1. Introduction
The concept of Spectro-Fluor™ technology is mainly based on the discovery of characteristic C-F fingerprint(s) in the spectral area of 500–900 cm
−1 allowing detection, characterization, imaging, monitoring, screening and measurement of fluoro-nanomaterials, fluoro-compounds, fluoro-molecules, fluoroorganic impurities or fluoro-degradation products [
1,
2,
3]. Spectro-Fluor™
aka Carbon-Fluorine Spectroscopy™ (CFS™) is one of the key tools of our in-house technological platform called PLIRFA™ (Pulsed Laser Isochronic Raman and Fluorescence Apparatus™), which provides alternative solutions and applications across the biomedical (e.g., molecular/cell/tissue imaging), life (e.g., unravelling of molecular interactions, structures, conformations), pharmaceutical (e.g., drug design, delivery, tracing, discovery), environmental (e.g., characterization of pesticides, other contaminants), and nano(bio)technological (e.g., characterization of nano-molecules/materials) fields [
1,
2]. Thereby, Spectro-Fluor™ represents an innovative, disruptive, green, affordable, cost-effective, reliable and flexible analytical tool of high resolution, sensitivity and specificity (
Figure 1) [
1,
2,
3]. Interestingly, this technology constitutes a major advance over conventional spectroscopy technologies (e.g., conventional Raman tools) because of its capability to: (i) highly specific and ultrasensitive detect (ppm-ppb level or <10 cells) C-F bonds (–C-F, =CF
2, –CF
3); (ii) detect and characterize any halo-organic bonds or unlabeled molecules, regardless their physical state (solid, liquid or gas), via glass and polymer containers, quartz vials; (iii) acquire data in real-time (0.1 second per data point from 1000 average pulses); (iv) quantitatively determine any fluorinated substance since the emitted C-F bond(s) signal is directly proportional to the analyte concentration; (v) unravel chemical structures (e.g., molecular length determination, molecular analogs differentiation with as low as one carbon resolution); (vi) preserve the sample integrity (
i.e., non-destructive and non-invasive technology) allowing re-using of the sample; (vii) analyze any unprepared sample in a solvent-less/green fashion; (viii) easily provide data interpretation thanks to its interconnection with powerful software and in-house developed databases; (ix) be used with low maintenance/calibration requirements; (x) operate using adequate time-gating and time-delays with a single laser source functioning in three modes: continuous detection, pulsed detection and two photons excitation/time resolved fluorescence; (xi) be conveniently employed anywhere due to new generation of portable/compact devices incorporating fiber optics; (xii) be versatile, flexible, progressive and hyphenated opening possibilities to be coupled/hybridized to diverse technology platforms such as high-throughput screening (HTS) (e.g., arrays, multiplex assays systems), microscopy and other imaging systems (e.g., confocal laser scanning microscopy, atomic force microscopy (AFM), positron emission tomography (PET), magnetic resonance imaging (MRI), computed tomography (CT)), chromatography (e.g., high-performance liquid chromatography (HPLC), mass spectrometry (MS)), sorting instruments (e.g., fluorescence-activated cell sorting (FACS) or magnetic-activated cell sorting (MACS)).
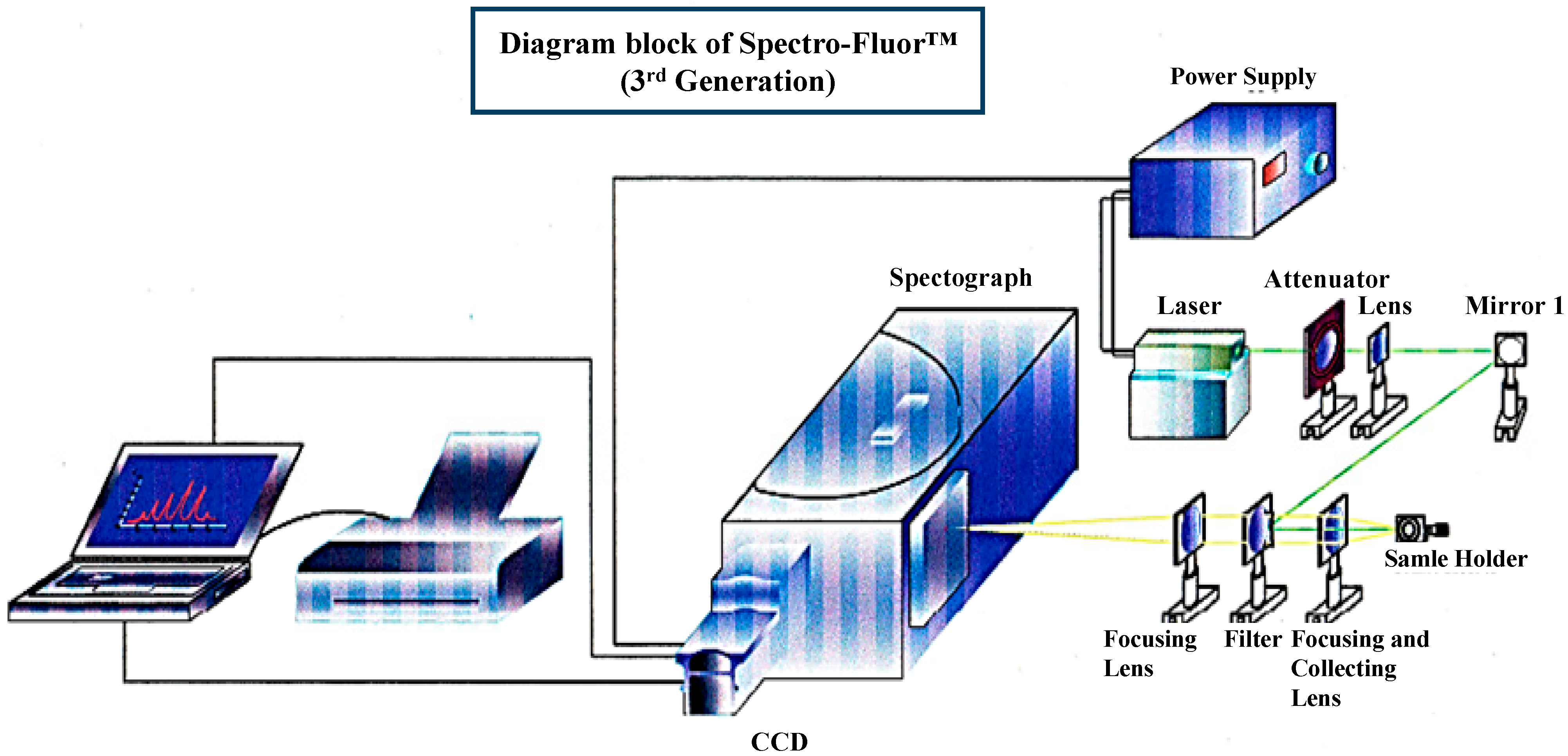
Figure 1.
3D-diagram of the Spectro-Fluor™ technology. This patented apparatus represents the third generation of the device, and includes a laser with power unit, a spectrograph, an ICCD camera with image intensifier, a desktop computer with appropriate software for data analysis. The next generation will represent a compact and portable device with focusing lenses replaced by fiber optics.
Figure 1.
3D-diagram of the Spectro-Fluor™ technology. This patented apparatus represents the third generation of the device, and includes a laser with power unit, a spectrograph, an ICCD camera with image intensifier, a desktop computer with appropriate software for data analysis. The next generation will represent a compact and portable device with focusing lenses replaced by fiber optics.
Besides, the specific features of fluorine in the formation of fluorocarbons make it attractive in the design of non-viscous but polar organic compounds, with a polarity limited to influencing the intra-molecular nature and inter-molecular interactions with the microenvironment [
4,
5]. Interestingly, carbon-fluorination displays the additional following properties [
6,
7]: (i) enhancement of thermal stability (10
7 Kcal/mol); (ii) increase of lipophilicity, hydrophobicity and solubility; (iii) improvement of the molecular bioavailability (e.g., diminution of the basicity of neighboring amines); and (iv) size comparable to H (
i.e., 1.47 Å
versus 1.20 Å) capable of mimicking enzyme substrates. Importantly, C-F bonds are [
1,
2,
3,
4]: (i) unique in nature; (ii) smaller and more stable than fluorescent dyes due to their covalent interactions; (iii) much less toxic than radioisotopes avoiding handling and logistic problems; (iv) less harmful at long-term exposure than radio-waves; and (v) inexpensively incorporated into molecules, compounds, materials or cells. Therefore, C-F bond(s) can be used in a number of industrial and academic areas including pharmacy, medicine and nanotechnology to characterize them, monitor them or confer some enhanced physicochemical features [
1,
2,
3,
4]. In terms of applications, a C-F bond can be employed as [
1,
2,
3,
4,
8,
9,
10,
11]: (i) a pharmaceutical security label to enhance the drug safety, thereby preventing and banning illegitimate drugs (
i.e., counterfeited or sub-standard drugs); (ii) a nano-label to enable drug trace during the whole pharmaceutical development process (
i.e., discovery, synthesis and production cycles, clinical and post-approval stages), subsequently bringing the product faster to the market; (iii) an
ex vivo,
in situ or
in vivo label for biologics (e.g., peptides, proteins including antibodies, nucleic acids) and cells/tissues (e.g., stem cells, tumor cells); (iv) a functional chemical group for nanomaterials (e.g., nanoparticles, nanocarbon tubes, nanogels, nanoemulsions, nanoporous silica glasses).
Nowadays, although several techniques and technologies are employed to detect and characterize proteins and biomarkers of disease are available in the market (e.g., enzyme-linked immunosorbent assay (ELISA), Western blot), we propose here an original molecular method to quickly, reliably and cost-effectively detect a protein of interest (e.g., biomarker of disease) from cancer cells using the unique Spectro-Fluor™ biotechnology.
3. Results and Discussion
We have developed an innovative photonic biotool called Spectro-Fluor™ Technology (Fluorotronics, Inc.) as well as a new
in vitro method based on
ex vivo incorporation of fluorinated amino acids into cells in order to detect lowly-expressed molecules such as biomarkers of disease. (
Figure 1 and
Figure 2). Indeed, the advantages of Spectro-Fluor™ Technology (
aka Carbon-Fluorine spectroscopy) over routinely used laboratory methods (e.g., Western blots, ELISA, protein dot blots) are represented by the rapid, safe, reliable, highly sensitive and specifique detection of C-F bond(s) formed into molecules [
1].
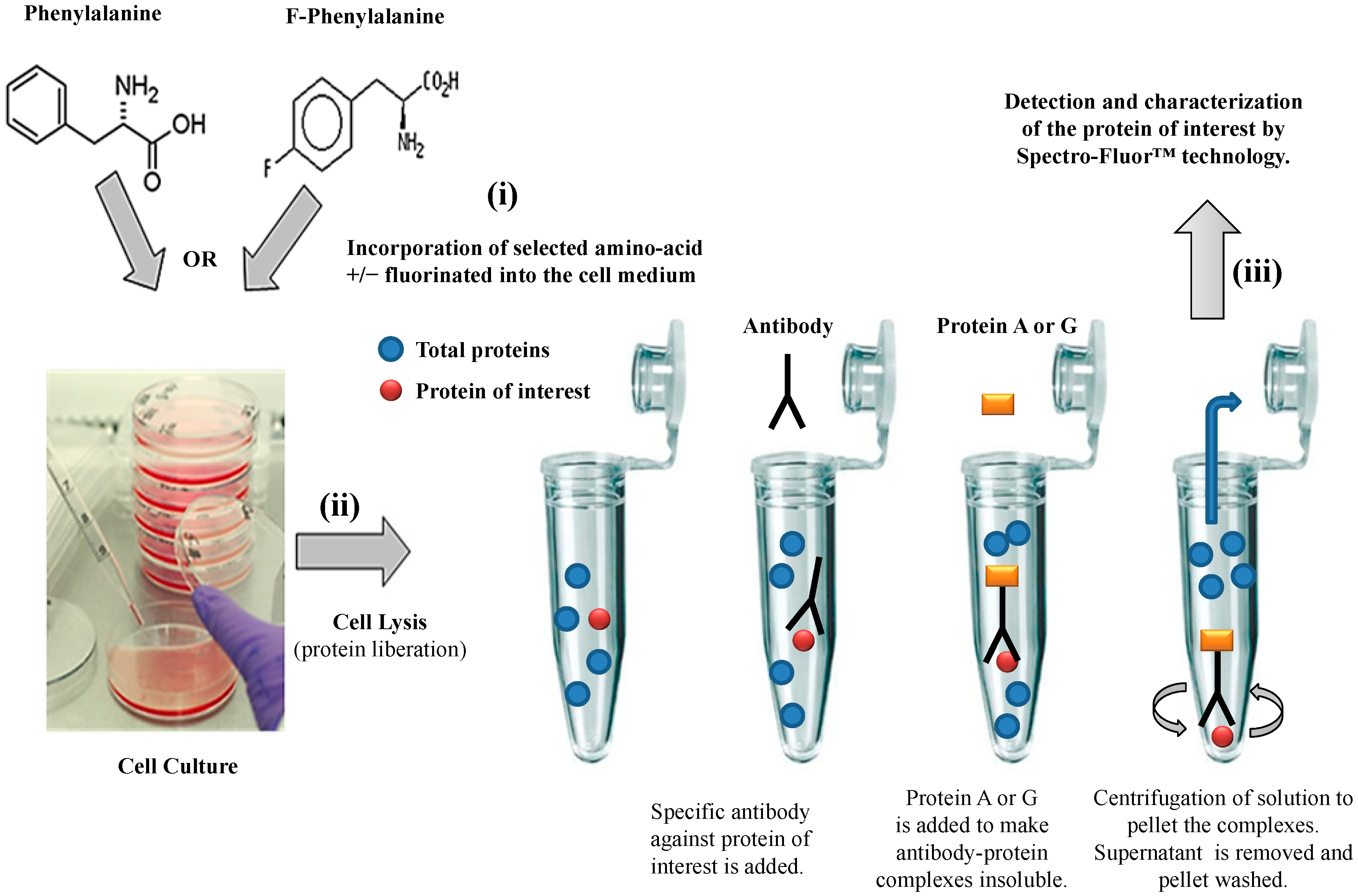
We have established a valuable
ex vivo protein labeling protocol that consisted of three steps (
Figure 2): (i) addition in excess (e.g., in this study, 10 mg/mL as non-limiting concentration) of a selected aromatic fluoro-aminoacid (e.g., in this study, 4-fluoro-
l-phenylalanine) into an adequate medium-containing cells (e.g., in this study, human colon carcinoma HCT-116 cells). In parallel, the corresponding non-modified amino-acid (e.g., in this study,
l-phenylalanine) was used as control; (ii) immediately after the amino-acid addition (control) and after a convenient period of time (e.g., in this study, 24 h), simultaneous isolation of both F-p21 and p21 (control) was performed by immune-precipitation (IP). IP is a procedure by which proteins or peptides that react specifically with an antibody are removed from solution and examined for quantity or physical characteristics. The expression of the two p21 protein forms (
i.e., F-p21 and p21) was routinely checked by Western blot, a widely used biochemical technique albeit long and expensive (data not shown due to low basal expression detectable with Western blot); (iii) comparative examination of the respective spectra obtained by Spectro-Fluor™ technology obtained in same experimental conditions.
Figure 2.
Flow chart of the experimental methodology. The three-step process is resumed by: (i) Ex-vivo protein labeling by incorporation of either 4-fluoro-l-phenylalanine. l-phenylalanine is used as a control; (ii) immune-precipitation by using only one antibody against the antigen/epitope of the protein of interest; (iii) detection and characterization of the protein of interest by Spectro-Fluor™ technology.
Figure 2.
Flow chart of the experimental methodology. The three-step process is resumed by: (i) Ex-vivo protein labeling by incorporation of either 4-fluoro-l-phenylalanine. l-phenylalanine is used as a control; (ii) immune-precipitation by using only one antibody against the antigen/epitope of the protein of interest; (iii) detection and characterization of the protein of interest by Spectro-Fluor™ technology.
This protocol could have been reduced in two steps (i.e., steps 2 and 3) if a fluorinated antibody against our protein of interest was available at the time of our experiments (pending patent).
The reasons why we opted for the phenylanine (Phe/F) aminoacid are the following: (i)
l-Phe is an aromatic aminoacid, and it is known that molecules containing aromatic rings better scatter the light [
1]; (ii)
l-Phe is present twice more (
n = 6) in the p21 sequence compared to the two other aromatic aminoacids which are tyrosine (Tyr/Y) and tryptophane (Trp/W) at the number of three (
Figure 3A,B and
Table 1). Even so, we also realized a parallel work with tyrosine (
i.e., 5-fluoro-
dl-tyrosine) or and tryptophane (
i.e., 5-fluoro-
dl-tryptophane), which led to same conclusions, albeit the best Spectro-Fluor™ signal was obtained with phenylalanine (data not shown).
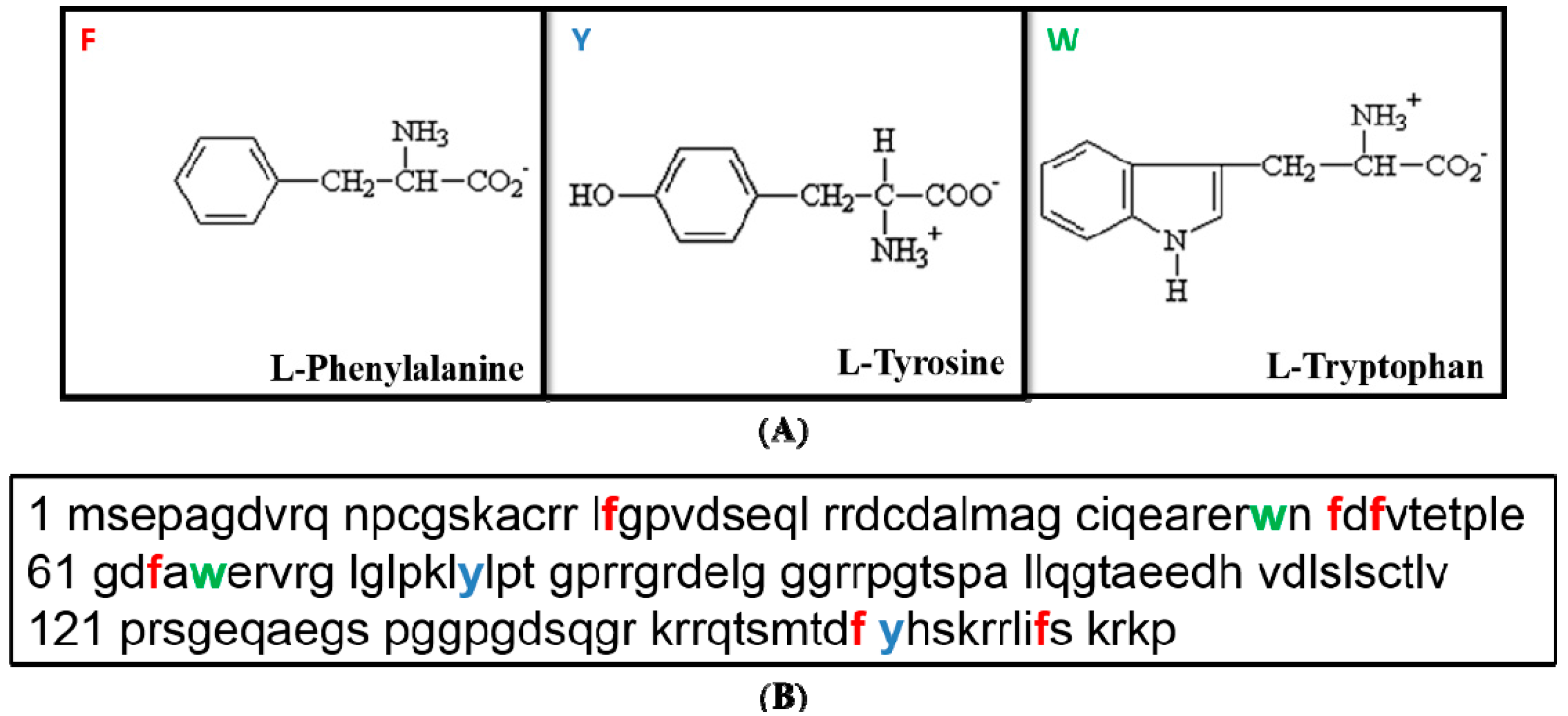
Figure 3.
Tertiary sequence of p21. (
A) Structure of the three aromatic acids,
i.e., Phe, Tyr and Trp. Phe contains only hydrophobic group whereas Tyr and Trp have both hydrophobic and hydrophilic side groups. (
B) Relative frequency of the three aromatic amino acids in the p21 primary structure,
i.e., Phenylalanine/Phe/F (in red) is at number of 6, Tyrosine/Tyr/Y (in blue) and Tryptophane/Trp/W (in green) are at number of three [
12].
Figure 3.
Tertiary sequence of p21. (
A) Structure of the three aromatic acids,
i.e., Phe, Tyr and Trp. Phe contains only hydrophobic group whereas Tyr and Trp have both hydrophobic and hydrophilic side groups. (
B) Relative frequency of the three aromatic amino acids in the p21 primary structure,
i.e., Phenylalanine/Phe/F (in red) is at number of 6, Tyrosine/Tyr/Y (in blue) and Tryptophane/Trp/W (in green) are at number of three [
12].
Table 1.
Single-letter amino-acid code, In accordance to the international genetic system.
Table 1.
Single-letter amino-acid code, In accordance to the international genetic system.
| Full Name of Amino-Acid | Single-Letter Code | Three-Letters Code |
|---|
| Glycine | G | Gly |
| Alanine | A | Ala |
| Leucine | L | Leu |
| Methionine | M | Met |
| Phenylalanine | F | Phe |
| Tryptophan | W | Trp |
| Lysine | K | Lys |
| Glutamine | Q | Gln |
| Glutamic Acid | E | Glu |
| Serine | S | Ser |
| Proline | P | Pro |
| Valine | V | Val |
| Isoleucine | I | Ile |
| Cysteine | C | Cys |
| Tyrosine | Y | Tyr |
| Histidine | H | His |
| Arginine | R | Arg |
| Asparagine | N | Asn |
| Aspartic Acid | D | Asp |
| Threonine | T | Thr |
The choice of the p21 protein (
aka WAF1, CAP20, Cip1 or Sdi1) is explained by the important following facts: (i) p21 is a key transcriptional factor, a cyclin kinase inhibitor (CKI), and a downstream target of p53, the so-called “guardian of the genome” potentially acting as an immune-suppressor. Thereby, p21 elicits pleiotropic biological functions of major biomedical interest (e.g., cell cycle arrest, gene expression regulation, DNA replication, DNA response to DNA damage, p53-dependent or -independent apoptosis (
aka programmed cell death), tumor suppression) [
13,
14,
15,
16,
17,
18,
19,
20,
21]; (ii) p21 is a small protein (21 kDa, 164 base pairs) and its basal expression under unstressed conditions is barely detectable, rending its characterization difficult albeit certain genotoxic conditions, such as ionizing radiation, can be a good way to see it up-regulated [
21]. In this regard, Spectro-Fluor™ technology, along with our molecular approach, constitutes a robust biotechnology to qualitatively and quantitatively determine p21. Further, the whole process is cost-effectively notably due to: (i) the use of only one antibody; (ii) the possibility to re-use the samples; (iii) the capability of Spectro-Fluor™ technology to reliably confirm the tissue-specific presence of p21 when coupled to conventional or advanced imaging systems.
Interestingly, the introduction of fluorine, a chemical element with the highest electronegativity, is known to affect the optical property of amino acids in both frequency and intensity [
4,
22]. Thereby, the main vibrational assignments of F-p21 obtained by Spectro-Fluor™ are selectively represented by a weaker band between 635–670 cm
−1, a C-F optical signature detected around 770 cm
−1, and an intense band detected at 1002–1060 cm
−1 including a slightly weaker band at 1039 cm
−1 indicative of a mono-substituted benzene ring vibration due to successful incorporation of phenylalanine residues into p21 (
Figure 4A,B,
Figure 5 and
Table 2).
Figure 4.
Spectro-Fluor™ spectra of the purified and enriched protein of interest. (A) Spectra in RIPA buffer of immune-precipitated p21 protein forms “before” (Time 0) and after (Time 24 h) effective incorporation of F-Phe (red spectrum) or non-labeled Phe (blue spectrum). 10 mg/mL of either amino-acid was added into McCoy’s 5a medium-containing HCT-116 colon cancer cells. (B) The F-p21 protein detection was performed after several washes with PBS 1X to significantly reduce the background generated by the RIPA buffer. The C-F signal is detected around 770 cm−1 and the aromatic bond optical assignment of the incorporated phenylalanine is expectedly observed at 1002 cm−1.
Figure 4.
Spectro-Fluor™ spectra of the purified and enriched protein of interest. (A) Spectra in RIPA buffer of immune-precipitated p21 protein forms “before” (Time 0) and after (Time 24 h) effective incorporation of F-Phe (red spectrum) or non-labeled Phe (blue spectrum). 10 mg/mL of either amino-acid was added into McCoy’s 5a medium-containing HCT-116 colon cancer cells. (B) The F-p21 protein detection was performed after several washes with PBS 1X to significantly reduce the background generated by the RIPA buffer. The C-F signal is detected around 770 cm−1 and the aromatic bond optical assignment of the incorporated phenylalanine is expectedly observed at 1002 cm−1.
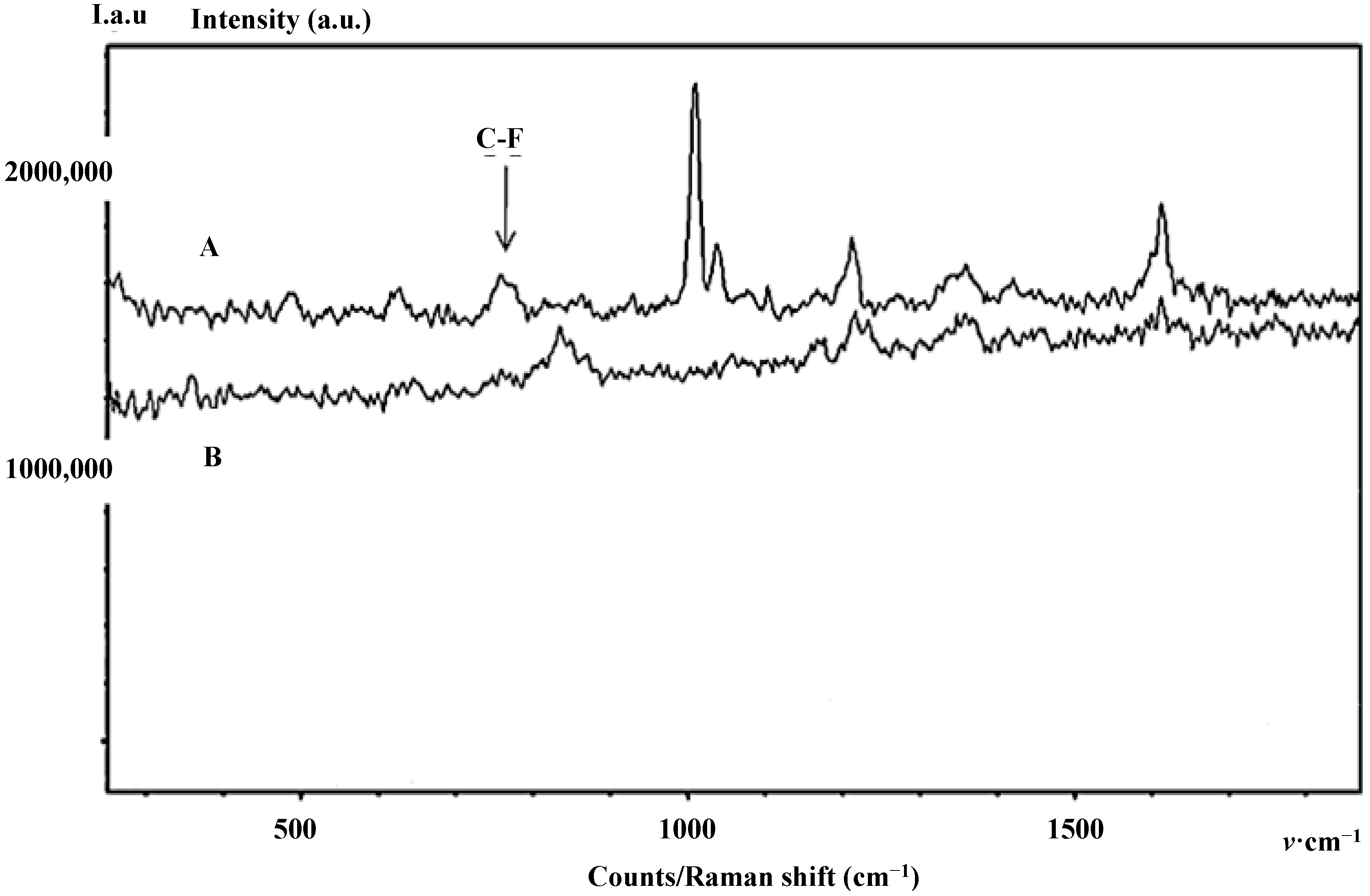
Figure 5.
Background-Free Spectro-Fluor™ spectra of the purified and enriched protein of interest. Spectra in RIPA buffer of immune-precipitated p21 protein forms after (Time 24 h) effective incorporation of F-Phe (A) or non-labeled Phe. (B) 10 mg/mL of either amino-acid was added into McCoy’s 5a medium-containing HCT-116 colon cancer cells. In this experiment, the p21 protein forms detection was performed after several washes with PBS 1X to significantly reduce the background generated by the RIPA buffer. The C-F signal is detected around 770 cm−1 and the aromatic bond optical assignment of the incorporated F-Phe is expectedly observed at 1002 cm−1. It is interesting to note that, in our experimental conditions, the signal of F-Phe incorporated in p21 is very specific when compared with the control. Loss of sensitivity is certainly due to the few washings preformed to eliminate the fluorescence background generated by the RIPA buffer.
Figure 5.
Background-Free Spectro-Fluor™ spectra of the purified and enriched protein of interest. Spectra in RIPA buffer of immune-precipitated p21 protein forms after (Time 24 h) effective incorporation of F-Phe (A) or non-labeled Phe. (B) 10 mg/mL of either amino-acid was added into McCoy’s 5a medium-containing HCT-116 colon cancer cells. In this experiment, the p21 protein forms detection was performed after several washes with PBS 1X to significantly reduce the background generated by the RIPA buffer. The C-F signal is detected around 770 cm−1 and the aromatic bond optical assignment of the incorporated F-Phe is expectedly observed at 1002 cm−1. It is interesting to note that, in our experimental conditions, the signal of F-Phe incorporated in p21 is very specific when compared with the control. Loss of sensitivity is certainly due to the few washings preformed to eliminate the fluorescence background generated by the RIPA buffer.
Table 2.
Main Spectro-Fluor™ vibrational assignments of the p21 protein.
Table 2.
Main Spectro-Fluor™ vibrational assignments of the p21 protein.
| Band (cm-1) | Vibrational mode | Mainly observed in: | References |
|---|
| 670 | C–S stretching | Cys | [22] |
| 766 | Symmetric ring breathing | Amino-acids, Proteins | [22] |
| 855 | Ring breathing | Tyr, DNA | [22] |
| 925 | C–C stretching | Proline, Valine, Protein | [22] |
| 1060 | C–C stretching | Lipids, Nucleic acids, Proteins | [22] |
| 1250–1270 | C–N stretch amide III | Proteins | [22] |
| 1300 | C–H2 | Lipids, Proteins | [22] |
| 1440–1470 | CH2 deformation | Proteins | [22] |
| 2580 | S–H stretch | Thiol group of Cys, Methionine | [22] |
| 2884 | C–H2 | Lipids | [22] |
| 2940 | C–H | Lipids, Proteins | [22] |
Expectedly, similar and low spectrum intensity of both F-p21 and p21 were noticed after immediate incorporation of F-Phe or Phe into the medium-containing HCT-116 cells. In this context, the cells would not have sufficient time to incorporate the respective Phe aminoacid. Nevertheless, the difficult spectral discrimination at early time between the two p21 forms has been overcome after 24 h of cells culture. Indeed, the spectral intensity obtained with F-p21 was greater (
i.e., amplitude about 1.5×), compared to that of p21 (
Figure 4A).
In order to eliminate the potential background due to two fluoride constituents in the RIPA buffer (
i.e., NaF (Sodium Fluoride), a phosphatase inhibitor and PMSF (PhenylMethylSulfonyl Fluoride), a protease inhibitor), which were required to preserve the protein integrity during immune-precipitation, we washed the samples several times with 1× PBS. Further, to avoid any misinterpretations regarding the sensitivity levels, three independent experiments for the four samples have been conducted in the same conditions. In agreement with our expectations, the amplitude of the signal intensity differed much when between F-p21 and p21 than between Y-p21 and p21 or between W-p21 and p21 (data not shown). This effect can be easily explained by the greater number of phenylalanine residues (
n = 6) in the p21 protein sequence when compared with the equal twice-lower number of tyrosine and tryptophan residues (
n = 3) (
Figure 3B).
Taken together, our results are encouraging and strongly suggest that Spectro-Fluor™ technology could be reliably used for the detection and characterization of bioactive molecules. Although fluorination is a common process used to enhance properties of pharmaceutics [
1], our present study pioneered the possibility to F-label proteins, including antibodies and biomarkers of diseases, which could be assessed by or used with Spectro-Fluor™ technology to reach a number of clinical objectives.
4. Conclusions
Fluorinated molecules, widely used in medicinal chemistry (e.g., drug design), emerge in the biomedical field (e.g., fluorinated bio-probes and theranostics™ such as F-nucleic acids, F-peptides) because of their special features (e.g., enhanced bioavailability and stability). Pharmacological studies suggest that fluorine modification may significantly reduce undesirable drug side-effects meantime offering superior efficacy. Therefore, we postulated that fluorinated proteins (e.g., biomarkers of disease, antibodies) should also be beneficial. Within the PLIRFA™ technology platform, which includes progressive analytical devices, we developed a becoming gold-standard Spectro-Fluor™ technology as well as a panel of green methods, enabling rapid, cost-effective, sensitive and specific detection and characterization of F-molecules, F-compounds, F-cells, F- tissues or F-(nano-)biomaterials. In this study, we successively show the possibility to detect a fluorinated protein (i.e., p53-downstream target p21) by Spectro-Fluor™ technology. This strongly suggests that this innovative biotechnology could significantly offer alternative solutions for a wide number of applications (e.g., molecular design, molecular imaging, development of F-antibodies) and technologies routinely used in laboratories and hospitals (e.g., western-blot, enzyme-linked immune sorbent assay, mass-spectrometry, MRI, PET). Eventually, we report that Spectro-Fluor™ technology is able to reduce the troublesome detection associated with low-expressed molecules (e.g., p21).