LivSCP: Improving Liver Fibrosis Classification Through Supervised Contrastive Pretraining
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
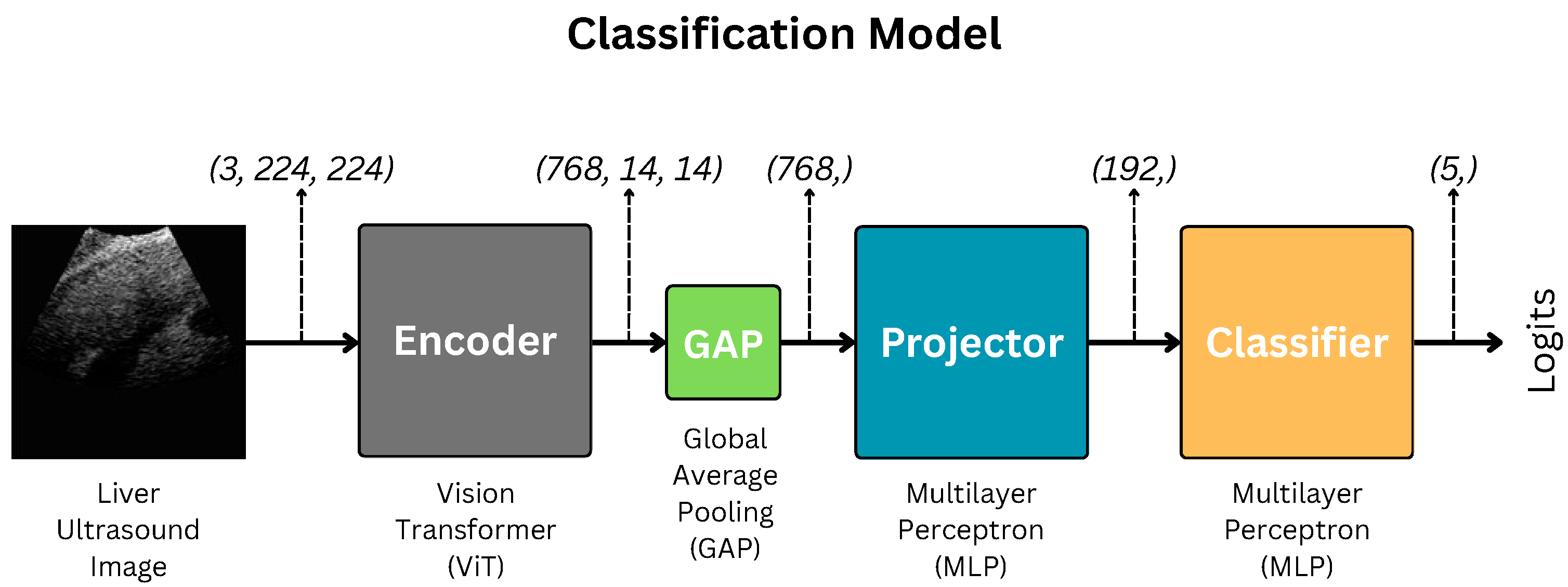
2.2. Encoder: Vision Transformer
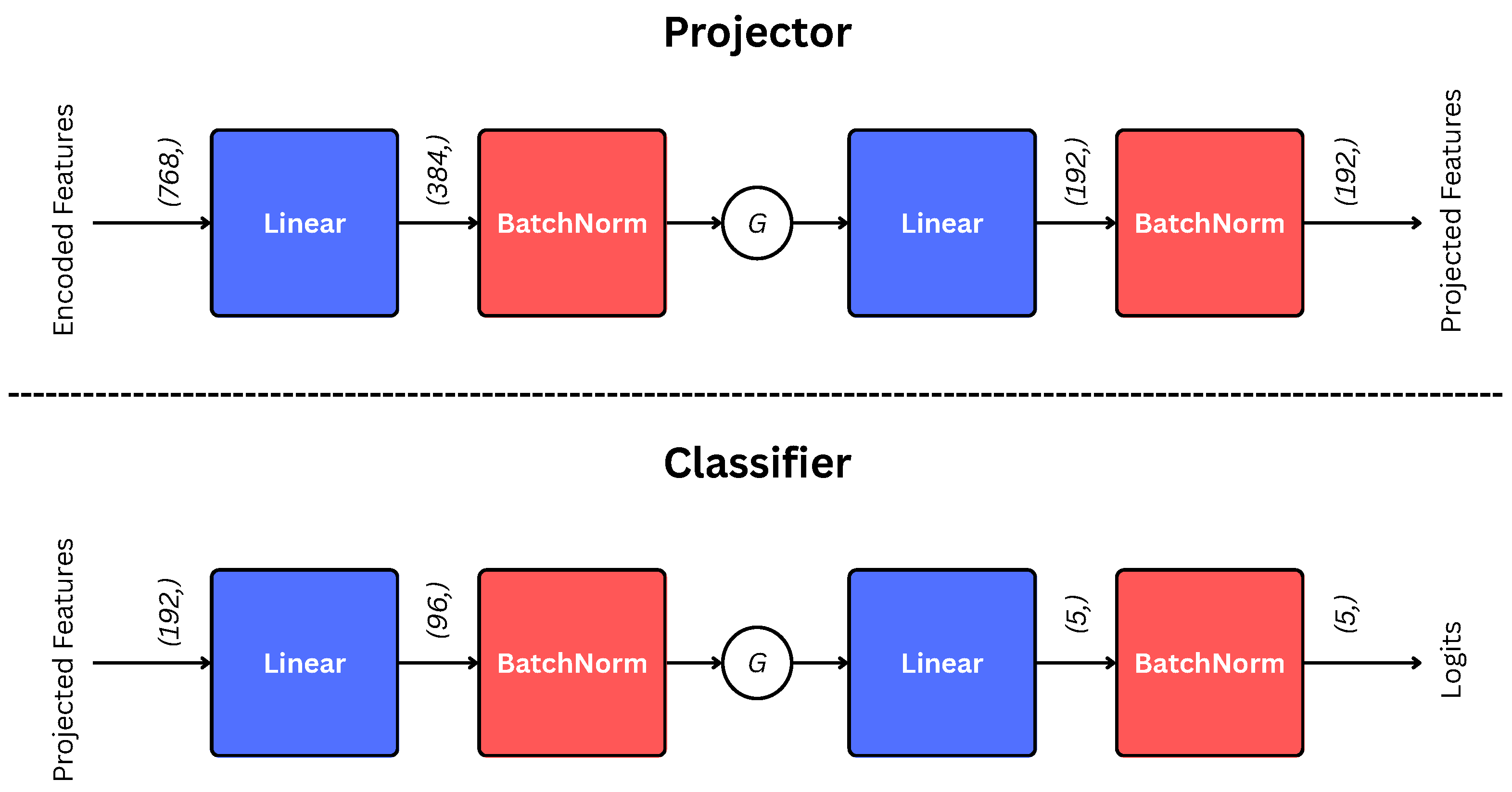
2.3. Projection and Classification Heads
2.4. Supervised Contrastive Learning
3. Results
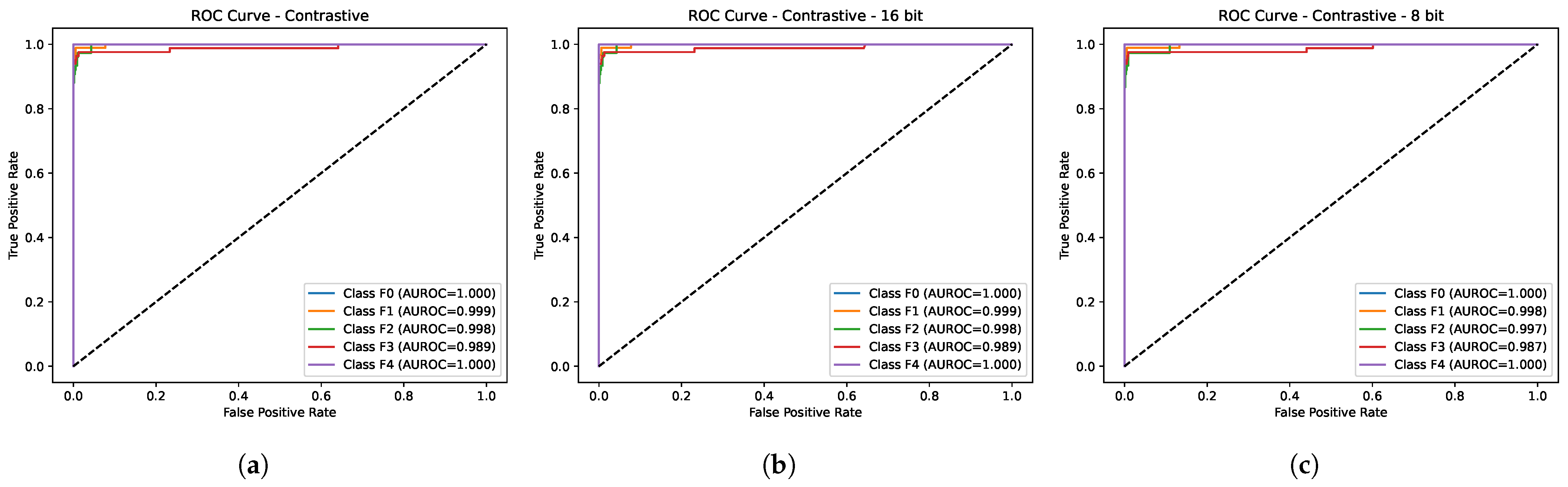
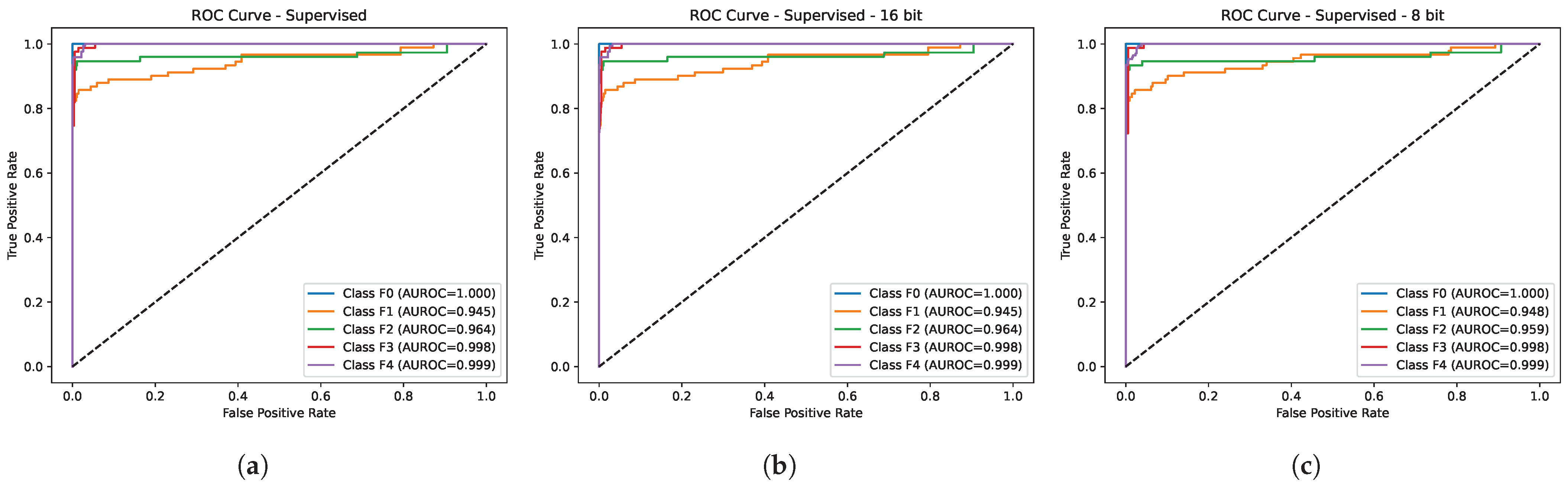
3.1. Quantitative Evaluation
3.2. Implementation Details
4. Discussion
4.1. Contribution
- Improved performance: As seen above, our method, LivSCP, improves classification performance without any changes to the network architecture of ViT. It outperforms all existing methods and our baseline (ViT with SL) consistently across all evaluation metrics.
- Solution to Low Data and Computation: Our method is successful in low-data and -computation settings. We have demonstrated that with a dataset of 6323 images in total (∼5058 images for training), and an NVIDIA P100 GPU, we found an absolute performance boost of up to 5.38% in accuracy with respect to our baseline and up to 14.93% with an existing method [22].
- Rigorous performance analysis: We conduct a rigorous quantitative analysis of the classification performance of existing methods, our baseline, and the proposed method at different precision (quantization) levels.
4.2. Comparison with Baseline
4.3. Comparison with Existing Methods
4.4. Implications
- Explainability: DL models used for automated disease classification are no less than unexplained black boxes. LivSCP, being one, will have similar characteristics, and we might question its interpretability. This creates immense scope for further research in developing solutions that are inherently explainable and do not need external methods to explain them, such as SHAP [44], GradCAM [45], etc.
- Case-specificity: LivSCP has been developed to address scenarios where both data and computational resources are limited. However, its performance may vary and may even improve when more data and greater computing capacity are available. Therefore, our method should not be generalized to broader studies without considering these constraints.
- Practical Errors: LivSCP has no solution to reduce the effect of imaging errors and noise, inconsistencies, etc. This creates another opportunity to develop methods that are more error-immune.
5. Conclusions and Future Scope
5.1. Conclusions
5.2. Future Scope
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NAFLD | Non-alcoholic Fatty Liver Disease |
| ARLD | Alcohol Related Liver Disease |
| DL | Deep Learning |
| CNN | Convolutional Neural Network |
| HCC | Hepatocellular Carcinoma |
| US | Ultrasound |
| SWE | Shear Wave Elastography |
| SL | Supervised Learning |
| UL | Unsupervised Learning |
| RL | Reinforcement Learning |
| SSL | Self-Supervised Learning |
| SCP | Supervised Contrastive Pretraining |
| SFT | Supervised Fine-tuning |
| MLP | Multilayer Perceptron |
| ViT | Vision Transformer |
| GAP | Global Average Pooling |
| mAUROC | Mean Area Under the Receiver Operating Characteristic Curve |
| CW-SCL | Class Weighted Supervised Contrastive Loss |
Appendix A
Quantizing Models
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Teng, M.L.; Ng, C.H.; Huang, D.Q.; Chan, K.E.; Tan, D.J.; Lim, W.H.; Yang, J.D.; Tan, E.; Muthiah, M.D. Global incidence and prevalence of nonalcoholic fatty liver disease. Clin. Mol. Hepatol. 2022, 29, S32. [Google Scholar] [CrossRef]
- Niu, X.; Zhu, L.; Xu, Y.; Zhang, M.; Hao, Y.; Ma, L.; Li, Y.; Xing, H. Global prevalence, incidence, and outcomes of alcohol related liver diseases: A systematic review and meta-analysis. BMC Public Health 2023, 23, 859. [Google Scholar]
- Amini-Salehi, E.; Letafatkar, N.; Norouzi, N.; Joukar, F.; Habibi, A.; Javid, M.; Sattari, N.; Khorasani, M.; Farahmand, A.; Tavakoli, S.; et al. Global prevalence of nonalcoholic fatty liver disease: An updated review Meta-Analysis comprising a population of 78 million from 38 countries. Arch. Med. Res. 2024, 55, 103043. [Google Scholar] [CrossRef] [PubMed]
- Elhence, A.; Bansal, B.; Gupta, H.; Anand, A.; Singh, T.P.; Goel, A.; Shalimar. Prevalence of non-alcoholic fatty liver disease in India: A systematic review and meta-analysis. J. Clin. Exp. Hepatol. 2022, 12, 818–829. [Google Scholar] [PubMed]
- Anton, M.C.; Shanthi, B.; Sridevi, C. Prevalence of non-alcoholic fatty liver disease in urban adult population in a tertiary care center, Chennai. Indian J. Community Med. 2023, 48, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Ufere, N.N.; Satapathy, N.; Philpotts, L.; Lai, J.C.; Serper, M. Financial burden in adults with chronic liver disease: A scoping review. Liver Transplant. 2022, 28, 1920–1935. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, M.; Ming, Y. Global burden of cirrhosis and other chronic liver diseases caused by specific etiologies from 1990 to 2021. BMC Gastroenterol. 2025, 25, 641. [Google Scholar] [CrossRef]
- Bravo, A.A.; Sheth, S.G.; Chopra, S. Liver biopsy. N. Engl. J. Med. 2001, 344, 495–500. [Google Scholar] [CrossRef]
- Rockey, D.C.; Caldwell, S.H.; Goodman, Z.D.; Nelson, R.C.; Smith, A.D. Liver biopsy. Hepatology 2009, 49, 1017–1044. [Google Scholar] [CrossRef]
- Schuppan, D.; Afdhal, N.H. Liver cirrhosis. Lancet 2008, 371, 838–851. [Google Scholar] [CrossRef] [PubMed]
- Castera, L.; Forns, X.; Alberti, A. Non-invasive evaluation of liver fibrosis using transient elastography. J. Hepatol. 2008, 48, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Joo, I.; Kang, T.W.; Paik, Y.H.; Sinn, D.H.; Ha, S.Y.; Kim, K.; Choi, C.; Lee, G.; Yi, J.; et al. Deep learning with ultrasonography: Automated classification of liver fibrosis using a deep convolutional neural network. Eur. Radiol. 2020, 30, 1264–1273. [Google Scholar] [CrossRef] [PubMed]
- Anteby, R.; Klang, E.; Horesh, N.; Nachmany, I.; Shimon, O.; Barash, Y.; Kopylov, U.; Soffer, S. Deep learning for noninvasive liver fibrosis classification: A systematic review. Liver Int. 2021, 41, 2269–2278. [Google Scholar] [CrossRef]
- Decharatanachart, P.; Chaiteerakij, R.; Tiyarattanachai, T.; Treeprasertsuk, S. Application of artificial intelligence in chronic liver diseases: A systematic review and meta-analysis. BMC Gastroenterol. 2021, 21, 10. [Google Scholar] [CrossRef]
- Kagadis, G.C.; Drazinos, P.; Gatos, I.; Tsantis, S.; Papadimitroulas, P.; Spiliopoulos, S.; Karnabatidis, D.; Theotokas, I.; Zoumpoulis, P.; Hazle, J.D. Deep learning networks on chronic liver disease assessment with fine-tuning of shear wave elastography image sequences. Phys. Med. Biol. 2020, 65, 215027. [Google Scholar] [CrossRef]
- Sun, C.; Xu, A.; Liu, D.; Xiong, Z.; Zhao, F.; Ding, W. Deep learning-based classification of liver cancer histopathology images using only global labels. IEEE J. Biomed. Health Inform. 2019, 24, 1643–1651. [Google Scholar] [CrossRef]
- Che, H.; Brown, L.G.; Foran, D.J.; Nosher, J.L.; Hacihaliloglu, I. Liver disease classification from ultrasound using multi-scale CNN. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1537–1548. [Google Scholar] [CrossRef]
- Lin, Y.S.; Huang, P.H.; Chen, Y.Y. Deep learning-based hepatocellular carcinoma histopathology image classification: Accuracy versus training dataset size. IEEE Access 2021, 9, 33144–33157. [Google Scholar] [CrossRef]
- Al-Hasani, M.; Sultan, L.R.; Sagreiya, H.; Cary, T.W.; Karmacharya, M.B.; Sehgal, C.M. Ultrasound radiomics for the detection of early-stage liver fibrosis. Diagnostics 2022, 12, 2737. [Google Scholar] [CrossRef]
- Duan, Y.Y.; Qin, J.; Qiu, W.Q.; Li, S.Y.; Li, C.; Liu, A.S.; Chen, X.; Zhang, C.X. Performance of a generative adversarial network using ultrasound images to stage liver fibrosis and predict cirrhosis based on a deep-learning radiomics nomogram. Clin. Radiol. 2022, 77, e723–e731. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.; Park, H.C.; Lee, O.J.; Yoon, C.; Choi, M.H.; Choi, C. Classification of liver fibrosis from heterogeneous ultrasound image. IEEE Access 2023, 11, 9920–9930. [Google Scholar] [CrossRef]
- Antony Asir Daniel, V.; Jeha, J. An optimal modified faster region CNN model for diagnosis of liver diseases from ultrasound images. IETE J. Res. 2024, 70, 3572–3589. [Google Scholar] [CrossRef]
- Ai, H.; Huang, Y.; Tai, D.I.; Tsui, P.H.; Zhou, Z. Ultrasonic Assessment of Liver Fibrosis Using One-Dimensional Convolutional Neural Networks Based on Frequency Spectra of Radiofrequency Signals with Deep Learning Segmentation of Liver Regions in B-Mode Images: A Feasibility Study. Sensors 2024, 24, 5513. [Google Scholar] [CrossRef]
- Park, H.C.; Joo, Y.; Lee, O.J.; Lee, K.; Song, T.K.; Choi, C.; Choi, M.H.; Yoon, C. Automated classification of liver fibrosis stages using ultrasound imaging. BMC Med. Imaging 2024, 24, 36. [Google Scholar] [CrossRef]
- Le, M.H.N.; Kha, H.Q.; Tran, N.M.; Nguyen, P.K.; Huynh, H.H.; Huynh, P.K.; Lam, H.; Le, N.Q.K. Radiomics in liver research: A paradigm shift in disease detection and staging. Eur. J. Radiol. Artif. Intell. 2025, 2, 100016. [Google Scholar] [CrossRef]
- Xia, F.; Wei, W.; Wang, J.; Wang, Y.; Wang, K.; Zhang, C.; Zhu, Q. Ultrasound radiomics-based logistic regression model for fibrotic NASH. BMC Gastroenterol. 2025, 25, 66. [Google Scholar] [CrossRef]
- Balestriero, R.; Lecun, Y. How Learning by Reconstruction Produces Uninformative Features For Perception. In Proceedings of the 41st International Conference on Machine Learning, Vienna, Austria, 21–27 July 2024; Volume 235, pp. 2566–2585. [Google Scholar]
- Gupta, V. Liver Histopathology (Fibrosis) Ultrasound Images. Kaggle. 2024. Available online: https://www.kaggle.com/datasets/vibhingupta028/liver-histopathology-fibrosis-ultrasound-images (accessed on 1 January 2025).
- Bicubic Interpolation. Wikipedia Article. Available online: https://en.wikipedia.org/wiki/Bicubic_interpolation (accessed on 6 December 2025).
- Khosla, P.; Teterwak, P.; Wang, C.; Sarna, A.; Tian, Y.; Isola, P.; Maschinot, A.; Liu, C.; Krishnan, D. Supervised Contrastive Learning. In Proceedings of the Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: New York, NY, USA, 2020; Volume 33, pp. 18661–18673. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Shlens, J.; Le, Q. RandAugment: Practical Automated Data Augmentation with a Reduced Search Space. In Proceedings of the Advances in Neural Information Processing Systems; Larochelle, H., Ranzato, M., Hadsell, R., Balcan, M., Lin, H., Eds.; Curran Associates, Inc.: New York, NY, USA, 2020; Volume 33, pp. 18613–18624. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Vienna, Austria, 4 May 2021. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.U.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: New York, NY, USA, 2017; Volume 30. [Google Scholar]
- Wightman, R. Vision Transformer (ViT) Base Patch16 224 - AugReg + ImageNet-21k + Fine-Tuned on ImageNet-1k. Model Version: Augreg2_in21k_ft_in1k. Available online: https://huggingface.co/timm/vit_base_patch16_224.augreg2_in21k_ft_in1k (accessed on 3 March 2025).
- Wang, Z.; Liu, L.; Duan, Y.; Kong, Y.; Tao, D. Continual Learning with Lifelong Vision Transformer. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 171–181. [Google Scholar] [CrossRef]
- Xue, M.; Zhang, H.; Song, J.; Song, M. Meta-attention for ViT-backed Continual Learning. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 150–159. [Google Scholar] [CrossRef]
- Li, D.; Cao, G.; Xu, Y.; Cheng, Z.; Niu, Y. Technical Report for ICCV 2021 Challenge SSLAD-Track3B: Transformers Are Better Continual Learners. arXiv 2022, arXiv:2201.04924. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2023, arXiv:1606.08415. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; Volume 9, pp. 249–256. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- AskariHemmat, M.; Hemmat, R.A.; Hoffman, A.; Lazarevich, I.; Saboori, E.; Mastropietro, O.; Sah, S.; Savaria, Y.; David, J.P. QReg: On Regularization Effects of Quantization. arXiv 2022, arXiv:2206.12372. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A unified approach to interpreting model predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Red Hook, NY, USA, 4–9 December 2017; NIPS’17, pp. 4768–4777. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int. J. Comput. Vis. 2020, 128, 336–359. [Google Scholar] [CrossRef]
- PyTorch Contributors. torch.ao.quantization.quantize_dynamic. PyTorch Documentation. 2025. Available online: https://docs.pytorch.org/docs/stable/generated/torch.ao.quantization.quantize_dynamic.html (accessed on 2 October 2025).

| Stage | # Images |
|---|---|
| F0 | 2114 |
| F1 | 861 |
| F2 | 793 |
| F3 | 857 |
| F4 | 1698 |
| Method | Accuracy | Precision | Recall | F1 Score | mAUROC |
|---|---|---|---|---|---|
| SL-FP32 | 0.9272 | 0.9422 | 0.9272 | 0.9248 | 0.9812 |
| LivSCP-FP32 † | 0.9810 | 0.9810 | 0.9810 | 0.9810 | 0.9972 |
| SL-FP16 | 0.9272 | 0.9422 | 0.9272 | 0.9298 | 0.9811 |
| LivSCP-FP16 | 0.9810 | 0.9810 | 0.9810 | 0.9810 | 0.9972 |
| SL-INT8 | 0.9287 | 0.9432 | 0.9287 | 0.9312 | 0.9807 |
| LivSCP-INT8 | 0.9810 | 0.9810 | 0.9810 | 0.9810 | 0.9964 |
| Method | Accuracy (in %) | AUROC |
|---|---|---|
| Lee et al. [13] | 83.5 & 76.4 | 0.901 & 0.857 (F4) |
| Joo et al. [22] | 84.37 | – |
| Park et al. [25] | 94 | 0.95 |
| Proposed | 98.10 | 0.9972 |
| Network | Accuracy (in %) |
|---|---|
| VGGNet | 83.17 |
| ResNet | 85.92 |
| DenseNet | 84.17 |
| EfficientNet | 85.17 |
| ViT (SL) | 83.42 |
| Proposed | 98.10 |
| Network | F0 | F1 | F2 | F3 | F4 |
|---|---|---|---|---|---|
| VGGNet | 0.96 | 0.96 | 0.98 | 0.94 | 0.96 |
| ResNet | 0.96 | 0.96 | 0.97 | 0.93 | 0.94 |
| DenseNet | 0.95 | 0.96 | 0.95 | 0.94 | 0.96 |
| EfficientNet | 0.96 | 0.96 | 0.97 | 0.94 | 0.96 |
| ViT (SL) | 0.97 | 0.94 | 0.96 | 0.94 | 0.96 |
| Proposed | 1 | 0.999 | 0.9983 | 0.98915 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dubey, Y.; Bhongade, A.; Fuzele, P. LivSCP: Improving Liver Fibrosis Classification Through Supervised Contrastive Pretraining. Diagnostics 2025, 15, 3226. https://doi.org/10.3390/diagnostics15243226
Dubey Y, Bhongade A, Fuzele P. LivSCP: Improving Liver Fibrosis Classification Through Supervised Contrastive Pretraining. Diagnostics. 2025; 15(24):3226. https://doi.org/10.3390/diagnostics15243226
Chicago/Turabian StyleDubey, Yogita, Aditya Bhongade, and Punit Fuzele. 2025. "LivSCP: Improving Liver Fibrosis Classification Through Supervised Contrastive Pretraining" Diagnostics 15, no. 24: 3226. https://doi.org/10.3390/diagnostics15243226
APA StyleDubey, Y., Bhongade, A., & Fuzele, P. (2025). LivSCP: Improving Liver Fibrosis Classification Through Supervised Contrastive Pretraining. Diagnostics, 15(24), 3226. https://doi.org/10.3390/diagnostics15243226






