The Role of ctDNA for Diagnosis and Histological Prediction in Early Stage Non-Small-Cell Lung Cancer: A Narrative Review
Abstract
1. Introduction
2. Methods
3. Literature Research Outcome
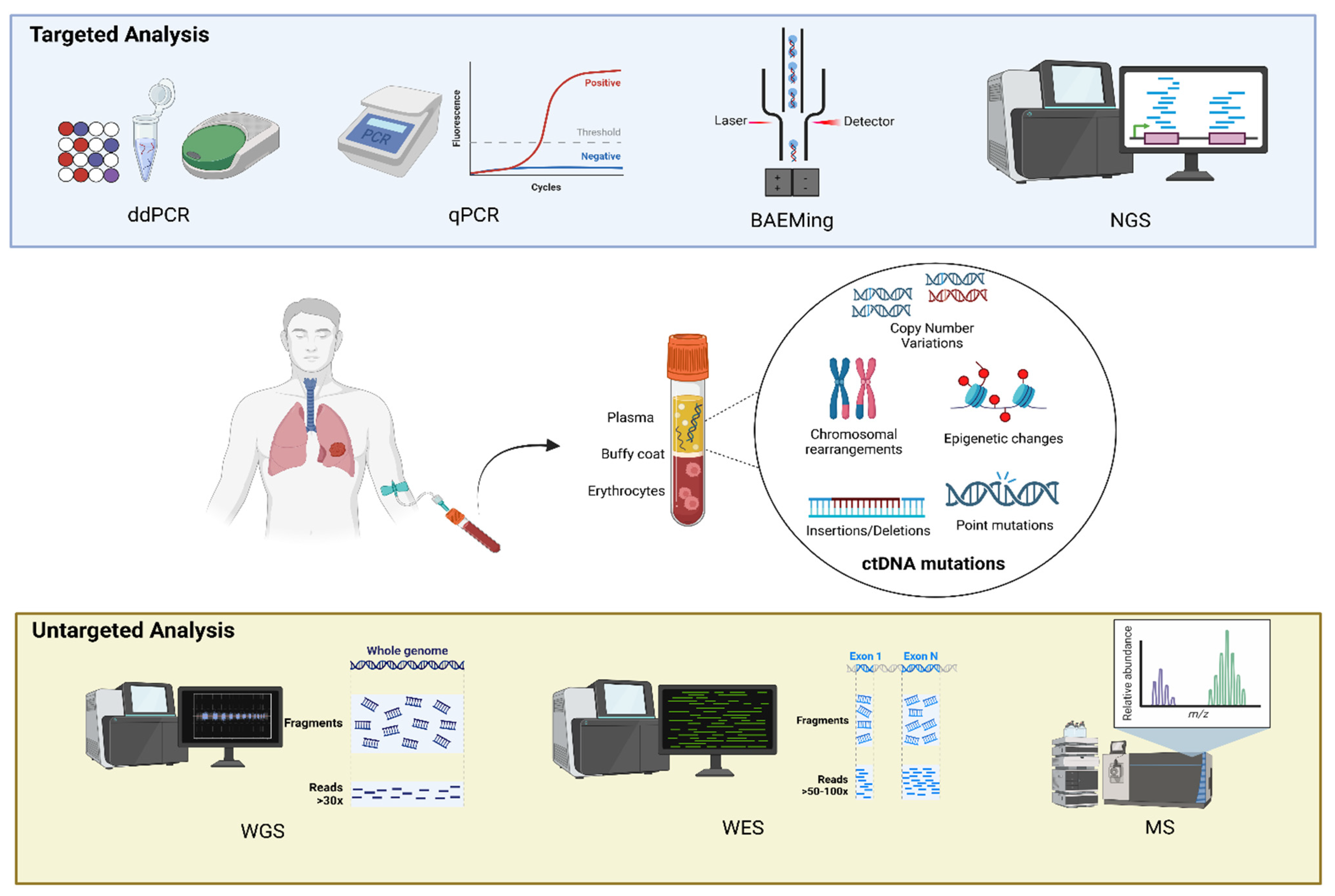
3.1. Methods of Identification of ctDNA
3.2. Baseline ctDNA, Lung Cancer Screening, and Diagnosis of Suspected Malignancy
3.3. Baseline ctDNA and Prediction of Histology, Pathological Features, and Staging
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marinello, A.; Tagliamento, M.; Pagliaro, A.; Conci, N.; Cella, E.; Vasseur, D.; Remon, J.; Levy, A.; Dall’Olio, F.G.; Besse, B. Circulating tumor DNA to guide diagnosis and treatment of localized and locally advanced non-small cell lung cancer. Cancer Treat Rev. 2024, 129, 102791. [Google Scholar] [CrossRef] [PubMed]
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Krebs, M.G.; Sloane, R.; Priest, L.; Lancashire, L.; Hou, J.M.; Greystoke, A.; Ward, T.H.; Ferraldeschi, R.; Ward, T.H.; Ferraldeschi, R.; et al. Evaluation and prognostic significance of circulating tumor cells in patients with non-small-cell lung cancer. J. Clin. Oncol. 2011, 29, 1556–1563. [Google Scholar] [CrossRef]
- Aggarwal, C.; Meropol, N.J.; Punt, C.J.; Iannotti, N.; Saidman, B.H.; Sabbath, K.D.; Gabrail, N.Y.; Picus, J.; Morse, M.A.; Mitchell, E.; et al. Relationship among circulating tumor cells, CEA and overall survival in patients with metastatic colorectal cancer. Ann. Oncol. 2013, 24, 420–428. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Morabito, A.; Rachiglio, A.M.; Sforza, V.; Landi, L.; Bria, E.; Delmonte, A.; Cappuzzo, F.; De Luca, A. Circulating tumour DNA in early stage and locally advanced NSCLC: Ready for clinical implementation? Nat. Rev. Clin. Oncol. 2025, 22, 215–231. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Han, D.S.C.; Jiang, P.; Chiu, R.W.K. Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372, eaaw3616. [Google Scholar]
- Krebs, M.G.; Malapelle, U.; André, F.; Paz-Ares, L.; Schuler, M.; Thomas, D.M.; Vainer, G.; Yoshino, T.; Rolfo, C. Practical Considerations for the Use of Circulating Tumor DNA in the Treatment of Patients With Cancer: A Narrative Review. JAMA Oncol. 2022, 8, 1830–1839. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Frankell, A.M.; Harrison, T.; Kisistok, J.; Garnett, A.; Johnson, L.; Veeriah, S.; Moreau, M.; Chesh, A.; Chaunzwa, T.L.; et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 2023, 616, 553–562. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hasenleithner, S.O.; Speicher, M.R. A clinician’s handbook for using ctDNA throughout the patient journey. Mol. Cancer 2022, 21, 81. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Santis, G.; Angell, R.; Nickless, G.; Quinn, A.; Herbert, A.; Cane, P.; Spicer, J.; Breen, R.; McLean, E.; Tobal, K. Screening for EGFR and KRAS mutations in endobronchial ultrasound derived transbronchial needle aspirates in non-small cell lung cancer using COLD-PCR. PLoS ONE 2011, 6, e25191. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Li, K.; Zhang, N.; Xu, B.; Liu, Z.; Zhao, D.; Dong, Y.; Mu, J.; Lin, H.; Shan, G.; Gao, S.; et al. Utility of Circulating Tumor DNA Assay in Identifying Mutations and Guiding Matched Targeted Therapy in Lung Cancers. Clin. Med. Insights Oncol. 2024, 18, 11795549241285238. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dang, D.K.; Park, B.H. Circulating tumor DNA: Current challenges for clinical utility. J. Clin. Investig. 2022, 132, e154941. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Murciano-Goroff, Y.R.; Hui, A.B.; Araujo Filho, J.A.; Hamilton, E.G.; Chabon, J.J.; Moding, E.J.; Bonilla, R.F.; Lebow, E.S.; Gomez, D.; Rimner, A.; et al. Early Circulating Tumor DNA Shedding Kinetics for Prediction of Platinum Sensitivity in Patients With Small Cell Lung Cancer. JCO Precis. Oncol. 2024, 8, e2400216. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guo, N.; Lou, F.; Ma, Y.; Li, J.; Yang, B.; Chen, W.; Ye, H.; Zhang, J.B.; Zhao, M.Y.; Wu, W.J.; et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci. Rep. 2016, 6, 33519. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, K.; Zhang, J.; Guan, T.; Yang, F.; Lou, F.; Chen, W.; Zhao, M.; Zhang, J.; Chen, S.; Wang, J. Comparison of plasma to tissue DNA mutations in surgical patients with non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2017, 154, 1123–1131.e2. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Uchida, J.; Kato, K.; Kukita, Y.; Kumagai, T.; Nishino, K.; Daga, H.; Nagatomo, I.; Inoue, T.; Kimura, M.; Oba, S.; et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin. Chem. 2015, 61, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeon, S.; Jun, H.R.; Sung, C.O.; Jang, S.J.; Choi, C.M.; Chun, S.M. Revolutionizing Non-Small Cell Lung Cancer Diagnosis: Ultra-High-Sensitive ctDNA Analysis for Detecting Hotspot Mutations with Long-term Stored Plasma. Cancer Res. Treat. 2024, 56, 484–501. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mosko, M.J.; Nakorchevsky, A.A.; Flores, E.; Metzler, H.; Ehrich, M.; van den Boom, D.J.; Sherwood, J.L.; Nygren, A.O. Ultrasensitive Detection of Multiplexed Somatic Mutations Using MALDI-TOF Mass Spectrometry. J. Mol. Diagn. 2016, 18, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Underhill, H.R.; Kitzman, J.O.; Hellwig, S.; Welker, N.C.; Daza, R.; Baker, D.N.; Gligorich, K.M.; Rostomily, R.C.; Bronner, M.P.; Shendure, J. Fragment Length of Circulating Tumor DNA. PLoS Genet. 2016, 12, e1006162. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Tran, H.T.; Heeke, S.; Sujit, S.; Vokes, N.; Zhang, J.; Aminu, M.; Lam, V.K.; Vaporciyan, A.; Swisher, S.G.; Godoy, M.C.B.; et al. Circulating tumor DNA and radiological tumor volume identify patients at risk for relapse with resected, early-stage non-small-cell lung cancer. Ann. Oncol. 2024, 35, 183–189. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hong, T.H.; Hwang, S.; Dasgupta, A.; Abbosh, C.; Hung, T.; Bredno, J.; Walker, J.; Shi, X.; Milenkova, T.; Horn, L.; et al. Clinical Utility of Tumor-Naïve Presurgical Circulating Tumor DNA Detection in Early-Stage NSCLC. J. Thorac. Oncol. 2024, 19, 1512–1524. [Google Scholar] [CrossRef] [PubMed]
- Bossé, Y.; Dasgupta, A.; Abadier, M.; Guthrie, V.; Song, F.; Saavedra Armero, V.; Gaudreault, N.; Orain, M.; Lamaze, F.C.; Melton, C.; et al. Prognostic implication of methylation-based circulating tumor DNA detection prior to surgery in stage I non-small cell lung cancer. Cancer Lett. 2024, 594, 216984. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Jiang, J.; Adams, H.P.; Yao, L.; Yaung, S.; Lal, P.; Balasubramanyam, A.; Fuhlbrück, F.; Tikoo, N.; Lovejoy, A.F.; Froehler, S.; et al. Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Cell-Free DNA in Stage I-IV Non-Small Cell Lung Cancer. J. Mol. Diagn. 2020, 22, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Sorber, L.; Zwaenepoel, K.; Deschoolmeester, V.; Van Schil, P.E.; Van Meerbeeck, J.; Lardon, F.; Rolfo, C.; Pauwels, P. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017, 107, 100–107. [Google Scholar] [CrossRef] [PubMed]
- Bredno, J.; Venn, O.; Chen, X.; Freese, P.; Ofman, J.J. Circulating Tumor DNA Allele Fraction: A Candidate Biological Signal for Multicancer Early Detection Tests to Assess the Clinical Significance of Cancers. Am. J. Pathol. 2022, 192, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Melton, C.A.; Freese, P.; Zhou, Y.; Shenoy, A.; Bagaria, S.; Chang, C.; Kuo, C.C.; Scott, E.; Srinivasan, S.; Cann, G.; et al. A Novel Tissue-Free Method to Estimate Tumor-Derived Cell-Free DNA Quantity Using Tumor Methylation Patterns. Cancers 2023, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Rickles-Young, M.; Tinoco, G.; Tsuji, J.; Pollock, S.; Haynam, M.; Lefebvre, H.; Glover, K.; Owen, D.H.; Collier, K.A.; Ha, G.; et al. Assay Validation of Cell-Free DNA Shallow Whole-Genome Sequencing to Determine Tumor Fraction in Advanced Cancers. J. Mol. Diagn. 2024, 26, 413–422. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zviran, A.; Schulman, R.C.; Shah, M.; Hill, S.T.K.; Deochand, S.; Khamnei, C.C.; Maloney, D.; Patel, K.; Liao, W.; Widman, A.J.; et al. Genome-wide cell-free DNA mutational integration enables ultra-sensitive cancer monitoring. Nat. Med. 2020, 26, 1114–1124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lissa, D.; Robles, A.I. Methylation analyses in liquid biopsy. Transl. Lung Cancer Res. 2016, 5, 492–504. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lockwood, C.M.; Borsu, L.; Cankovic, M.; Earle, J.S.L.; Gocke, C.D.; Hameed, M.; Jordan, D.; Lopategui, J.R.; Pullambhatla, M.; Reuther, J.; et al. Recommendations for Cell-Free DNA Assay Validations: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2023, 25, 876–897. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V.; et al. The IASLC Lung Cancer Staging Project: Proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar]
- Abbosh, C.; Birkbak, N.J.; Swanton, C. Early stage NSCLC—Challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018, 15, 577–586. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Zhang, B.; Zhou, C.; Zhao, Q.; Wang, Y.; Fang, Y.; Hu, Z.; Lv, P.; Miao, L.; Yang, R.; et al. A combined model of circulating tumor DNA methylated SHOX2/SCT/HOXA7 and clinical features facilitates the discrimination of malignant from benign pulmonary nodules. Lung Cancer 2024, 199, 108064. [Google Scholar] [CrossRef] [PubMed]
- Xu-Welliver, M.; Carbone, D.P. Blood-based biomarkers in lung cancer: Prognosis and treatment decisions. Transl. Lung Cancer Res. 2017, 6, 708–712. [Google Scholar] [CrossRef]
- Xu, J.; Pu, Y.; Lin, R.; Xiao, S.; Fu, Y.; Wang, T. PEAC: An Ultrasensitive and Cost-Effective MRD Detection System in Non-small Cell Lung Cancer Using Plasma Specimen. Front. Med. 2022, 9, 822200. [Google Scholar] [CrossRef]
- McKelvey, B.A.; Andrews, H.S.; Baehner, F.L.; Chen, J.; Espenschied, C.R.; Fabrizio, D.; Gorton, V.; Gould, C.; Guinney, J.; Jones, G.; et al. Advancing Evidence Generation for Circulating Tumor DNA: Lessons Learned from A Multi-Assay Study of Baseline Circulating Tumor DNA Levels across Cancer Types and Stages. Diagnostics 2024, 14, 912. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.Z.; Lou, F.; Yang, F.; Zhang, J.B.; Ye, H.; Chen, W.; Guan, T.; Zhao, M.Y.; Su, X.X.; Shi, R.; et al. Circulating Tumor DNA Detection in Early-Stage Non-Small Cell Lung Cancer Patients by Targeted Sequencing. Sci. Rep. 2016, 6, 31985. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- National Lung Screening Trial Research. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- Zhu, S.; Wu, R.; Liu, X.; Xie, B.; Xie, C.; Li, S.; Wu, Z.; Zhang, Z.; Tang, Z.; Gu, L. Clinical application of ctDNA in early diagnosis, treatment and prognosis of patients with non-small cell lung cancer. Future Oncol. 2024, 20, 2213–2224. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhang, B.; Niu, X.; Zhang, Q.; Wang, C.; Liu, B.; Yue, D.; Li, C.; Giaccone, G.; Li, S.; Gao, L.; et al. Circulating tumor DNA detection is correlated to histologic types in patients with early-stage non-small-cell lung cancer. Lung Cancer 2019, 134, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451, Erratum in Nature 2018, 554, 264. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Guo, Q.; Wang, J.; Xiao, J.; Wang, L.; Hu, X.; Yu, W.; Song, G.; Lou, J.; Chen, J. Heterogeneous mutation pattern in tumor tissue and circulating tumor DNA warrants parallel NGS panel testing. Mol. Cancer 2018, 17, 131. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ding, H.; Yuan, M.; Yang, Y.; Xu, X.S. Identifying key circulating tumor DNA parameters for predicting clinical outcomes in metastatic non-squamous non-small cell lung cancer after first-line chemoimmunotherapy. Nat. Commun. 2024, 15, 6862. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Stadler, J.C.; Belloum, Y.; Deitert, B.; Sementsov, M.; Heidrich, I.; Gebhardt, C.; Keller, L.; Pantel, K. Current and Future Clinical Applications of ctDNA in Immuno-Oncology. Cancer Res. 2022, 82, 349–358. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lococo, F.; Boldrini, L.; Diepriye, C.-D.; Evangelista, J.; Nero, C.; Flamini, S.; Minucci, A.; De Paolis, E.; Vita, E.; Cesario, A.; et al. Lung cancer multi-omics digital human avatars for integrating precision medicine into clinical practice: The LANTERN study. BMC Cancer 2023, 23, 540, Erratum in BMC Cancer 2023, 23, 1082. [Google Scholar]

| Item | Specification |
|---|---|
| Date of search | 10 December 2024 |
| Databases and other sources searched | PubMed |
| Search terms used | “ctDNA” and “early stage” and “NSCLC” |
| Time frame | 1 January 2000 to 30 November 2024 |
| Inclusion criteria | Original article, English language, and clinical trial (randomized, prospective, or retrospective) |
| Selection process | Two authors (C.S. and S.R.S.) independently reviewed the abstracts identified with this search, while a third author (F.L) was consulted in the case of discrepancies |
| Author | Period | Country | N of Patients | Histology and Stage | ctDNA Search Strategy | Analyzed Mutations on ctDNA |
|---|---|---|---|---|---|---|
| Santis [12] | 2011 | Austria | 132 | Adenocarcinoma (71.2%), squamous-cell carcinoma (12.8%), not specified (13.7%) Advanced stage | quantitative real-time polymerase chain reaction (qPCR) | EGFR, KRAS |
| Mosko [21] | 2015 | USA | 122 | Test performed on melanoma patients | UltraSEEK Oncogene Panel | BRAFV600E, EGFRG719S, KRASG13D, PIK3CA |
| Underhill [22] | 2016 | USA | N/A | N/A | Illumina sequencing | EGFR |
| Zviran [34] | 2020 | USA | N/A | N/A | ichorCNA test | circulating tumor allele fraction |
| Tran [23] | 2023 | USA | 117 | All NSCLC histologies Early stage | 500 kb plasma-only assay prototype | ALK, BRAF, EGFR, ERBB2, KRAS, MET, NF1, NRAS, PIK3CA, PTEN RB1, STK11, TP53 |
| Melton [32] | 2023 | USA | N/A | N/A | multicancer early detection (MCED) test | small-variant allele fraction |
| Lee [20] | 2024 | Korea | 104 | All NSCLC histologies Early stage | ULV1 panel and targeted next-generation sequencing (CT-ULTRA) | EGFR |
| Hong [24] | 2024 | Korea | 414 | Adenocarcinoma Stage I | methylation-based multicancer early detection (MCED) assay | EGFR, ALK |
| Bossè [25] | 2024 | Canada | 260 | All NSCLC histologies Stage I | Oncomine Precision Assay on the Ion Torrent Genexus System | EGFR, ALK, KRAS, BRAF, ROS1, MET, RET, NTRK, TP53, ERBB2 |
| Rickles-Young [33] | 2024 | USA | 34 | N/A | Qiagen Circulating DNA Kit on the QIAsymphony liquid handling system |
| Author | Period | Country | Kind of Study | Outcome in ctDNA Detection |
|---|---|---|---|---|
| Newman [18] | 2014 | USA | Prospective | 100% of stage II–IV and 50% of stage I NSCLC patients |
| Chen [43] | 2016 | China | Prospective | NSCLC stage IA, IB, IIA: concordance of 50.4%, sensitivity of 53.8%, specificity of 47.3% |
| He [39] | 2024 | China | Prospective | AUC of 0.87 and an accuracy of 0.75 in detecting primary lung cancer |
| McKelvey [42] | 2024 | USA | Prospective | 87.9% early stage lung cancer identified |
| Author | Period | Country | Kind of Study | Outcome in ctDNA Detection |
|---|---|---|---|---|
| Chen [43] | 2016 | China | Prospective | GGO-predominant tumors had a cfDNA concentration more than 10-times lower that in solid tumors |
| Abbosh [47] | 2017 | UK | Prospective | No association between driver events in KRAS, EGFR, or TP53 and ctDNA detection |
| Zhang [46] | 2019 | China | Prospective | 18% lung adenocarcinoma detection vs. 50% squamous-cell carcinoma detection |
| Hong [24] | 2024 | Korea | Prospective | 22% lung adenocarcinoma identification vs. 81% squamous-cell carcinoma identification |
| Clinical trial.gov Reference | Year of Registration | Kind of Study | Status | Primary Endpoint |
|---|---|---|---|---|
| NCT04585477 | 2021-04-08 | Interventional | Recruiting | Change in ctDNA from trial enrollment to after two cycles of adjuvant durvalumab in stage I–III NSCLC patients who had a positive ctDNA status after surgery |
| NCT05921474 | 2023-04-03 | Observational | Recruiting | To assess whether liquid biopsy for molecular residual disease during follow up can predict a recurrence of lung cancer |
| NCT06284317 | 2025-02 | Interventional | Recruiting | To determine whether additional adjuvant immunotherapy with durvalumab after neoadjuvant chemoimmunotherapy has an effect on disease-free survival (DFS) in patients who do not achieve complete pathological response (pCR) as per local assessment according to the IASLC |
| NCT04712877 | 2022-06-15 | Observational | Recruiting | Feasibility of comprehensive molecular profiling to detect actionable oncogenic drivers in patients with suspected early stage lung cancers scheduled to undergo biopsies to establish the diagnosis of lung cancer |
| NCT04638582 | 2022-08-28 | Interventional | Recruiting | To predict the occurrence of a pCR based on the resolution of ctDNA detectability in early stage NSCLC |
| NCT03791034 | 2017-08-28 | Observational | Recruiting | To evaluate whether peripheral circulating cell-free tumor DNA (cfDNA) can aid the screening of recurrence after complete resection of early stage non-small-cell lung cancer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sassorossi, C.; Evangelista, J.; Stefani, A.; Chiappetta, M.; Martino, A.; Campanella, A.; De Paolis, E.; Nachira, D.; Del Re, M.; Guerrera, F.; et al. The Role of ctDNA for Diagnosis and Histological Prediction in Early Stage Non-Small-Cell Lung Cancer: A Narrative Review. Diagnostics 2025, 15, 904. https://doi.org/10.3390/diagnostics15070904
Sassorossi C, Evangelista J, Stefani A, Chiappetta M, Martino A, Campanella A, De Paolis E, Nachira D, Del Re M, Guerrera F, et al. The Role of ctDNA for Diagnosis and Histological Prediction in Early Stage Non-Small-Cell Lung Cancer: A Narrative Review. Diagnostics. 2025; 15(7):904. https://doi.org/10.3390/diagnostics15070904
Chicago/Turabian StyleSassorossi, Carolina, Jessica Evangelista, Alessio Stefani, Marco Chiappetta, Antonella Martino, Annalisa Campanella, Elisa De Paolis, Dania Nachira, Marzia Del Re, Francesco Guerrera, and et al. 2025. "The Role of ctDNA for Diagnosis and Histological Prediction in Early Stage Non-Small-Cell Lung Cancer: A Narrative Review" Diagnostics 15, no. 7: 904. https://doi.org/10.3390/diagnostics15070904
APA StyleSassorossi, C., Evangelista, J., Stefani, A., Chiappetta, M., Martino, A., Campanella, A., De Paolis, E., Nachira, D., Del Re, M., Guerrera, F., Boldrini, L., Urbani, A., Margaritora, S., Minucci, A., Bria, E., & Lococo, F. (2025). The Role of ctDNA for Diagnosis and Histological Prediction in Early Stage Non-Small-Cell Lung Cancer: A Narrative Review. Diagnostics, 15(7), 904. https://doi.org/10.3390/diagnostics15070904












