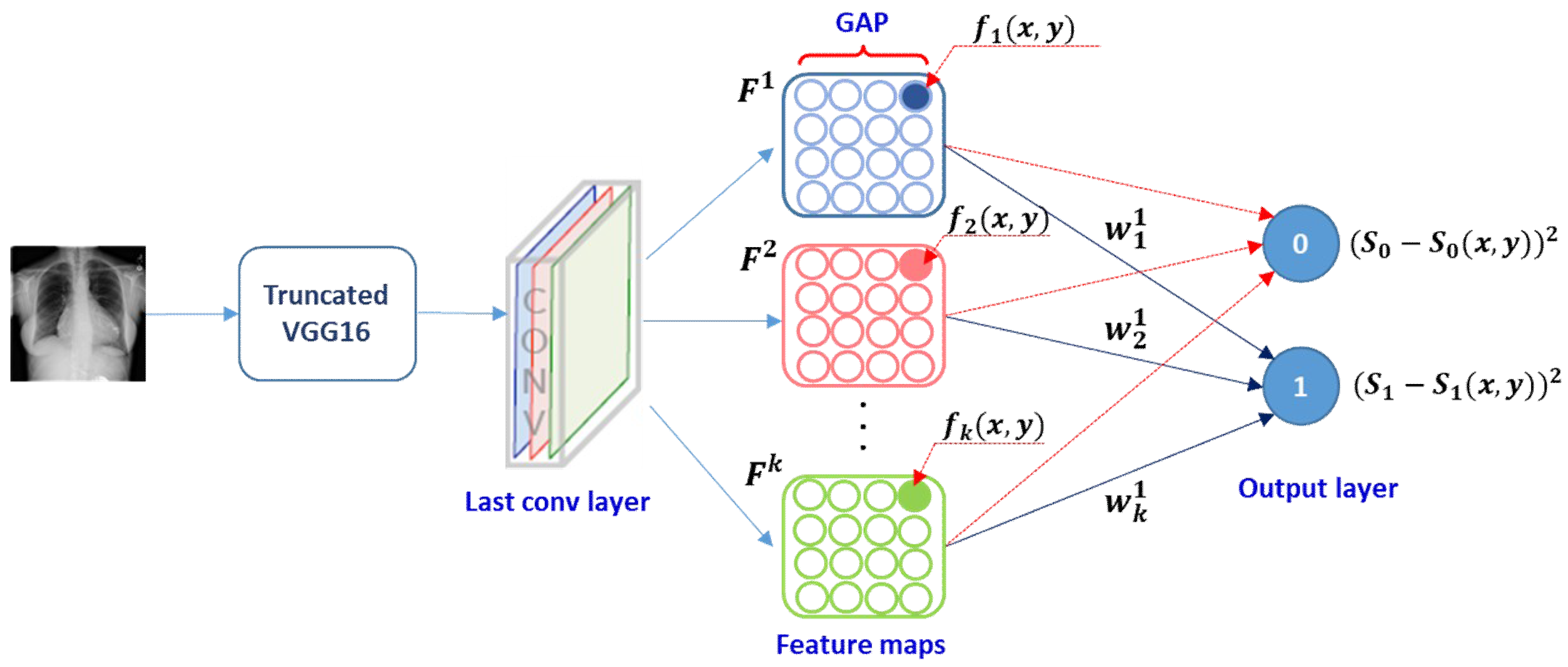
3.3. Class-Selective Relevance Map (CRM)
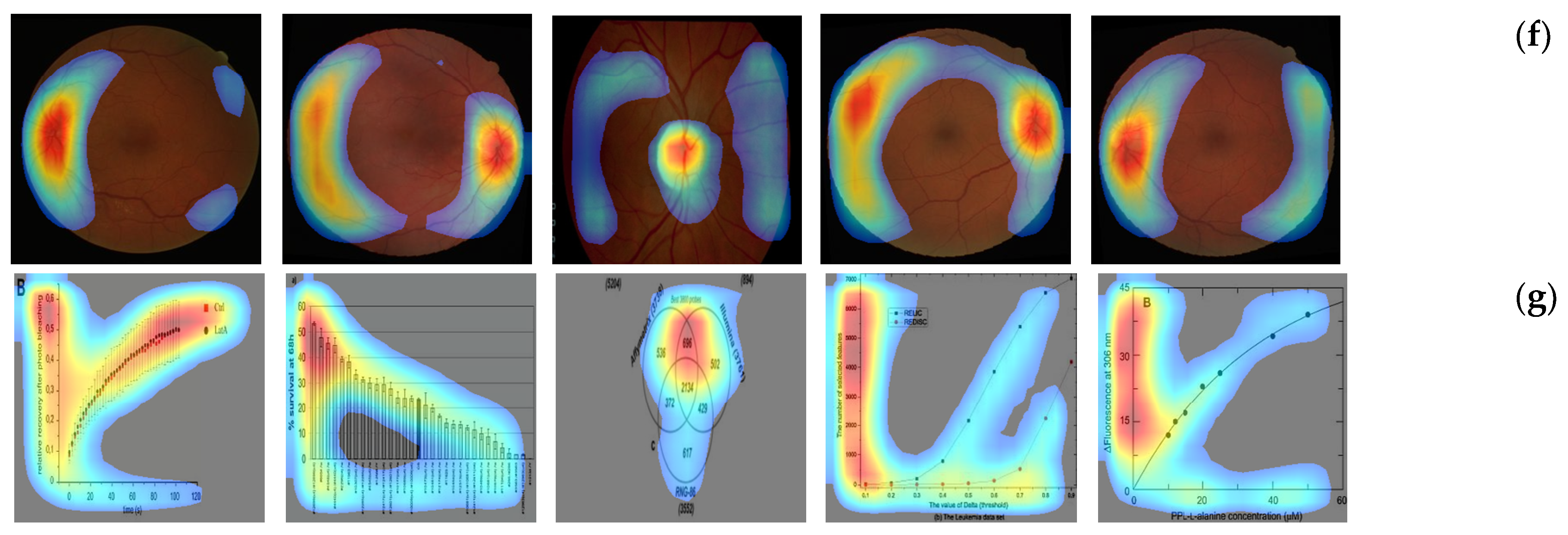
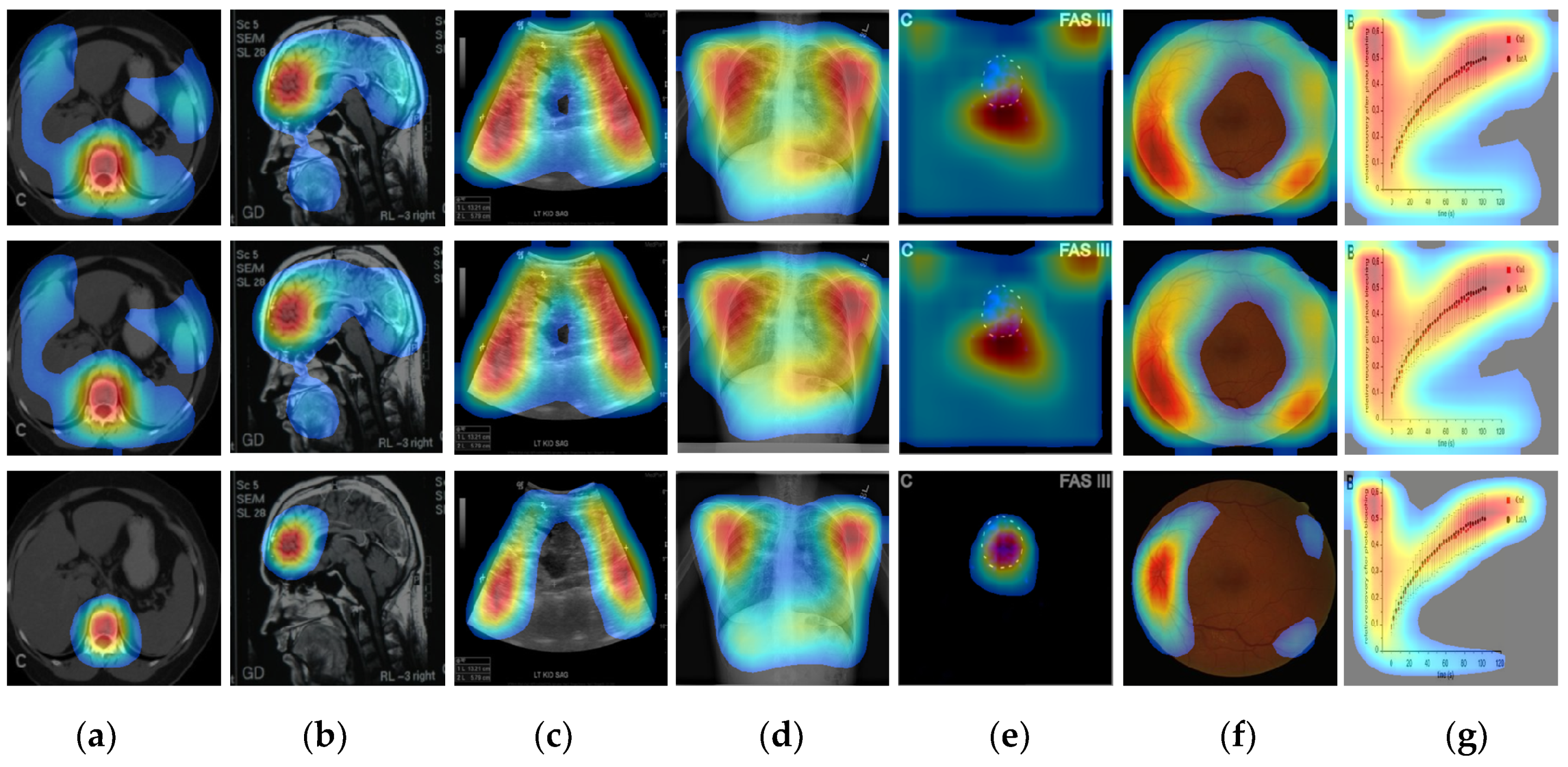
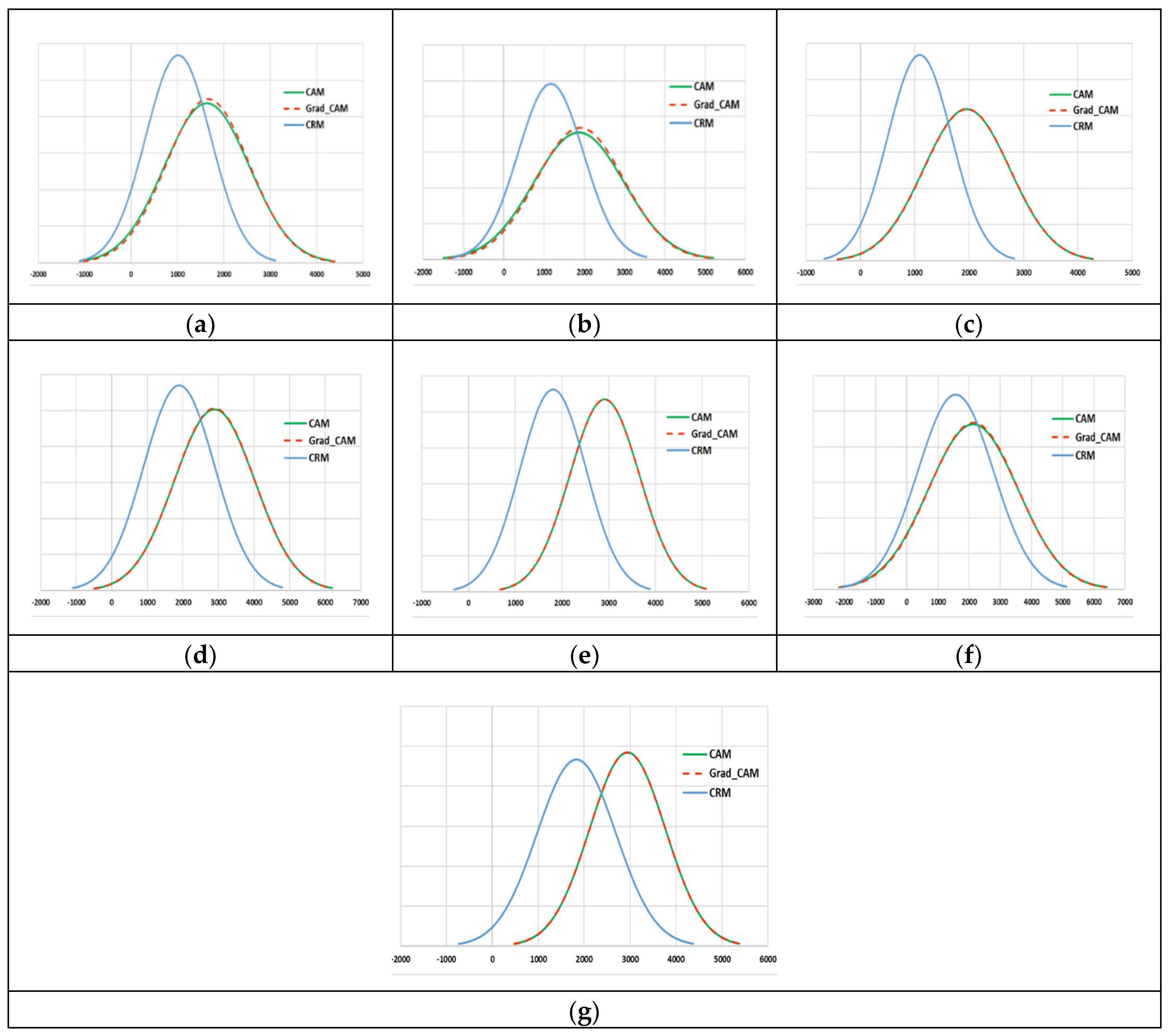
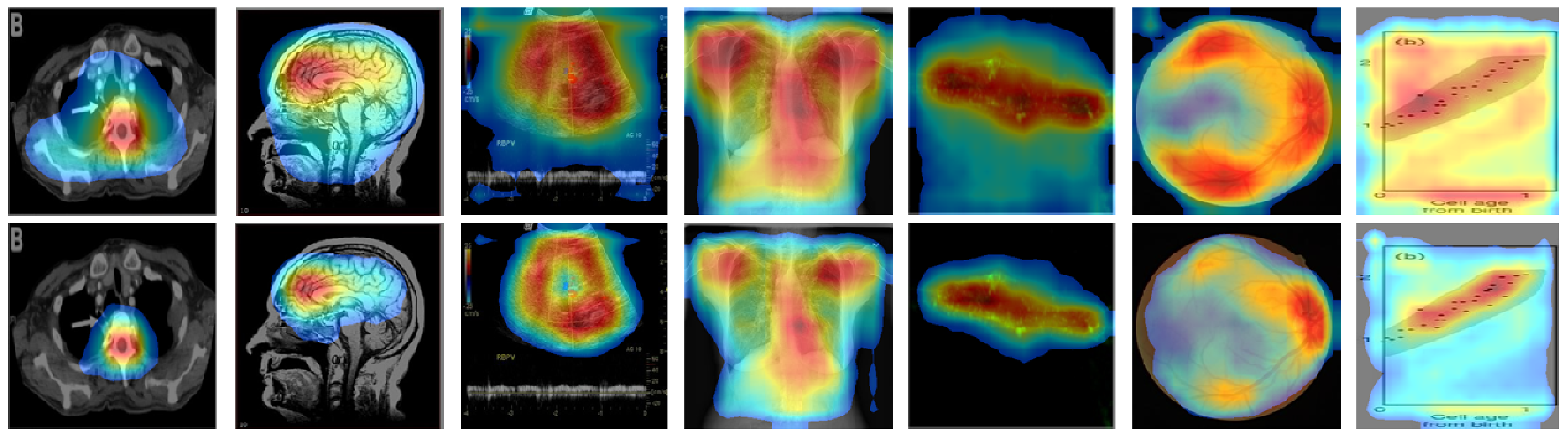
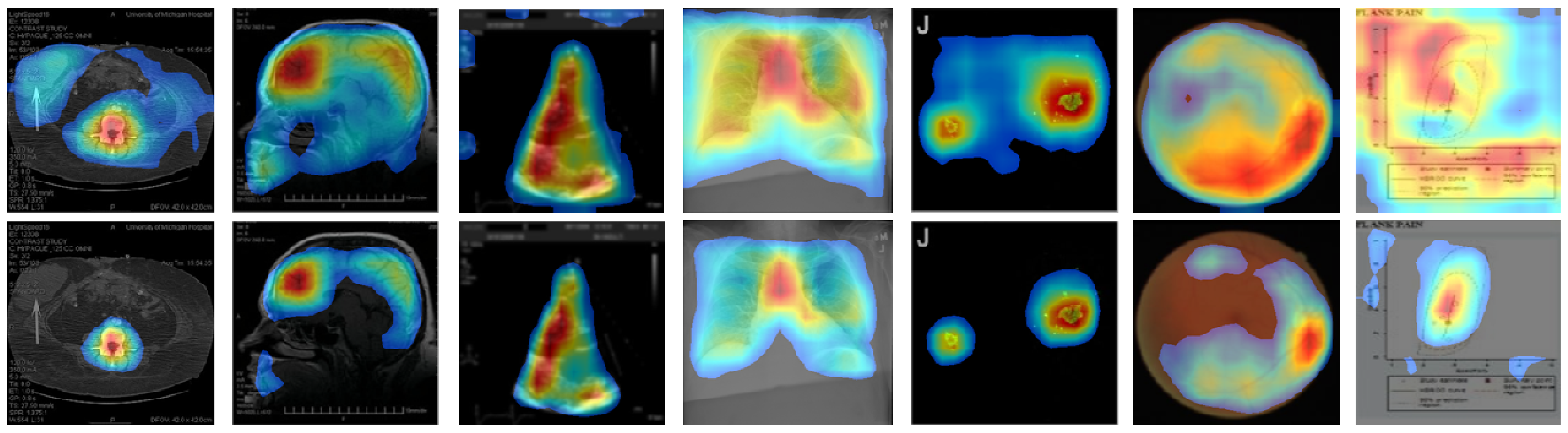
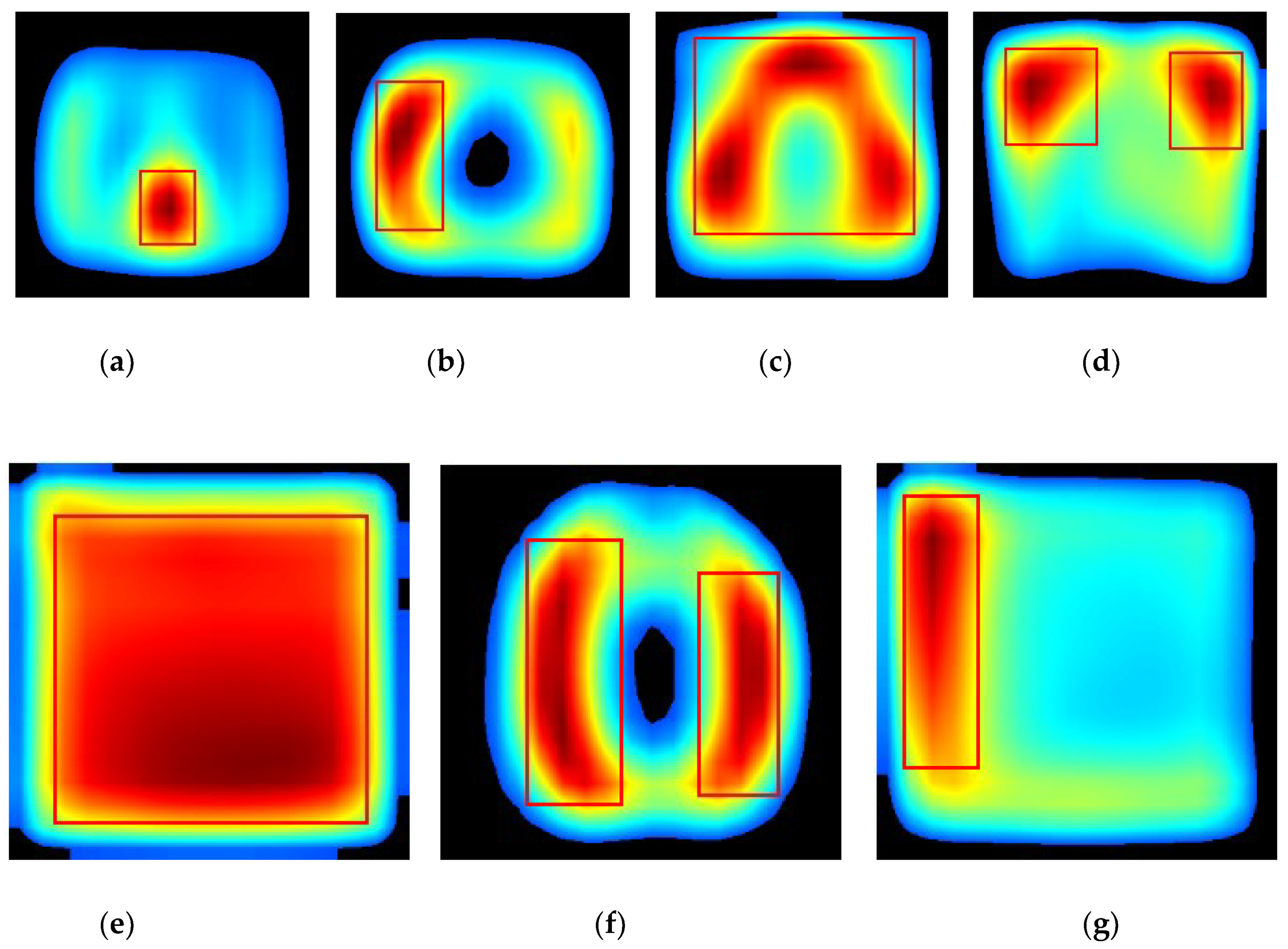
4. Results and Discussion
4.1. Performance Evaluation
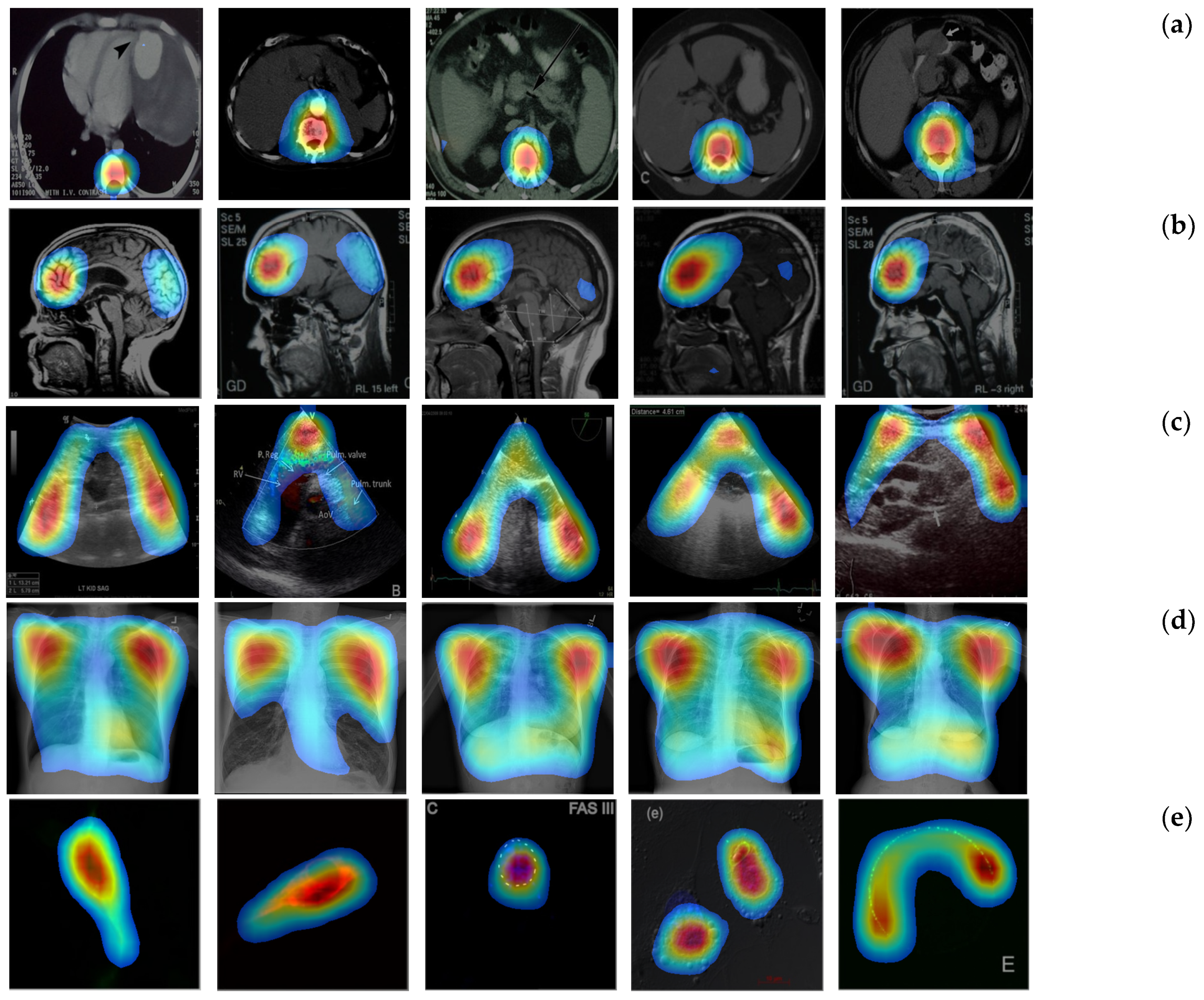
4.2. Localization Evaluation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Montani, S.; Bellazzi, R. Supporting decisions in medical applications: The knowledge management perspective. Int. J. Med. Inform. 2002, 68, 79–90. [Google Scholar] [CrossRef]
- Demner-Fushman, D.; Antani, S.; Thoma, G.R.; Simpson, M. Design and development of a multimodal biomedical Information retrieval system. J. Comput. Sci. Eng. 2012, 6, 168–177. [Google Scholar] [CrossRef]
- De Herrera, A.G.S.; Schaer, R.; Bromuri, S.; Müller, H. Overview of the imageCLEF 2016 medical task. In Proceedings of the CEUR Workshop, Évora, Portugal, 5–8 September 2016; Volume 1609, pp. 219–232. [Google Scholar]
- De Herrera, A.G.S.; Markonis, D.; Müller, H. Bag-of-colors for biomedical document image classification. In Proceedings of the MCBR-CDS 2012—MICCAI International Workshop on Medical Content-Based Retrieval for Clinical Decision Support, Nice, France, 1 October 2012; Lecture Notes in Computer Science. Greenspan, H., Müller, H., Syeda-Mahmood, T., Eds.; Springer; Volume 7723, pp. 110–121. [Google Scholar]
- Pelka, O.; Friedrich, C.M. FHDO biomedical computer science group at medical classification task of ImageCLEF 2015. In Proceedings of the CEUR Workshop, Toulouse, France, 8–11 September 2015; Volume 1391. [Google Scholar]
- Cirujeda, P.; Binefa, X. Medical image classification via 2D color feature based covariance descriptors. In Proceedings of the CEUR Workshop, Toulouse, France, 8–11 September 2015; Volume 1391. [Google Scholar]
- Li, P.; Sorensen, S.; Kolagunda, A.; Jiang, X.; Wang, X.; Kambhamettu, C.; Shatkay, H. UDEL CIS at imageCLEF medical task 2016. In Proceedings of the CEUR Workshop, Évora, Portugal, 5–8 September 2016; Volume 1609, pp. 334–346. [Google Scholar]
- De Herrera, A.G.S.; Kalpathy-Cramer, J.; Demner-Fushman, D.; Antani, S.; Müller, H. Overview of the ImageCLEF 2013 medical tasks. In Proceedings of the CEUR Workshop, Valencia, Spain, 23–26 September 2013; Volume 1179. [Google Scholar]
- Zhang, Y.D.; Dong, Z.; Chen, X.; Jia, W.; Du, S.; Muhammad, K.; Wang, S.H. Image based fruit category classification by 13-layer deep convolutional neural network and data augmentation. Multimed. Tools Appl. 2019, 78, 3613–3632. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Wang, S.H.; Muhammad, K.; Hong, J.; Sangaiah, A.K.; Zhang, Y.D. Alcoholism identification via convolutional neural network based on parametric ReLU, dropout, and batch normalization. Neural Comput. Appl. 2018, 1–16. [Google Scholar] [CrossRef]
- Yu, Y.; Lin, H.; Yu, Q.; Meng, J.; Zhao, Z.; Li, Y.; Zuo, L. Modality classification for medical images using multiple deep convolutional neural networks. J. Comput. Inf. Syst. 2015, 11, 5403–5413. [Google Scholar]
- Koitka, S.; Friedrich, C.M. Traditional feature engineering and deep learning approaches at medical classification task of imageCLEF 2016. In Proceedings of the CEUR Workshop, Évora, Portugal, 5–8 September 2016; Volume 1609, pp. 304–317. [Google Scholar]
- Kumar, A.; Kim, J.; Lyndon, D.; Fulham, M.; Feng, D. An ensemble of fine-tuned convolutional neural networks for medical image classification. IEEE J. Biomed. Heath Inform. 2017, 21, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Lin, H.; Meng, J.; Wei, X.; Guo, H.; Zhao, Z. Deep transfer learning for modality classification of medical images. Information 2017, 8, 91. [Google Scholar] [CrossRef]
- Zhang, J.; Xia, Y.; Wu, Q.; Xie, Y. Classification of medical images and illustrations in the biomedical literature using synergic deep learning. arXiv, 2017; arXiv:physics/1706.09092. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the 13th ECCV 2014—European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; Lecture Notes in Computer Science. Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer; Volume 8689, pp. 818–833. [Google Scholar]
- Mahendran, A.; Vedaldi, A. Understanding deep image representations by inverting them. In Proceedings of the IEEE Conference on Computer Vision Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 5188–5196. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2921–2929. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. In Proceedings of the International conference, Computer Vision Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 618–626. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Decencière, E.; Zhang, X.; Cazuguel, G.; Laÿ, B.; Cochener, B.; Trone, C.; Gain, P.; Ordóñez-Varela, J.R.; Massin, P.; Erginay, A.; et al. Feedback on a publicly distributed image database: The Messidor database. Image Anal. Stereol. 2014, 33, 231–234. [Google Scholar] [CrossRef]
- Staal, J.J.; Abramoff, M.D.; Niemeijer, M.; Viergever, M.A.; van Ginneken, B. Ridge based vessel segmentation in color images of the retina. IEEE Trans. Med. Imaging 2004, 23, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Bergstra, J.; Bengio, Y. Random search for hyper-parameter optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Abadi, M.; Agarwal, A.; Barham, P. Tensorflow: Large-scale Machine Learning on Heterogeneous Distributed Systems. Software. 2015. Available online: tensorflow.org (accessed on 10 December 2018).
- Chollet, F. Keras. GitHub. 2015. Available online: https://github.com/fchollet/keras (accessed on 10 December 2018).
- Mozer, M.C.; Smolensky, P. Using relevance to reduce network size automatically. Connect. Sci. 1989, 1, 3–16. [Google Scholar] [CrossRef]
- Erdogan, S.S.; Ng, G.S.; Patrick, K.H.C. Measurement criteria for neural network pruning. In Proceedings of the IEEE Conference TENCON, Perth, Australia, 26–29 November 1996; Volume 1, pp. 83–89. [Google Scholar]
- Kim, I.; Chien, S. Speed-up of error backpropagation algorithm with class-selective relevance. Neurocomputing 2002, 48, 1009–1014. [Google Scholar] [CrossRef]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception architecture for computer vision. In Proceedings of the IEEE Conference Computer Vision Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 2818–2826. [Google Scholar]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference Computer Vision Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
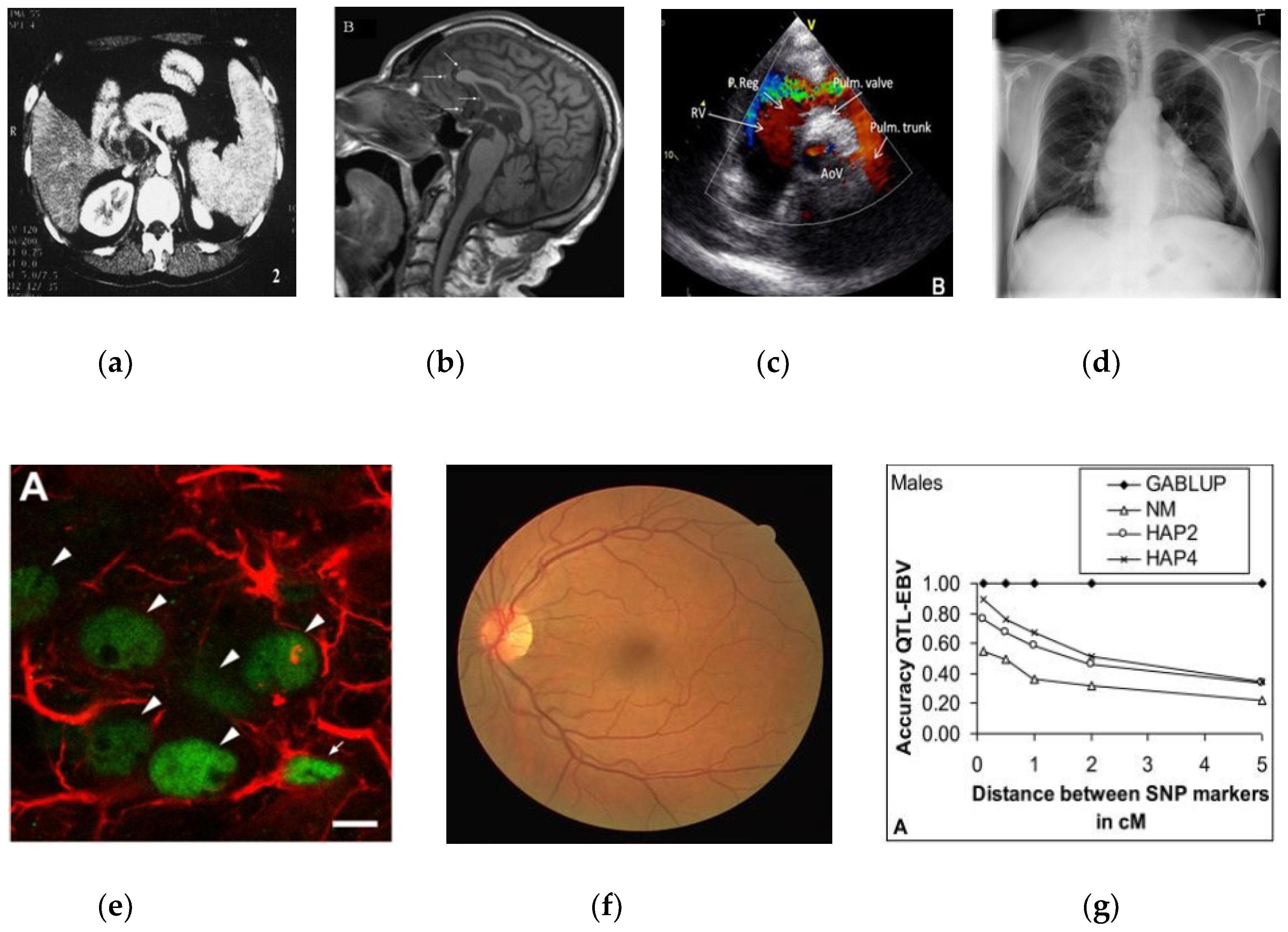
| Category | Samples | Training | Validation | Testing | File Type | Bit-Depth |
|---|---|---|---|---|---|---|
| Abdominal CT | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Brain MRI | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Chest X-ray | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Cardiac abdomen ultrasound | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Fluorescence microscopy | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Retinal fundoscopy | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Statistical graphs | 6000 | 5000 | 500 | 500 | JPG | 8-bit |
| Model | Accuracy | AUC | Recall | Precision | F1-score | MCC |
|---|---|---|---|---|---|---|
| VGG16 | 0.98 | 0.994 | 0.98 | 0.981 | 0.98 | 0.986 |
| Methods | Abdomen CT | Brain MRI | Cardiac Abdomen Ultrasound | Chest X-ray | Fluorescence Microscopy | Retinal Fundoscopy | Statistical Graphs |
|---|---|---|---|---|---|---|---|
| CAM | 49,477 (55.0) | 54,894 (61.0) | 56,972 (63.3) | 76,488 (85.0) | 79,900 (88.8) | 58,514 (65.0) | 82,444 (91.6) |
| Grad-CAM | 49,478 (55.0) | 54,896 (61.0) | 56,973 (63.3) | 76,488 (85.0) | 79,901 (88.8) | 58,515 (65.0) | 82,445 (91.6) |
| CRM | 26,596 (29.6) | 32,298 (35.9) | 28,966 (32.2) | 57,363 (63.7) | 52,448 (58.3) | 43,334 (48.1) | 52,932 (58.8) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, I.; Rajaraman, S.; Antani, S. Visual Interpretation of Convolutional Neural Network Predictions in Classifying Medical Image Modalities. Diagnostics 2019, 9, 38. https://doi.org/10.3390/diagnostics9020038
Kim I, Rajaraman S, Antani S. Visual Interpretation of Convolutional Neural Network Predictions in Classifying Medical Image Modalities. Diagnostics. 2019; 9(2):38. https://doi.org/10.3390/diagnostics9020038
Chicago/Turabian StyleKim, Incheol, Sivaramakrishnan Rajaraman, and Sameer Antani. 2019. "Visual Interpretation of Convolutional Neural Network Predictions in Classifying Medical Image Modalities" Diagnostics 9, no. 2: 38. https://doi.org/10.3390/diagnostics9020038
APA StyleKim, I., Rajaraman, S., & Antani, S. (2019). Visual Interpretation of Convolutional Neural Network Predictions in Classifying Medical Image Modalities. Diagnostics, 9(2), 38. https://doi.org/10.3390/diagnostics9020038







