3. Materials and Methods
3.1. Construction of the Impedance Plethysmography System
A custom-built circuit containing an instrument amplifier, a demodulation circuit, and a bandpass filter was developed to amplify and clarify the impedance signal captured from the armband electrodes.
Figure 1 presents a streamlined design of the Impedance Plethysmography (IPG) system used in this study, illustrating the flow of components from signal acquisition to display. The IPG part includes armband electrodes placed on the body, which receive a stable 5 mA current generated by a 95 kHz oscillator. The resulting impedance changes, indicative of blood volume variations, are amplified and processed through an instrument amplifier, a demodulation circuit, and a bandpass filter. This filtered signal is then further amplified for clarity. The processed data are sent to an Arduino UNO microcontroller.
Constructing this IPG system was the first essential step in this study, enabling the accurate and non-invasive measurement of cardiovascular parameters required for further analysis and model assessment. By integrating the IPG signal with the ECG signal, the impedance and ECG data were leveraged. By capturing both cardiovascular impedance changes and ECG timing, this setup can provide a non-invasive solution to continuous blood pressure tracking.
IPG Component
The IPG part (outlined in orange) includes several key elements that facilitate the detection of blood volume changes:
Human body interface: Armband electrodes are placed on the body to measure impedance changes related to blood flow.
Oscillator (95 kHz): A 95 kHz oscillator generates a high-frequency current, which is applied to the body through the electrodes. This frequency is selected to minimize interference from other physiological signals and to optimize sensitivity to impedance changes.
5 mA current source: A constant 5 mA current is applied via the armband electrodes to measure impedance changes due to varying blood volume within the body segment.
Signal amplification: The signal is first processed by an instrument amplifier to enhance weak impedance signals, followed by a demodulation circuit to extract relevant information.
Bandpass filtering: The bandpass filter employed in our study is a hardware-based implementation integrated within the custom-made circuit. It is designed to isolate the frequency range of 0.3 Hz to 5 Hz, which is critical for capturing cardiovascular dynamics. This range effectively excludes high-frequency noise and very low-frequency drift, which are not relevant to the physiological signals we intended to analyze. The filter’s hardware integration ensures minimal delay and high fidelity in real-time signal processing during data acquisition from the electrodes connected to the arm.
High-gain amplifier (×500): The filtered signal is further amplified to improve signal clarity, making it suitable for subsequent processing and analysis.
Arduino UNO: an Arduino UNO (Arduino SRL, Turin, Italy) microcontroller processes the IPG signals. It also communicates with the LCD screen.
LCD screen: It displays the status of the electrode connectivity.
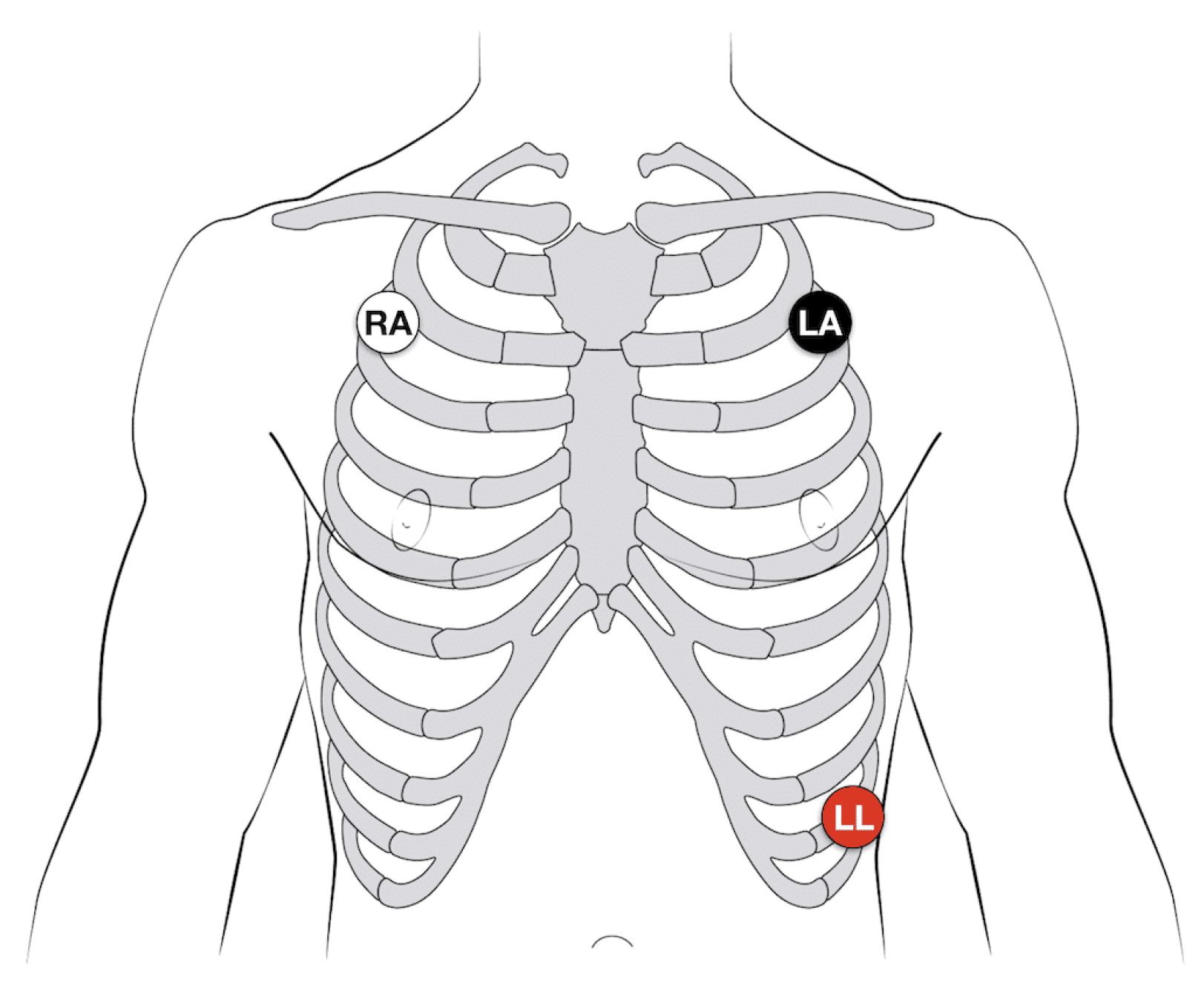
Using a 3-electrode configuration, the ECG electrodes are placed strategically on the body, as shown in
Figure 2.
3.2. Data Collection for System Comparison and Validation
To comprehensively study the relationship between Impedance Plethysmography (IPG) Pulse Transit Time (PTT) and blood pressure, with the aim of assessing the feasibility of predicting blood pressure from IPG PTT, data collection was conducted under two distinct physiological conditions: at rest and post-exercise. A rest mode of 5 min was established initially to obtain baseline measurements, after which subjects engaged in physical exercise until their heart rate increased by 20% above the resting rate. This increase was targeted to induce physiological changes in blood pressure, creating a controlled variation in cardiovascular response that facilitates a robust comparison of PTT across different blood pressure states. Following the exercise-induced increase, measurements were taken for an additional 5 min to capture the immediate post-exercise phase.
Data from the IPG system were collected concurrently with signals from the BIOPAC MP150 ECG system (BIOPAC Systems, Goleta, CA, USA), the Finapres® NOVA hemodynamic monitoring system (Finapres Medical Systems, Enschede, The Netherlands), and a Photoplethysmography (PPG) signal obtained from the Nellcor N600x Pulse Oximeter (Medtronic, Dublin, Ireland). This multi-system setup enabled a comprehensive analysis of PTT variability in relation to fluctuating blood pressure values, providing insights into the potential of IPG PTT as a non-invasive predictor of blood pressure.
A total of 100 healthy individuals participated in this study, representing a balanced sample of the adult population, composed of 50 males and 50 females. The average age of the participants was 24.2 ± 6.34 years, and the average BMI was 27.5 ± 4.6, reflecting data for the entire group. Participants were included in this study if they were adults aged 18–35, without any known chronic diseases. The exclusion criteria were specifically designed to omit individuals with any health conditions, such as hypertension, diabetes, or chronic pulmonary diseases, or those on medications that could affect blood pressure or oxygen saturation, such as beta-blockers, diuretics, or corticosteroids.
Measurements were collected before and after exercise for each participant, capturing data from the IPG system, BIOPAC ECG, the Nellcor oximeter, and the Finapres® NOVA system. Each device recorded analog signals at a sampling rate of 256 Hz, which were digitized using a National Instruments NI-USB-6128 analog-to-digital converter and stored on an HP ProBook laptop. Data acquisition was managed using LabVIEW 2024 (National Instruments, Austin, TX, USA), with subsequent offline analysis conducted in MATLAB R2022b (MathWorks, Natick, MA, USA).
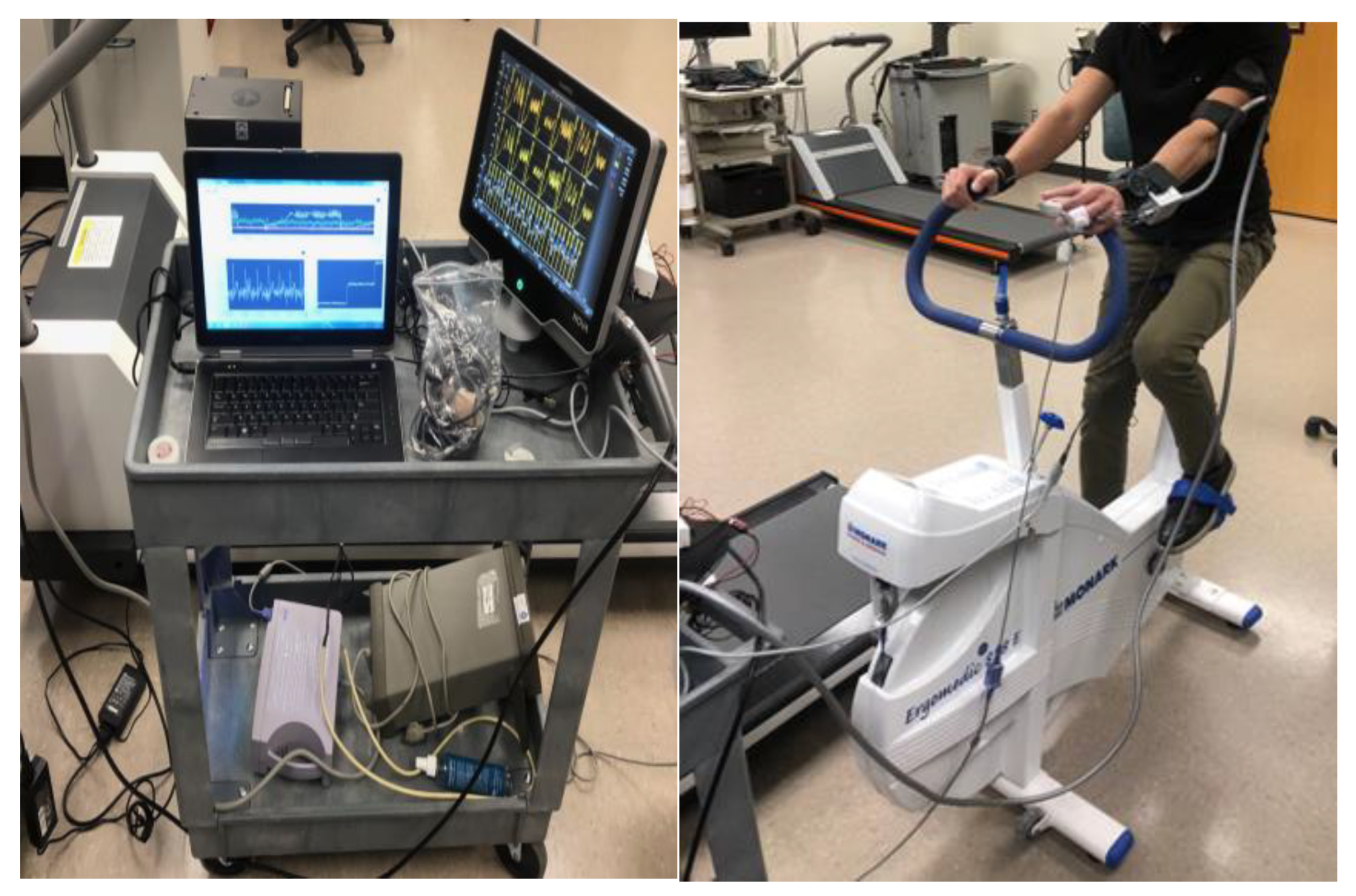
The exercise component involved participants using the Monark 828E Ergomedic Microfit Robobike (Monark Exercise AB, Vansbro, Sweden) as shown in
Figure 3, with measurements taken both at rest and after a controlled physical exertion designed to increase heart rate by approximately 20% above resting levels. The workload was adjusted to consistently achieve this heart rate increase, simulating a mild cardiovascular load and allowing for dynamic blood pressure responses.
This study adhered to ethical standards, with approval from the Institutional Review Board (IRB) of the Center of Excellence in Intelligent Engineering Systems at King Abdulaziz University (approval number 22-CEIES-Biomed-2024). All participants provided informed consent, ensuring compliance with ethical guidelines.
3.3. Understanding Blood Pressure and Related Cardiovascular Measurements
For classification purposes, blood pressure levels were organized into categories, including normal, elevated, and hypertensive stages, in accordance with established clinical guidelines. These categories allowed us to systematically analyze and predict blood pressure status, assessing the efficacy of machine learning algorithms in identifying and categorizing these distinct blood pressure ranges.
To clarify the categorization of blood pressure levels used in our study,
Table 1 presents the definitions of healthy and unhealthy blood pressure ranges. These categories align with the 2017 guidelines from the American Heart Association; however, it is important to note that the updated 2023 guidelines were not considered in this study [
25].
In addition to standard blood pressure readings, this study incorporated several supplementary cardiovascular measurements to provide a more comprehensive view of cardiovascular health. Heart rate (HR), measured as the number of heart beats per minute, served as a key parameter reflecting cardiac function. Two types of PTT were also used: PTT from Photoplethysmography, which captures the time it takes for the pulse wave to travel between two points using optical methods, and PTT from Impedance Plethysmography (PTT IPG), which measures the pulse transit through impedance changes. Both PTT PPG and PTT IPG are important indicators for estimating blood pressure, with PTT IPG providing an alternative approach to PTT PPG for more nuanced readings. By incorporating these parameters, we aimed to enhance the predictive capabilities of this study’s models and to better understand how these physiological signals relate to blood pressure changes.
Furthermore, Pulse Wave Velocity (PWV) values from both Photoplethysmography (PWV PPG) and Impedance Plethysmography (PWV IPG) were used as critical features to assess the performance of machine learning models in predicting blood pressure categories. PWV, being a key indicator of arterial stiffness, provided valuable information on cardiovascular health, allowing the models to better classify blood pressure status based on physiological characteristics.
Table 2 provides a comprehensive summary of the demographic data and physiological variables measured both before and after exercise.
3.4. Assessing Machine Learning Algorithms for Blood Pressure Prediction Using PTT
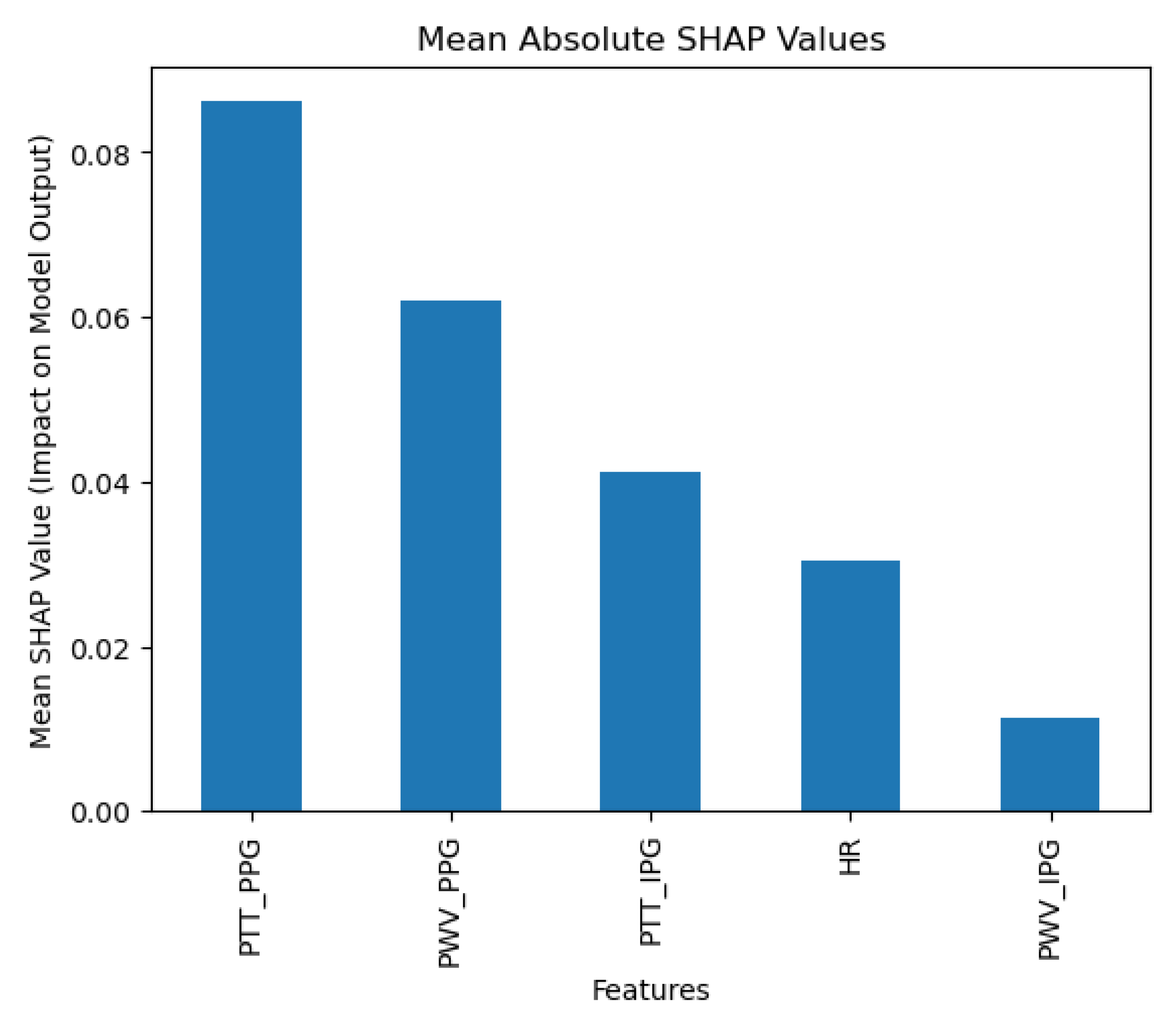
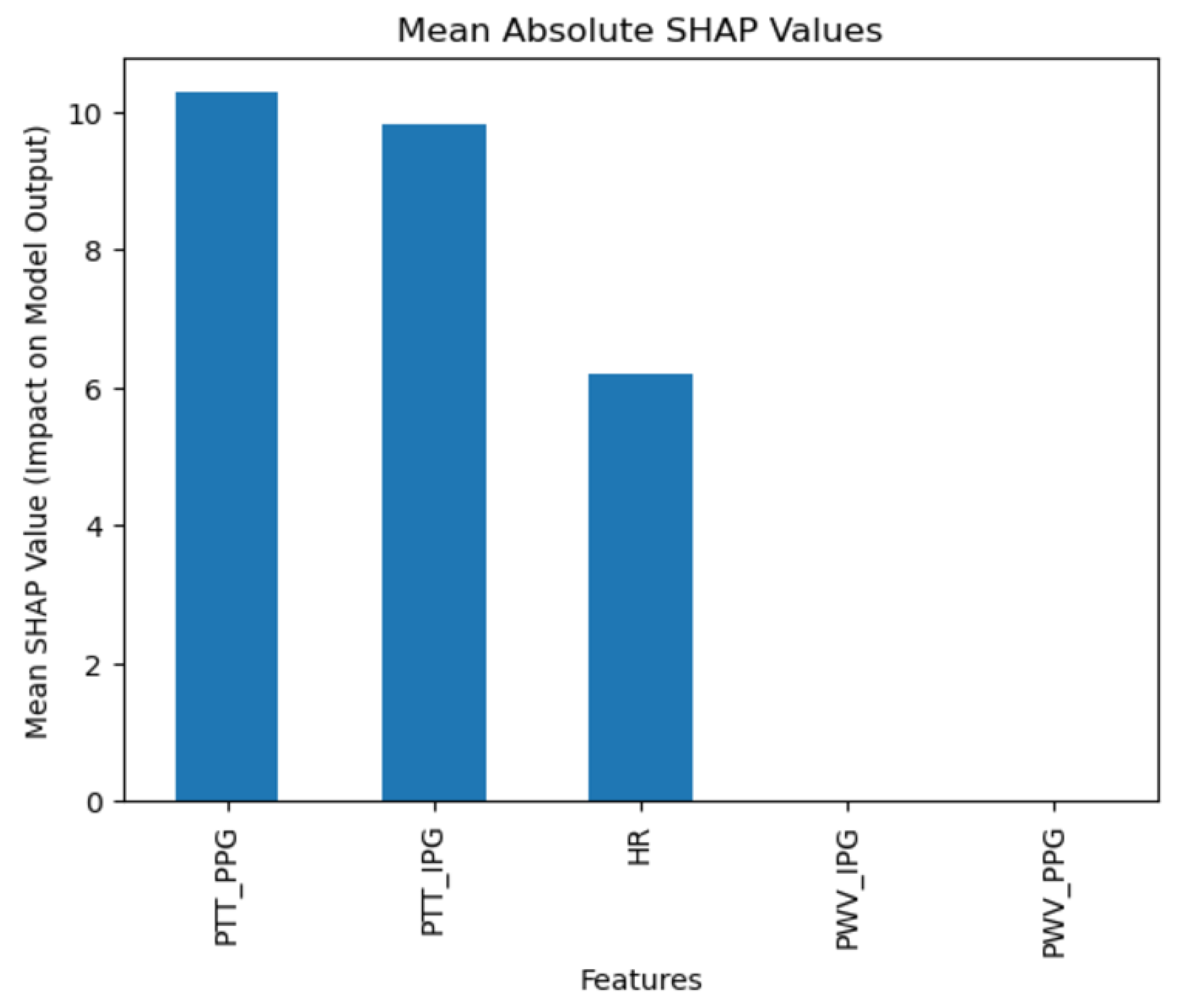
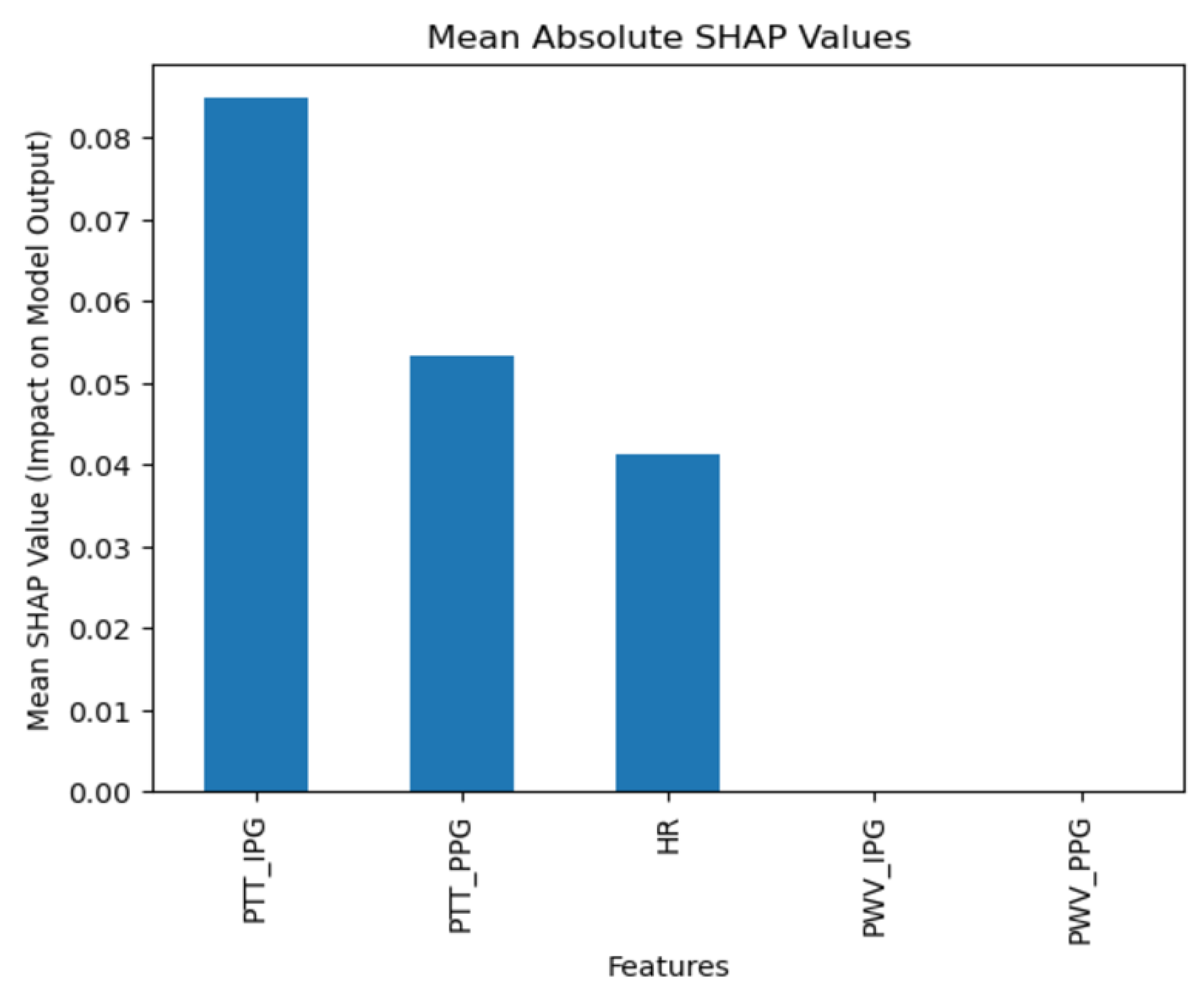
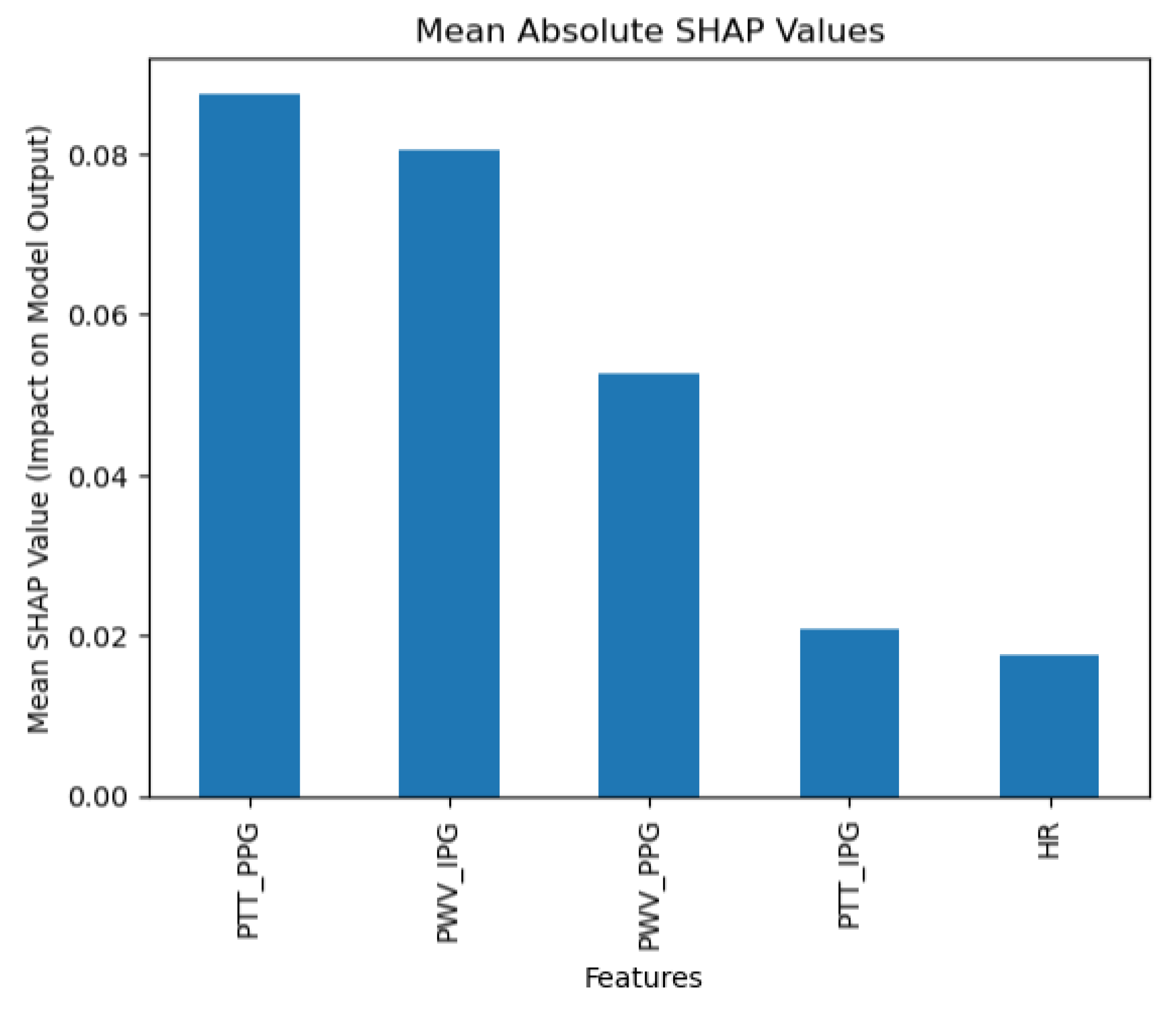
The collected data were processed using various machine learning classification algorithms to assess the efficacy of predicting blood pressure categories based on PTT measurements and the other cardiovascular parameters. The analysis primarily focused on two outputs for each algorithm: (1) the overall prediction accuracy, as indicated by performance metrics such as accuracy, precision, recall, and F1-score, and (2) feature importance analysis using SHAP (SHapley Additive exPlanations) values to interpret the impact of each feature on the model’s predictions. The analysis was conducted in Python version 3.11.5 (Python Software Foundation, Wilmington, DE, USA).
The dataset consisted of all previously mentioned features, including systolic and diastolic blood pressure values. These features were selected as predictive inputs, with the target variable representing blood pressure status, categorized as either normal or elevated. A supervised machine learning approach was applied in this analysis as labeled data were available to train the model on the relationship between input features and blood pressure categories.
The data were split into training and testing sets, with 70% of the data used for training and 30% reserved for testing, ensuring an unbiased model evaluation. Standard scaling and Min-Max scaling were applied to normalize feature distributions based on the specific requirements of each model, enhancing the model’s performance and accuracy in predicting blood pressure status.
To evaluate the efficacy of different machine learning models for predicting blood pressure status, we tested four classification algorithms: Random Forest, Logistic Regression, Support Vector Classifier (SVC), K-Neighbors Classifier (KNN), and Naïve Bayes.
The Random Forest classifier, an ensemble model, was configured with 150 estimators, a maximum depth of 10, and a minimum sample split of 5. Standard scaling was applied to this model to ensure consistent feature scaling across the trees, enhancing the stability of the model’s predictions.
The Logistic Regression model was configured as a linear classifier, was trained with a maximum iteration limit of 1000 to support model convergence, and used a regularization parameter C=0.5 to prevent overfitting. This model employed the ‘liblinear’ solver, which is suitable for binary classification problems, and Min-Max scaling was applied to normalize feature values between 0 and 1, improving model performance by aligning the feature range.
The Support Vector Classifier (SVC) was configured with a linear kernel to create a clear decision boundary, a degree of 4, gamma set to ‘auto’ to automatically scale based on feature variance, and a regularization parameter C = 1.0 to balance margin maximization with error minimization. This model was also standardized using Standard scaling, which ensured data centering and scaling, facilitating robust model performance and enabling probability estimates for improved interpretability.
The K-Neighbors Classifier (KNN) was configured with 5 neighbors, using ‘distance’ weighting to assign more influence to closer points in classification decisions. Min-Max scaling was applied to normalize the feature range, ensuring that the distance-based algorithm could more accurately assess feature relationships across the scaled dataset.
Finally, the Naïve Bayes classifier was implemented using Gaussian Naïve Bayes (GaussianNB), which assumes that continuous features, such as HR, PTT, and PWV, follow a Gaussian distribution. This probabilistic approach is effective for handling clinical datasets with continuous variables and imbalanced classes. Using Stratified K-Fold cross-validation with five splits ensured balanced representation across blood pressure categories. Key metrics, including precision, recall, F1-score, accuracy, and AUC, were calculated for each fold, with 95% confidence intervals to evaluate reliability. GaussianNB efficiently leveraged feature relationships for classification, making it suitable for the dataset’s continuous and variable nature.
4. Data Analysis
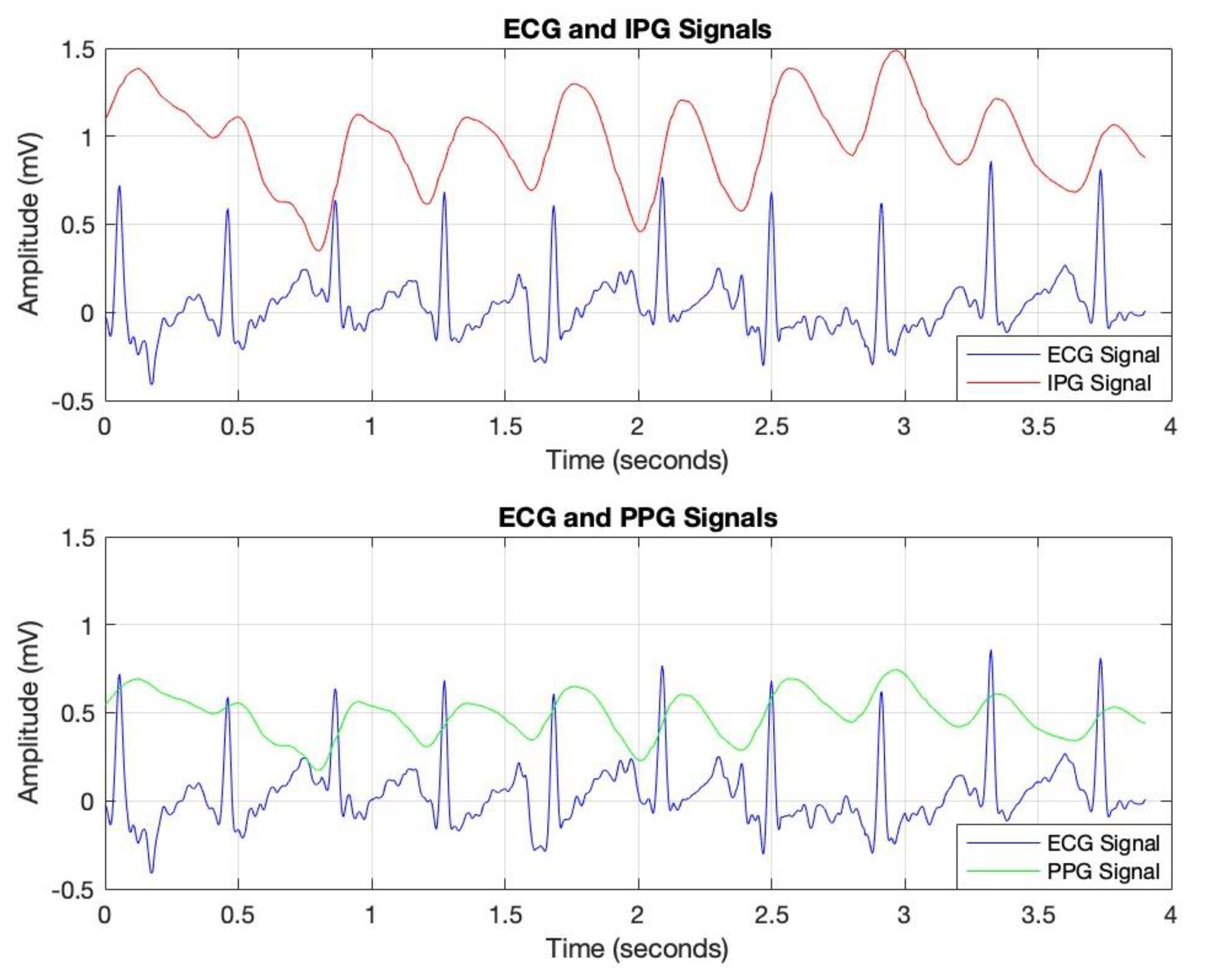
The data analysis commenced with the processing of IPG, ECG, and PPG signals, sampled at a rate of 256 Hz, as depicted in
Figure 4. This sampling rate was selected to ensure accurate calculation of Pulse Transit Time (PTT), as outlined in the methodology section. Data acquisition was conducted under both resting and post-exercise conditions to introduce controlled variability in cardiovascular responses, thereby enhancing the robustness of the analysis.
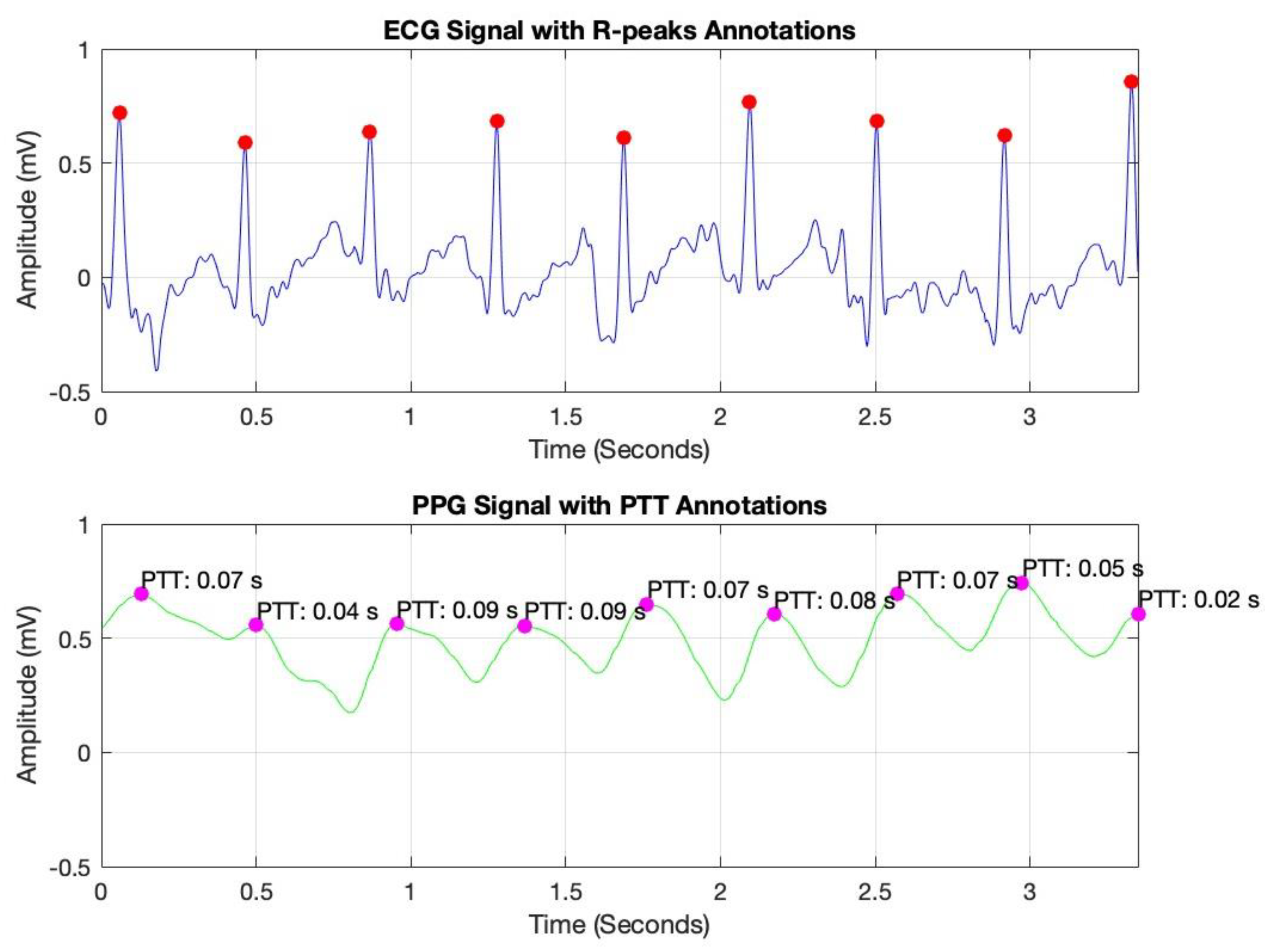
To optimize signal quality, a bandpass filter was applied to each signal, isolating the relevant frequency ranges associated with cardiovascular dynamics. This filtering process effectively attenuated noise and extraneous frequencies, thereby improving the clarity and reliability of the data for subsequent PTT calculations. In
Figure 5, peak detection was conducted using MATLAB’s findpeaks command, identifying R-peaks in the ECG signal (marked with red dots) and corresponding peaks in the PPG signal (marked with magenta dots). By measuring the time interval between these peaks, we derived the PTT values, which are essential for non-invasive blood pressure estimation.
Pulse Transit Time (PTT) is typically calculated as the time difference between the R-peak in the ECG signal (indicating the onset of the cardiac cycle) and the corresponding peak in a peripheral signal, such as the PPG or IPG signal (indicating the arrival of the pulse wave at a distal site) [
10]. The formula for PTT can be expressed as [
10]:
where
tPPG/IPG is the time of the peak in the PPG or IPG signal, marking the arrival of the pulse wave at the peripheral measurement site.
tECG is the time of the R-peak in the ECG signal, representing the start of the cardiac cycle.
These annotated plots validate the system’s accuracy in capturing key cardiovascular parameters, supporting the reliability of PTT as a foundation for continuous blood pressure monitoring.
Furthermore, the heart rate (HR) calculation was based on the R-R interval, which represents the time between consecutive R-peaks in the ECG signal.
Given a sampling rate of 256 Hz, the
R-R interval was determined by measuring the difference between the indices (
i) of consecutive R-peaks and then dividing by the sampling rate to convert this value into seconds, as follows:
The HR in beats per minute was then calculated as the reciprocal of the R-R interval, multiplied by 60:
As previously discussed, the performance of the machine learning models was evaluated using key metrics, namely, accuracy, precision, recall, and F1-score, as outlined below.
Accuracy: This metric assesses the overall correctness of the model by calculating the ratio of correctly classified instances (both true positives and true negatives) to the total number of instances. It provides a general measure of the model’s effectiveness in making accurate predictions across all classes [
26].
Precision: This metric emphasizes the accuracy of the model’s positive predictions by calculating the proportion of true positives among all instances predicted as positive. Precision reflects the model’s ability to avoid false positives, indicating how reliable its positive predictions are [
26].
Recall (or sensitivity): This metric evaluates the model’s ability to identify all relevant instances in the dataset. It is calculated as the proportion of true positives out of all actual positive instances, indicating how effectively the model captures the true positives and minimizes false negatives [
26].
where for Equations (2)–(4), TP: True Positive, TN: True Negative, FP: False Positive, and FN: False Negative.
F1-score: This metric provides a balanced evaluation of a model’s performance across individual classes, offering a more nuanced assessment than overall accuracy alone. The F1-score combines precision and recall, calculating their harmonic mean to deliver a single metric that accounts for both false positives and false negatives. This makes it particularly useful for models where balancing precision and recall is essential, providing a comprehensive measure of the model’s predictive ability [
26,
27].
Additionally, feature importance analysis was conducted using SHAP (SHapley Additive exPlanations) to interpret the contributions of each feature to the machine learning model’s predictions. SHAP values quantify the impact of each feature on the final prediction, highlighting both the individual influence of each feature and its relative importance compared to others. This approach also reveals how interactions between features affect model outcomes, providing a comprehensive understanding of the model’s reliance on specific input variables [
28].
where:
ϕ(f) represents the SHAP values for a particular feature.
ϕ0 is the expected value of the model’s prediction.
M is the total number of features.
f(xi) is the model’s prediction when feature i is included.
E[f(xi)] is the expected prediction when feature i is excluded.
6. Discussion
In our study, we integrated Impedance Plethysmography (IPG) with Photoplethysmography (PPG) to explore the potential for enhancing blood pressure prediction through non-invasive methods. This combination of modalities aimed to capitalize on the complementary strengths of each technique: IPG’s sensitivity to volumetric blood flow changes and PPG’s ability to accurately track pulse wave dynamics. This approach sought to provide a comprehensive analysis that might overcome some of the limitations faced by current cuffless monitoring technologies, which typically rely on single-signal data acquisition.
Reflecting on similar studies, such as those by Stergiou et al. [
15] and Liu et al. [
16], our methodology offers a different angle by employing dual-modality data. These studies have validated the efficacy of wearable technologies using primarily electrocardiograms and photoplethysmograms, focusing on single-signal techniques. In contrast, our work attempted to merge the data richness from both IPG and PPG, potentially offering a more detailed physiological snapshot across various states of cardiac activity.
Our analysis employed a diverse array of machine learning algorithms, from ensemble methods like Random Forest to Logistic Regression, Support Vector Machines, K-Nearest Neighbors, and Naïve Bayes. By evaluating metrics such as accuracy, precision, recall, and F1-score, alongside their macro and weighted averages, we gained a comprehensive understanding of each model’s capability to handle both typical and atypical cardiovascular events.
The performance of the machine learning models was assessed across various evaluation metrics, including accuracy, area under the curve (AUC), and macro and weighted averages of precision, recall, and F1-score. These metrics provide a comprehensive understanding of each model’s ability to classify blood pressure levels effectively, and the results highlight significant differences in model performance.
Among the models, K-Nearest Neighbors (KNN) achieved the highest accuracy at 0.93, followed closely by Random Forest at 0.90. Random Forest also recorded the highest AUC of 0.96, indicating its superior ability to differentiate between blood pressure classes. KNN and Naïve Bayes demonstrated strong AUC values of 0.88 and 0.92, respectively, while Support Vector Machine (SVM) and Logistic Regression showed relatively lower AUCs of 0.86 and 0.89.
Random Forest excelled in macro precision (0.95) and macro recall (0.81), showcasing balanced performance across all classes. Its weighted precision, recall, and F1-score also remained consistently high (0.92, 0.90, and 0.88, respectively). KNN performed exceptionally well in macro averages, particularly in macro precision (0.97) and F1-score (0.90), indicating its strong capability in distinguishing between classes. Naïve Bayes showed moderate performance, with slightly lower macro averages but comparable weighted averages.
Confidence intervals for the metrics further confirm the robustness of Random Forest and KNN. Random Forest displayed the narrowest confidence intervals across metrics, such as precision (0.80–0.98), recall (0.81–0.95), and AUC (0.90–1.00), underscoring its stability and reliability. KNN and Naïve Bayes also showed reasonable intervals, although KNN exhibited slightly wider intervals for some metrics, suggesting variability across data splits.
AUC analysis by class revealed that Random Forest consistently outperformed other models, particularly for hypertension stages (AUC = 1.00 for hypertension stages 1 and 2) (
Table 5). This indicates its exceptional ability to distinguish between these critical classes. KNN also achieved perfect AUCs for the hypertension stages but struggled with the elevated class (AUC = 0.61). Naïve Bayes demonstrated solid performance across most classes but did not achieve the same level of distinction as Random Forest for critical hypertension categories.
The results underscore Random Forest’s overall superiority, attributed to its ability to handle class imbalances and complex relationships between variables effectively. Its ensemble nature likely contributed to its consistent performance across multiple evaluation metrics and BP ranges. KNN’s high performance, particularly in macro precision and F1-score, can be linked to its distance-weighted approach, which allows it to better classify data points with clear proximity-based patterns. However, KNN’s limitations in handling the elevated class highlight potential issues with unbalanced datasets and subtle class distinctions.
The comparative analysis highlights Random Forest as the most reliable model for non-invasive BP classification, supported by its high accuracy, AUC, and stability across metrics. However, further refinement, particularly for models like Logistic Regression and SVM, could improve their ability to distinguish between blood pressure classes, especially in cases of elevated BP, which remains a challenge across models.
This study highlights the importance of selecting and tuning the right machine learning algorithms based on their ability to accurately detect and send alerts upon abnormal blood pressure levels. An ensemble approach, leveraging the strengths of various models, might offer the most robust solution for developing advanced, reliable, and responsive health monitoring systems. This strategy ensures that users receive timely alerts that could lead to prompt medical interventions, thereby enhancing overall health outcomes.
Moreover, the use of SHAP values in our analysis helped elucidate the contribution of individual features to the models’ predictions, highlighting the significant impact of combining IPG and PPG data. This deeper level of analysis was intended to help pinpoint areas where sensor technology and algorithmic approaches could be refined for better performance.
Despite these contributions, our study is not without its limitations. The dual-modality approach, while offering rich data, also introduces complexity in data processing and analysis. The accuracy of such systems heavily relies on the precision of signal capture and the synchronization between IPG and PPG data streams, which can be challenging to maintain consistently outside of controlled environments. Moreover, our study’s sample was relatively homogeneous and limited to a specific age group, which may affect the generalizability of the findings to other populations or clinical settings. The absence of hypertensive crisis values stems from the inclusion criteria, which focused on healthy individuals aged 18–35 years without chronic conditions or medications affecting blood pressure. Consequently, the dataset predominantly captured normal, elevated, and hypertensive Stage 1 and Stage 2 blood pressure ranges, leaving a critical data gap for the hypertensive crisis category.
This limitation underscores the need for future studies to incorporate a broader population with diverse blood pressure levels, including those in the hypertensive crisis range. Such an approach would enable a more comprehensive evaluation of machine learning models, ensuring their applicability and robustness across the full spectrum of blood pressure conditions. Addressing this limitation will enhance the clinical relevance of these models, particularly in detecting and managing severe hypertension cases.
In comparison to the existing literature, our study adds to the ongoing dialogue about the feasibility and challenges of non-invasive blood pressure monitoring. The integration of IPG and PPG for blood pressure estimation presented here offers a potential pathway for future research, particularly in enhancing the accuracy and reliability of these measurements for broader clinical use. Future work should aim to address the noted limitations by expanding this study’s demographic diversity and conducting tests in more varied real-world environments.
Yavarimanesh et al. evaluated the stability of cuffless BP calibration models over one year, identifying the toe PAT-SBP model as the most reliable, with an RMSE of 9.6 ± 0.8 mmHg and a requirement of annual recalibration [
11]. Their study, however, was limited to 10 participants and emphasized long-term recalibration needs [
11]. In contrast, our study applied machine learning to classify BP categories in a larger sample of 100 participants, achieving 90% accuracy (Random Forest) with an AUC of 0.96. While Yavarimanesh et al. focused on feature stability over time, our work highlights the robustness of machine learning models for BP classification across pre- and post-exercise conditions. Both studies underscore the importance of feature selection, with toe PAT and PTT PPG emerging as critical predictors.
Zhang et al. demonstrated that pulse arrival time (PAT) was an inadequate surrogate for Pulse Transit Time (PTT) in blood pressure (BP) prediction, with PAT yielding errors ∼80% higher than PTT and failing to meet FDA error limits [
12]. PTT was found to be more reliable for BP tracking, particularly during dynamic changes induced by hemodynamic interventions [
12].
In contrast, our study focuses on leveraging PTT-based features for everyday BP monitoring in healthy users rather than clinical settings. Using machine learning models, such as Random Forest, we achieved 90% accuracy and a high AUC of 0.96 for BP classification. These results highlight the practicality and robustness of PTT-derived features for non-invasive and cuffless BP monitoring in daily life, aligning with the need for simple and accurate solutions for healthy individuals.
Table 6 compares our study’s findings with key related studies discussed in
Section 3.4 (Assessing Machine Learning Algorithms for Blood Pressure Prediction Using PTT), highlighting the focus, performance, limitations, and potential of machine learning in predicting and monitoring blood pressure.
The proposed technology, therefore, should not be viewed as a replacement for traditional blood pressure monitoring devices that provide precise measurements critical for clinical diagnosis and treatment. Instead, it should be seen as a supplementary tool that offers a convenient and non-invasive way to keep a regular check on one’s cardiovascular health, encouraging proactive health management in everyday life settings.