Next-Generation Sequencing for Bloodstream Infections: Shaping the Future of Rapid Diagnostics and Precision Medicine
Abstract
1. Introduction
2. Search Strategy and Selection Criteria
3. Technologies and Workflow for Sequencing in BSIs
3.1. From Short-Read NGS to Real-Time Long Reads
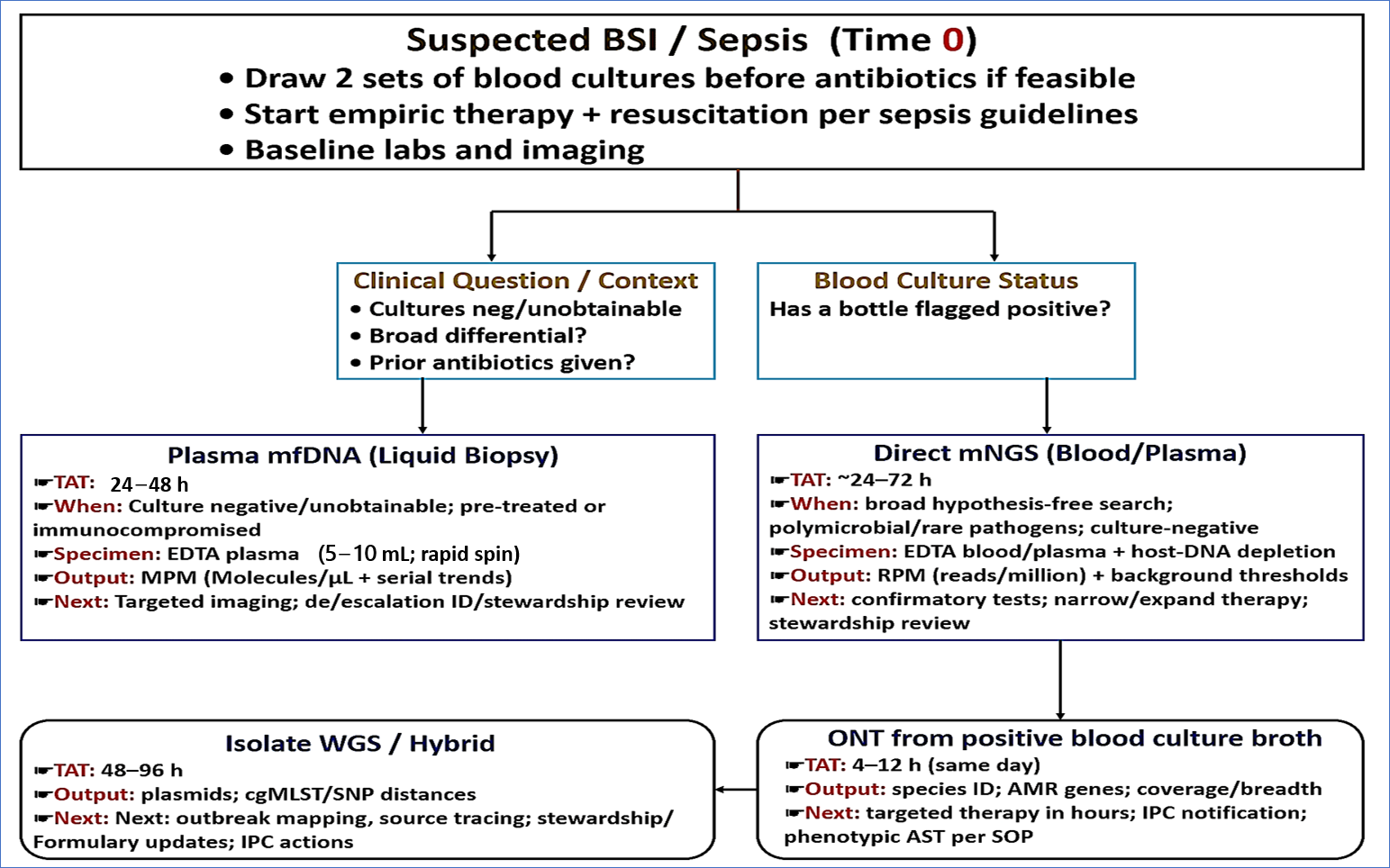
3.2. Sample to Answer Routes in Suspected BSI
3.2.1. mcfDNA (Liquid Biopsy)
3.2.2. Direct Metagenomics from Blood or Plasma
3.2.3. Rapid Sequencing of Positive BCs
3.3. Targeted Sequencing: Focused Speed and Depth
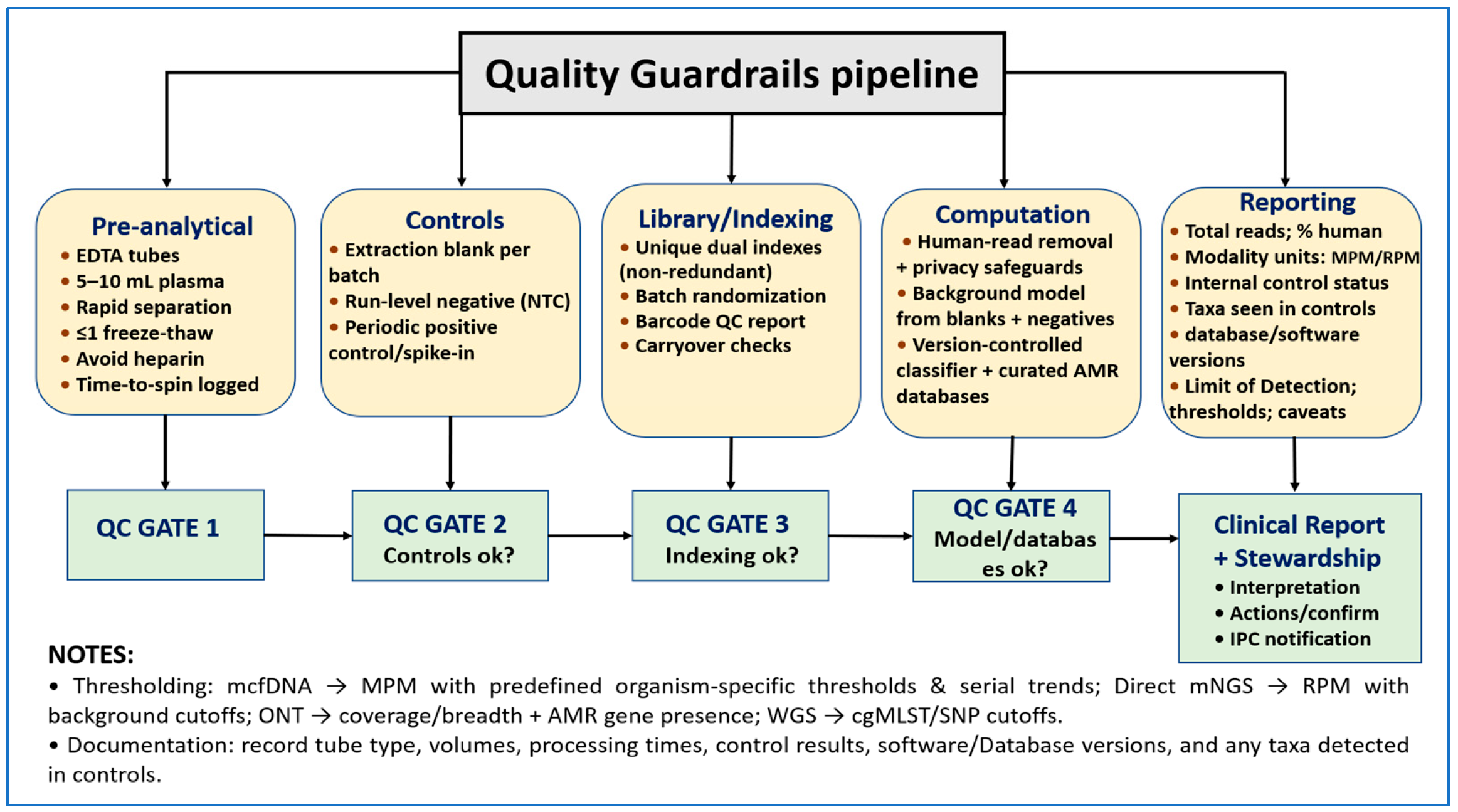
3.4. Bioinformatics, Reporting, and Quality Safeguards
3.5. TAT and Where Each Method Fits
4. Applications of NGS in BSIs
4.1. Bacterial BSIs: Rapid Pathogen ID and AMR Profiling
4.2. Viral BSIs
4.3. Fungal BSIs
4.4. Polymicrobial and Culture-Negative Infections
5. Technical and Translational Challenges
5.1. Sample Preparation (Low Microbial Load, Host-DNA Depletion, and Preanalytics)
5.2. Bioinformatics Variability (Classifier Choice, AMR Annotation, and Contamination Control)
5.3. Interpretation Hurdles (Infection vs. Colonization vs. Background)
5.4. Economic and Regulatory Barriers (Cost, TAT, Reimbursement)
6. Future Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Holmes, C.L.; Albin, O.R.; Mobley, H.L.; Bachman, M.A. Bloodstream infections: Mechanisms of pathogenesis and opportunities for intervention. Nat. Rev. Microbiol. 2025, 23, 210–224. [Google Scholar] [CrossRef]
- Costa, S.P.; Carvalho, C.M. Burden of bacterial bloodstream infections and recent advances for diagnosis. Pathog. Dis. 2022, 80, ftac027. [Google Scholar] [CrossRef]
- Santella, B.; Folliero, V.; Pirofalo, G.M.; Serretiello, E.; Zannella, C.; Moccia, G.; Santoro, E.; Sanna, G.; Motta, O.; De Caro, F. Sepsis—A retrospective cohort study of bloodstream infections. Antibiotics 2020, 9, 851. [Google Scholar] [CrossRef]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef]
- Fernando, S.M.; Rochwerg, B.; Seely, A.J. Clinical implications of the third international consensus definitions for sepsis and septic shock (Sepsis-3). CMAJ 2018, 190, E1058–E1059. [Google Scholar] [CrossRef]
- Verboom, D.M.; Frencken, J.F.; Ong, D.S.; Horn, J.; van der Poll, T.; Bonten, M.J.; Cremer, O.L.; Klein Klouwenberg, P.M. Robustness of sepsis-3 criteria in critically ill patients. J. Intensive Care 2019, 7, 46. [Google Scholar] [CrossRef]
- WHO. Sepsis. Available online: https://www.who.int/news-room/fact-sheets/detail/sepsis (accessed on 13 September 2025).
- Opota, O.; Croxatto, A.; Prod’Hom, G.; Greub, G. Blood culture-based diagnosis of bacteraemia: State of the art. Clin. Microbiol. Infect. 2015, 21, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Gharib, A.A.; Abd El-Aziz, N.K.; Hussein, A.; Mohammed, E.A. Identification of Pyogenic Bacteria in Human and Animals Conventionally and Non-nucleic Acid Molecularly Using MALDI-TOF MS. Benha Vet. Med. J. 2018, 35, 263–273. [Google Scholar] [CrossRef]
- Scheer, C.; Fuchs, C.; Gründling, M.; Vollmer, M.; Bast, J.; Bohnert, J.; Zimmermann, K.; Hahnenkamp, K.; Rehberg, S.; Kuhn, S.-O. Impact of antibiotic administration on blood culture positivity at the beginning of sepsis: A prospective clinical cohort study. Clin. Microbiol. Infect. 2019, 25, 326–331. [Google Scholar] [CrossRef]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Aguilar, G.R.; Mestrovic, T.; Smith, G.; Han, C. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Elbehiry, A.; Abalkhail, A. Metagenomic Next-Generation Sequencing in Infectious Diseases: Clinical Applications, Translational Challenges, and Future Directions. Diagnostics 2025, 15, 1991. [Google Scholar] [CrossRef] [PubMed]
- Kamau, E.; Yang, S. Metagenomic sequencing of positive blood culture fluid for accurate bacterial and fungal species identification: A pilot study. Microorganisms 2023, 11, 1259. [Google Scholar] [CrossRef]
- Doan, T.; Acharya, N.R.; Pinsky, B.A.; Sahoo, M.K.; Chow, E.D.; Banaei, N.; Budvytiene, I.; Cevallos, V.; Zhong, L.; Zhou, Z. Metagenomic DNA sequencing for the diagnosis of intraocular infections. Ophthalmology 2017, 124, 1247–1248. [Google Scholar] [CrossRef]
- Chiu, C.Y.; Miller, S.A. Clinical metagenomics. Nat. Rev. Genet. 2019, 20, 341–355. [Google Scholar] [CrossRef]
- Batool, M.; Galloway-Peña, J. Clinical metagenomics—Challenges and future prospects. Front. Microbiol. 2023, 14, 1186424. [Google Scholar] [CrossRef]
- Wang, L.; Li, S.; Qin, J.; Tang, T.; Hong, J.; Tung, T.-H.; Xu, C.; Yu, S.; Qian, J. Clinical diagnosis application of metagenomic next-generation sequencing of plasma in suspected sepsis. Infect. Drug Resist. 2023, 16, 891–901. [Google Scholar] [CrossRef]
- Blauwkamp, T.A.; Thair, S.; Rosen, M.J.; Blair, L.; Lindner, M.S.; Vilfan, I.D.; Kawli, T.; Christians, F.C.; Venkatasubrahmanyam, S.; Wall, G.D. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat. Microbiol. 2019, 4, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Chang, E.J.; Ledeboer, N.; Messacar, K.; Lindner, M.S.; Venkatasubrahmanyam, S.; Wilber, J.C.; Vaughn, M.L.; Bercovici, S.; Perkins, B.A. Plasma microbial cell-free DNA sequencing from over 15,000 patients identified a broad spectrum of pathogens. J. Clin. Microbiol. 2023, 61, e01855-22. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhang, S.; Zhao, Y.; Ding, X.; Yang, F.; Zhao, Y. Diagnostic value of metagenomic next-generation sequencing in sepsis and bloodstream infection. Front. Cell. Infect. Microbiol. 2023, 13, 1117987. [Google Scholar]
- Harris, P.N.; Bauer, M.J.; Lüftinger, L.; Beisken, S.; Forde, B.M.; Balch, R.; Cotta, M.; Schlapbach, L.; Raman, S.; Shekar, K. Rapid nanopore sequencing and predictive susceptibility testing of positive blood cultures from intensive care patients with sepsis. Microbiol. Spectr. 2024, 12, e03065-23. [Google Scholar] [CrossRef]
- Sheka, D.; Alabi, N.; Gordon, P.M. Oxford nanopore sequencing in clinical microbiology and infection diagnostics. Brief. Bioinform. 2021, 22, bbaa403. [Google Scholar]
- Govender, K.N.; Street, T.L.; Sanderson, N.D.; Leach, L.; Morgan, M.; Eyre, D.W. Rapid clinical diagnosis and treatment of common, undetected, and uncultivable bloodstream infections using metagenomic sequencing from routine blood cultures with Oxford Nanopore. medRxiv 2025. medRxiv:2025.2001.2008.25320182. [Google Scholar]
- Garvey, M. Hospital acquired sepsis, disease prevalence, and recent advances in sepsis mitigation. Pathogens 2024, 13, 461. [Google Scholar] [CrossRef]
- Mestrovic, T.; Aguilar, G.R.; Swetschinski, L.R.; Ikuta, K.S.; Gray, A.P.; Weaver, N.D.; Han, C.; Wool, E.E.; Hayoon, A.G.; Hay, S.I. The burden of bacterial antimicrobial resistance in the WHO European region in 2019: A cross-country systematic analysis. Lancet Public Health 2022, 7, e897–e913. [Google Scholar]
- Lipworth, S.; Matlock, W.; Shaw, L.; Vihta, K.-D.; Rodger, G.; Chau, K.; Barker, L.; George, S.; Kavanagh, J.; Davies, T. The plasmidome associated with Gram-negative bloodstream infections: A large-scale observational study using complete plasmid assemblies. Nat. Commun. 2024, 15, 1612. [Google Scholar] [CrossRef]
- Hammerum, A.M.; Karstensen, K.T.; Roer, L.; Kaya, H.; Lindegaard, M.; Porsbo, L.J.; Kjerulf, A.; Pinholt, M.; Holzknecht, B.J.; Worning, P. Surveillance of vancomycin-resistant enterococci reveals shift in dominating clusters from vanA to vanB Enterococcus faecium clusters, Denmark, 2015 to 2022. Eurosurveillance 2024, 29, 2300633. [Google Scholar]
- Lohde, M.; Wagner, G.E.; Dabernig-Heinz, J.; Viehweger, A.; Braun, S.D.; Monecke, S.; Diezel, C.; Stein, C.; Marquet, M.; Ehricht, R. Accurate bacterial outbreak tracing with Oxford Nanopore sequencing and reduction of methylation-induced errors. Genome Res. 2024, 34, 2039–2047. [Google Scholar] [CrossRef]
- Sajib, M.S.I.; Brunker, K.; Oravcova, K.; Everest, P.; Murphy, M.E.; Forde, T. Advances in Host Depletion and Pathogen Enrichment Methods for Rapid Sequencing–Based Diagnosis of Bloodstream Infection. J. Mol. Diagn. 2024, 26, 741–753. [Google Scholar] [CrossRef]
- Eisenhofer, R.; Minich, J.J.; Marotz, C.; Cooper, A.; Knight, R.; Weyrich, L.S. Contamination in low microbial biomass microbiome studies: Issues and recommendations. Trends Microbiol. 2019, 27, 105–117. [Google Scholar] [CrossRef]
- Camargo, J.F.; Ahmed, A.; Morris, M.I.; Anjan, S.; Prado, C.E.; Martinez, O.V.; Dalai, S.C.; Komanduri, K.V. Next generation sequencing of microbial cell-free DNA for rapid noninvasive diagnosis of infectious diseases in immunocompromised hosts. Biol. Blood Marrow Transplant. 2019, 25, S356–S357. [Google Scholar] [CrossRef]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Crit. Care Med. 2021, 49, e1063–e1143. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef]
- Taxt, A.M.; Avershina, E.; Frye, S.A.; Naseer, U.; Ahmad, R. Rapid identification of pathogens, antibiotic resistance genes and plasmids in blood cultures by nanopore sequencing. Sci. Rep. 2020, 10, 7622. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.-Y.; Wu, H.-C.; Li, Y.-L.; Cheng, H.-W.; Liou, C.-H.; Chen, F.-J.; Liao, Y.-C. Comprehensive pathogen identification and antimicrobial resistance prediction from positive blood cultures using nanopore sequencing technology. Genome Med. 2024, 16, 141. [Google Scholar] [CrossRef]
- Dabernig-Heinz, J.; Lohde, M.; Hölzer, M.; Cabal, A.; Conzemius, R.; Brandt, C.; Kohl, M.; Halbedel, S.; Hyden, P.; Fischer, M.A. A multicenter study on accuracy and reproducibility of nanopore sequencing-based genotyping of bacterial pathogens. J. Clin. Microbiol. 2024, 62, e00628-24. [Google Scholar] [CrossRef] [PubMed]
- Wenger, A.M.; Peluso, P.; Rowell, W.J.; Chang, P.-C.; Hall, R.J.; Concepcion, G.T.; Ebler, J.; Fungtammasan, A.; Kolesnikov, A.; Olson, N.D. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 2019, 37, 1155–1162. [Google Scholar] [CrossRef]
- Han, Y.; He, J.; Li, M.; Peng, Y.; Jiang, H.; Zhao, J.; Li, Y.; Deng, F. Unlocking the Potential of Metagenomics with the PacBio High-Fidelity Sequencing Technology. Microorganisms 2024, 12, 2482. [Google Scholar] [CrossRef]
- Espinosa, E.; Bautista, R.; Larrosa, R.; Plata, O. Advancements in long-read genome sequencing technologies and algorithms. Genomics 2024, 116, 110842. [Google Scholar] [CrossRef]
- Mahmoud, M.; Huang, Y.; Garimella, K.; Audano, P.A.; Wan, W.; Prasad, N.; Handsaker, R.E.; Hall, S.; Pionzio, A.; Schatz, M.C. Utility of long-read sequencing for All of Us. Nat. Commun. 2024, 15, 837. [Google Scholar] [CrossRef]
- Khezri, A.; Avershina, E.; Ahmad, R. Hybrid assembly provides improved resolution of plasmids, antimicrobial resistance genes, and virulence factors in Escherichia coli and Klebsiella pneumoniae clinical isolates. Microorganisms 2021, 9, 2560. [Google Scholar] [CrossRef]
- Fung, M.; Patel, N.; DeVoe, C.; Ryan, C.N.; McAdams, S.; Pamula, M.; Dwivedi, A.; Teraoka, J.; Smollin, M.; Sam, S. Utility of serial microbial cell-free DNA sequencing for inpatient and outpatient pathogen surveillance among allogeneic hematopoietic stem cell transplant recipients. Open Forum Infect. Dis. 2024, 11, ofae330. [Google Scholar] [CrossRef]
- Bergin, S.P.; Chemaly, R.F.; Dadwal, S.S.; Hill, J.A.; Lee, Y.J.; Haidar, G.; Luk, A.; Drelick, A.; Chin-Hong, P.V.; Benamu, E. Plasma microbial cell-free DNA sequencing in immunocompromised patients with pneumonia: A prospective observational study. Clin. Infect. Dis. 2024, 78, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Shah, J.R.; Sohail, M.R.; Lasco, T.; Goss, J.A.; Mohajer, M.A.; Khalil, S. Clinical utility of plasma microbial cell-free DNA sequencing in determining microbiologic etiology of infectious syndromes in solid organ transplant recipients. Ther. Adv. Infect. Dis. 2024, 11, 20499361241308643. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; de Vries, C.R.; Ruffin, F.; Sharma-Kuinkel, B.; Park, L.; Hong, D.; Scott, E.R.; Blair, L.; Degner, N.; Hollemon, D.H. Microbial cell-free DNA identifies etiology of bloodstream infections, persists longer than conventional blood cultures, and its duration of detection is associated with metastatic infection in patients with Staphylococcus aureus and gram-negative bacteremia. Clin. Infect. Dis. 2022, 74, 2020–2027. [Google Scholar]
- Han, D.; Li, R.; Shi, J.; Tan, P.; Zhang, R.; Li, J. Liquid biopsy for infectious diseases: A focus on microbial cell-free DNA sequencing. Theranostics 2020, 10, 5501. [Google Scholar] [CrossRef] [PubMed]
- Toldi, G.; Majid, A. The role of microbial cell free DNA sequencing in sepsis detection in the neonate. Pediatr. Res. 2025, 97, 464–465. [Google Scholar] [CrossRef] [PubMed]
- Sutton, A.J.; Lupu, D.S.; Bergin, S.P.; Holland, T.L.; McAdams, S.A.; Dadwal, S.S.; Nguyen, K.; Nolte, F.S.; Tremblay, G.; Perkins, B.A. Cost-Effectiveness of Plasma Microbial Cell-Free DNA Sequencing When Added to Usual Care Diagnostic Testing for Immunocompromised Host Pneumonia. PharmacoEconomics 2024, 42, 1029–1045. [Google Scholar] [CrossRef]
- Shean, R.C.; Garrett, E.; Malleis, J.; Lieberman, J.A.; Bradley, B.T. A retrospective observational study of mNGS test utilization to examine the role of diagnostic stewardship at two academic medical centers. J. Clin. Microbiol. 2024, 62, e00605–e00624. [Google Scholar] [CrossRef]
- Graf, E.H.; Bryan, A.; Bowers, M.; Grys, T.E. One size fits small: The narrow utility for plasma metagenomics. J. Appl. Lab. Med. 2025, 10, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Yu, X.; Gai, W.; Liu, Y.; Qi, Y.; Zheng, Y.; Zhang, N.; Cai, X.; Li, G.; Chen, B. Diagnostic value of plasma and blood cells metagenomic next-generation sequencing in patients with sepsis. Biochem. Biophys. Res. Commun. 2023, 683, 149079. [Google Scholar] [CrossRef]
- Peng, H.; Pan, M.; Zhou, Z.; Chen, C.; Xing, X.; Cheng, S.; Zhang, S.; Zheng, H.; Qian, K. The impact of preanalytical variables on the analysis of cell-free DNA from blood and urine samples. Front. Cell Dev. Biol. 2024, 12, 1385041. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Woo, H.I.; Kim, J.-W.; Kim, Y.; Lee, K.-A. Clinical practice guidelines for pre-analytical procedures of plasma epidermal growth factor receptor variant testing. Ann. Lab. Med. 2022, 42, 141–149. [Google Scholar] [CrossRef]
- García, M.E.; Blanco, J.L.; Caballero, J.s.; Gargallo-Viola, D. Anticoagulants interfere with PCR used to diagnose invasive aspergillosis. J. Clin. Microbiol. 2002, 40, 1567–1568. [Google Scholar] [CrossRef]
- Rajar, P.; Dhariwal, A.; Salvadori, G.; Junges, R.; Åmdal, H.A.; Berild, D.; Fugelseth, D.; Saugstad, O.D.; Lausten-Thomsen, U.; Greisen, G. Microbial DNA extraction of high-host content and low biomass samples: Optimized protocol for nasopharynx metagenomic studies. Front. Microbiol. 2022, 13, 1038120. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Jurasz, H.; Pawłowski, T.; Perlejewski, K. Contamination issue in viral metagenomics: Problems, solutions, and clinical perspectives. Front. Microbiol. 2021, 12, 745076. [Google Scholar] [CrossRef]
- Schlaberg, R.; Chiu, C.Y.; Miller, S.; Procop, G.W.; Weinstock, G.; Professional Practice Committee and Committee on Laboratory Practices of the American Society for Microbiology; Microbiology Resource Committee of the College of American Pathologists. Validation of metagenomic next-generation sequencing tests for universal pathogen detection. Arch. Pathol. Lab. Med. 2017, 141, 776–786. [Google Scholar] [CrossRef]
- Pang, F.; Xu, W.; Zhao, H.; Chen, S.; Tian, Y.; Fu, J.; You, Z.; Song, P.; Xian, Q.; Zhao, Q. Comprehensive evaluation of plasma microbial cell-free DNA sequencing for predicting bloodstream and local infections in clinical practice: A multicenter retrospective study. Front. Cell. Infect. Microbiol. 2024, 13, 1256099. [Google Scholar] [CrossRef]
- Benoit, P.; Brazer, N.; de Lorenzi-Tognon, M.; Kelly, E.; Servellita, V.; Oseguera, M.; Nguyen, J.; Tang, J.; Omura, C.; Streithorst, J. Seven-year performance of a clinical metagenomic next-generation sequencing test for diagnosis of central nervous system infections. Nat. Med. 2024, 30, 3522–3533. [Google Scholar] [CrossRef]
- Yuan, L.; Zhu, X.Y.; Lai, L.M.; Chen, Q.; Liu, Y.; Zhao, R. Clinical application and evaluation of metagenomic next-generation sequencing in pathogen detection for suspected central nervous system infections. Sci. Rep. 2024, 14, 16961. [Google Scholar] [CrossRef]
- Costello, M.; Fleharty, M.; Abreu, J.; Farjoun, Y.; Ferriera, S.; Holmes, L.; Granger, B.; Green, L.; Howd, T.; Mason, T. Characterization and remediation of sample index swaps by non-redundant dual indexing on massively parallel sequencing platforms. BMC Genom. 2018, 19, 332. [Google Scholar] [CrossRef] [PubMed]
- Clarridge, J.E., III. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 2004, 17, 840–862. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium; Fungal Barcoding Consortium Author List. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Sajib, M.S.I.; Oravcova, K.; Brunker, K.; Everest, P.; Galarion, M.J.H.; Fuentes, M.; Wilson, C.; Murphy, M.E.; Forde, T. MultiSeq-AMR: A modular amplicon-sequencing workflow for rapid detection of bloodstream infection and antimicrobial resistance markers. Microb. Genom. 2025, 11, 001383. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brandalise, D.; Coste, A.T.; Sanglard, D.; Lamoth, F. Exploration of novel mechanisms of azole resistance in Candida auris. Antimicrob. Agents Chemother. 2024, 68, e01265-24. [Google Scholar] [CrossRef]
- Quek, Z.R.; Ng, S.H. Hybrid-capture target enrichment in human pathogens: Identification, evolution, biosurveillance, and genomic epidemiology. Pathogens 2024, 13, 275. [Google Scholar] [CrossRef]
- STROBE-Metagenomics. A STROBE Extension Statement to Guide the Reporting of Metagenomics Studies. EQUATOR Network. Available online: https://www.equator-network.org/reporting-guidelines/strobe-metagenomics-a-strobe-extension-statement-to-guide-the-reporting-of-metagenomics-studies/ (accessed on 12 November 2025).
- Illumina. Minimize Index Hopping in Multiplexed Runs: Tips and Best Practices to Avoid Sequencing Read Misalignment Associated with Index Switching. Available online: https://www.illumina.com/techniques/sequencing/ngs-library-prep/multiplexing/index-hopping.html (accessed on 12 November 2025).
- Balks, J.; Grumaz, S.; Mazzitelli, S.; Neder, U.; Lemloh, L.; Melaku, T.; Glaser, K.; Mueller, A.; Kipfmueller, F. Microbial cell-free DNA-sequencing as an addition to conventional diagnostics in neonatal sepsis. Pediatr. Res. 2025, 97, 614–624. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, G.; Lau, H.C.-H.; Yu, J. Metagenomic sequencing for microbial DNA in human samples: Emerging technological advances. Int. J. Mol. Sci. 2022, 23, 2181. [Google Scholar] [CrossRef]
- Fierer, N.; Leung, P.M.; Lappan, R.; Eisenhofer, R.; Ricci, F.; Holland, S.I.; Dragone, N.; Blackall, L.L.; Dong, X.; Dorador, C. Guidelines for preventing and reporting contamination in low-biomass microbiome studies. Nat. Microbiol. 2025, 10, 1570–1580. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast identification and removal of sequence contamination from genomic and metagenomic datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef] [PubMed]
- Farouni, R.; Djambazian, H.; Ferri, L.E.; Ragoussis, J.; Najafabadi, H.S. Model-based analysis of sample index hopping reveals its widespread artifacts in multiplexed single-cell RNA-sequencing. Nat. Commun. 2020, 11, 2704. [Google Scholar] [CrossRef]
- Hornung, B.V.; Zwittink, R.D.; Kuijper, E.J. Issues and current standards of controls in microbiome research. FEMS Microbiol. Ecol. 2019, 95, fiz045. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Lu, J.; Rincon, N.; Wood, D.E.; Breitwieser, F.P.; Pockrandt, C.; Langmead, B.; Salzberg, S.L.; Steinegger, M. Metagenome analysis using the Kraken software suite. Nat. Protoc. 2022, 17, 2815–2839. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Bharucha, T.; Oeser, C.; Balloux, F.; Brown, J.R.; Carbo, E.C.; Charlett, A.; Chiu, C.Y.; Claas, E.C.; de Goffau, M.C.; de Vries, J.J. STROBE-metagenomics: A STROBE extension statement to guide the reporting of metagenomics studies. Lancet Infect. Dis. 2020, 20, e251–e260. [Google Scholar] [CrossRef]
- Hogan, C.A.; Miller, S.; Piantadosi, A.; Gaston, D.C.; Simner, P.J.; Nash, S.; Babady, N.E. Which trial do we need? Plasma metagenomic next-generation sequencing to diagnose infections in patients with haematological malignancies and febrile neutropenia: Proposal for a randomized-controlled trial. Clin. Microbiol. Infect. 2023, 29, 1474–1479. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. Peer J. Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Song, L.; Breitwieser, F.P.; Salzberg, S.L. Centrifuge: Rapid and sensitive classification of metagenomic sequences. Genome Res. 2016, 26, 1721–1729. [Google Scholar] [CrossRef]
- Davis, N.M.; Proctor, D.M.; Holmes, S.P.; Relman, D.A.; Callahan, B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 2018, 6, 226. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed]
- Wilm, A.; Aw, P.P.K.; Bertrand, D.; Yeo, G.H.T.; Ong, S.H.; Wong, C.H.; Khor, C.C.; Petric, R.; Hibberd, M.L.; Nagarajan, N. LoFreq: A sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012, 40, 11189–11201. [Google Scholar] [CrossRef]
- Agustinho, D.P.; Fu, Y.; Menon, V.K.; Metcalf, G.A.; Treangen, T.J.; Sedlazeck, F.J. Unveiling microbial diversity: Harnessing long-read sequencing technology. Nat. Methods 2024, 21, 954–966. [Google Scholar] [CrossRef]
- Kullar, R.; Chisari, E.; Snyder, J.; Cooper, C.; Parvizi, J.; Sniffen, J. Next-generation sequencing supports targeted antibiotic treatment for culture negative orthopedic infections. Clin. Infect. Dis. 2023, 76, 359–364. [Google Scholar]
- Harris, S.R.; Cartwright, E.J.; Török, M.E.; Holden, M.T.; Brown, N.M.; Ogilvy-Stuart, A.L.; Ellington, M.J.; Quail, M.A.; Bentley, S.D.; Parkhill, J. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: A descriptive study. Lancet Infect. Dis. 2013, 13, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Permana, B.; Harris, P.N.; Runnegar, N.; Lindsay, M.; Henderson, B.C.; Playford, E.; Paterson, D.L.; Beatson, S.A.; Forde, B.M. Using genomics to investigate an outbreak of vancomycin-resistant Enterococcus faecium ST78 at a large tertiary hospital in Queensland. Microbiol. Spectr. 2023, 11, e04204–e04222. [Google Scholar] [CrossRef]
- Gomez-Simmonds, A.; Annavajhala, M.K.; Seeram, D.; Hokunson, T.W.; Park, H.; Uhlemann, A.-C. Genomic epidemiology of carbapenem-resistant Enterobacterales at a New York City hospital over a 10-year period reveals complex plasmid-clone dynamics and evidence for frequent horizontal transfer of blaKPC. Genome Res. 2024, 34, 1895–1907. [Google Scholar] [CrossRef] [PubMed]
- Snitkin, E.S.; Zelazny, A.M.; Thomas, P.J.; Stock, F.; Program, N.C.S.; Henderson, D.K.; Palmore, T.N.; Segre, J.A. Tracking a hospital outbreak of carbapenem-resistant Klebsiella pneumoniae with whole-genome sequencing. Sci. Transl. Med. 2012, 4, ra116–ra148. [Google Scholar] [CrossRef]
- Spencer, M.D.; Winglee, K.; Passaretti, C.; Earl, A.M.; Manson, A.L.; Mulder, H.P.; Sautter, R.L.; Fodor, A.A. Whole genome sequencing detects inter-facility transmission of carbapenem-resistant Klebsiella pneumoniae. J. Infect. 2019, 78, 187–199. [Google Scholar] [CrossRef]
- Han, W.; Zhou, P.; Chen, C.; Wu, C.; Shen, L.; Wan, C.; Xiao, Y.; Zhang, J.; Wang, B.; Shi, J. Characteristic of KPC-12, a KPC variant conferring resistance to ceftazidime-avibactam in the carbapenem-resistant Klebsiella pneumoniae ST11-KL47 clone background. Infect. Drug Resist. 2024, 17, 2541–2554. [Google Scholar] [CrossRef]
- Marimuthu, K.; Venkatachalam, I.; Koh, V.; Harbarth, S.; Perencevich, E.; Cherng, B.P.Z.; Fong, R.K.C.; Pada, S.K.; Ooi, S.T.; Smitasin, N. Whole genome sequencing reveals hidden transmission of carbapenemase-producing Enterobacterales. Nat. Commun. 2022, 13, 3052. [Google Scholar] [CrossRef]
- Tsukada, M.; Miyazaki, T.; Aoki, K.; Yoshizawa, S.; Kondo, Y.; Sawa, T.; Murakami, H.; Sato, E.; Tomida, M.; Otani, M. The outbreak of multispecies carbapenemase-producing Enterobacterales associated with pediatric ward sinks: IncM1 plasmids act as vehicles for cross-species transmission. Am. J. Infect. Control 2024, 52, 801–806. [Google Scholar] [CrossRef]
- Kallonen, T.; Brodrick, H.J.; Harris, S.R.; Corander, J.; Brown, N.M.; Martin, V.; Peacock, S.J.; Parkhill, J. Systematic longitudinal survey of invasive Escherichia coli in England demonstrates a stable population structure only transiently disturbed by the emergence of ST131. Genome Res. 2017, 27, 1437–1449. [Google Scholar] [CrossRef] [PubMed]
- Rath, A.; Kieninger, B.; Fritsch, J.; Caplunik-Pratsch, A.; Blaas, S.; Ochmann, M.; Pfeifer, M.; Hartl, J.; Holzmann, T.; Schneider-Brachert, W. Whole-genome sequencing reveals two prolonged simultaneous outbreaks involving Pseudomonas aeruginosa high-risk strains ST111 and ST235 with resistance to quaternary ammonium compounds. J. Hosp. Infect. 2024, 145, 155–164. [Google Scholar] [CrossRef]
- Aranzamendi, M.; Xanthopoulou, K.; Sánchez-Urtaza, S.; Burgwinkel, T.; Arazo del Pino, R.; Lucaßen, K.; Perez-Vazquez, M.; Oteo-Iglesias, J.; Sota, M.; Marimón, J.M. Genomic surveillance uncovers a 10-year persistence of an OXA-24/40 Acinetobacter baumannii clone in a tertiary hospital in Northern Spain. Int. J. Mol. Sci. 2024, 25, 2333. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, F.; Yuan, D.; Zhai, W.; Liu, S.; Zhou, Y.; Yang, H. HIV-1 drug resistance detected by next-generation sequencing among ART-naïve individuals: A systematic review and meta-analysis. Viruses 2024, 16, 239. [Google Scholar] [CrossRef]
- Li, J.Z.; Paredes, R.; Ribaudo, H.J.; Svarovskaia, E.S.; Metzner, K.J.; Kozal, M.J.; Hullsiek, K.H.; Balduin, M.; Jakobsen, M.R.; Geretti, A.M.; et al. Low-Frequency HIV-1 Drug Resistance Mutations and Risk of NNRTI-Based Antiretroviral Treatment Failure: A Systematic Review and Pooled Analysis. JAMA 2011, 305, 1327–1335. [Google Scholar]
- El Bouzidi, K.; Datir, R.P.; Kwaghe, V.; Roy, S.; Frampton, D.; Breuer, J.; Ogbanufe, O.; Murtala-Ibrahim, F.; Charurat, M.; Dakum, P. Deep sequencing of HIV-1 reveals extensive subtype variation and drug resistance after failure of first-line antiretroviral regimens in Nigeria. J. Antimicrob. Chemother. 2022, 77, 474–482. [Google Scholar] [CrossRef]
- Terrault, N.A.; Lok, A.S.; McMahon, B.J.; Chang, K.M.; Hwang, J.P.; Jonas, M.M.; Brown Jr, R.S.; Bzowej, N.H.; Wong, J.B. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Hepatology 2018, 67, 1560–1599. [Google Scholar] [CrossRef] [PubMed]
- AASLD. Chronic Hepatitis B. Available online: https://www.aasld.org/practice-guidelines/chronic-hepatitis-b (accessed on 22 September 2025).
- Lowe, C.F.; Merrick, L.; Harrigan, P.R.; Mazzulli, T.; Sherlock, C.H.; Ritchie, G. Implementation of next-generation sequencing for hepatitis B virus resistance testing and genotyping in a clinical microbiology laboratory. J. Clin. Microbiol. 2016, 54, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Mokaya, J.; McNaughton, A.L.; Bester, P.A.; Goedhals, D.; Barnes, E.; Marsden, B.D.; Matthews, P.C. Hepatitis B virus resistance to tenofovir: Fact or fiction? A systematic literature review and structural analysis of drug resistance mechanisms. Wellcome Open Res. 2020, 5, 151. [Google Scholar] [CrossRef] [PubMed]
- AASLD. HCV Resistance Primer: HCV Guidance: Recommendations for Testing, Managing, and Treating Hepatitis C. Available online: https://www.hcvguidelines.org/evaluate/resistance (accessed on 12 September 2025).
- Panel, A.-I.H.C.G. Hepatitis C guidance 2019 update: AASLD-IDSA recommendations for testing, managing, and treating hepatitis C virus infection. Hepatology 2020, 71, 686. [Google Scholar]
- Sahoo, M.K.; Lefterova, M.I.; Yamamoto, F.; Waggoner, J.J.; Chou, S.; Holmes, S.P.; Anderson, M.W.; Pinsky, B.A. Detection of cytomegalovirus drug resistance mutations by next-generation sequencing. J. Clin. Microbiol. 2013, 51, 3700–3710. [Google Scholar] [CrossRef]
- López-Aladid, R.; Guiu, A.; Mosquera, M.M.; López-Medrano, F.; Cofán, F.; Linares, L.; Torre-Cisneros, J.; Vidal, E.; Moreno, A.; Aguado, J.M. Improvement in detecting cytomegalovirus drug resistance mutations in solid organ transplant recipients with suspected resistance using next generation sequencing. PLoS ONE 2019, 14, e0219701. [Google Scholar] [CrossRef]
- Mallory, M.A.; Hymas, W.C.; Simmon, K.E.; Pyne, M.T.; Stevenson, J.B.; Barker, A.P.; Hillyard, D.R.; Hanson, K.E. Development and validation of a next-generation sequencing assay with open-access analysis software for detecting resistance-associated mutations in CMV. J. Clin. Microbiol. 2023, 61, e00829-23. [Google Scholar] [CrossRef]
- Andreani, J.; Truffot, A.; Tilloy, V.; Jardin, H.; Lespinasse, M.; Usal, M.; Larrat, S.; Morand, P.; Lupo, J.; Hantz, S. Long-range PCRs and next-generation sequencing to detect cytomegalovirus drug resistance-associated mutations. Antimicrob. Agents Chemother. 2025, 69, e0014125. [Google Scholar] [CrossRef]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Hogan, C.A.; Stevens, B.A.; Sahoo, M.K.; Huang, C.; Garamani, N.; Gombar, S.; Yamamoto, F.; Murugesan, K.; Kurzer, J.; Zehnder, J. High frequency of SARS-CoV-2 RNAemia and association with severe disease. Clin. Infect. Dis. 2021, 72, e291–e295. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, C.; Takov, K.; Burnap, S.A.; Singh, B.; Ali, H.; Theofilatos, K.; Reed, E.; Hasman, M.; Nabeebaccus, A.; Fish, M. SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat. Commun. 2021, 12, 3406. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Serrano, D.A.; Roy-Vallejo, E.; Zurita Cruz, N.D.; Martín Ramírez, A.; Rodríguez-García, S.C.; Arevalillo-Fernández, N.; Galván-Román, J.M.; Fontán García-Rodrigo, L.; Vega-Piris, L.; Chicot Llano, M. Detection of SARS-CoV-2 RNA in serum is associated with increased mortality risk in hospitalized COVID-19 patients. Sci. Rep. 2021, 11, 13134. [Google Scholar] [CrossRef]
- Lockhart, S.R.; Etienne, K.A.; Vallabhaneni, S.; Farooqi, J.; Chowdhary, A.; Govender, N.P.; Colombo, A.L.; Calvo, B.; Cuomo, C.A.; Desjardins, C.A. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2017, 64, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Chow, N.A.; Muñoz, J.F.; Gade, L.; Berkow, E.L.; Li, X.; Welsh, R.M.; Forsberg, K.; Lockhart, S.R.; Adam, R.; Alanio, A. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. MBio 2020, 11, e03364-19. [Google Scholar] [CrossRef]
- Munoz, J.F.; Welsh, R.M.; Shea, T.; Batra, D.; Gade, L.; Howard, D.; Rowe, L.A.; Meis, J.F.; Litvintseva, A.P.; Cuomo, C.A. Clade-specific chromosomal rearrangements and loss of subtelomeric adhesins in Candida auris. Genetics 2021, 218, iyab029. [Google Scholar] [CrossRef]
- Kappel, D.; Gifford, H.; Brackin, A.; Abdolrasouli, A.; Eyre, D.W.; Jeffery, K.; Schlenz, S.; Aanensen, D.M.; Brown, C.S.; Borman, A. Genomic epidemiology describes introduction and outbreaks of antifungal drug-resistant Candida auris. npj Antimicrob. Resist. 2024, 2, 26. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Kim, Y.-J.; Yong, D.; Byun, J.-H.; Kim, T.S.; Chang, Y.S.; Choi, M.J.; Byeon, S.A.; Won, E.J.; Kim, S.H. Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg. Infect. Dis. 2018, 24, 1768. [Google Scholar] [CrossRef]
- McTaggart, L.R.; Eshaghi, A.; Hota, S.; Poutanen, S.M.; Johnstone, J.; De Luca, D.G.; Bharat, A.; Patel, S.N.; Kus, J.V. First Canadian report of transmission of fluconazole-resistant Candida parapsilosis within two hospital networks confirmed by genomic analysis. J. Clin. Microbiol. 2024, 62, e01161-23. [Google Scholar] [CrossRef]
- Alexander, B.D.; Johnson, M.D.; Pfeiffer, C.D.; Jiménez-Ortigosa, C.; Catania, J.; Booker, R.; Castanheira, M.; Messer, S.A.; Perlin, D.S.; Pfaller, M.A. Increasing echinocandin resistance in Candida glabrata: Clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013, 56, 1724–1732. [Google Scholar] [CrossRef]
- Beyda, N.D.; John, J.; Kilic, A.; Alam, M.J.; Lasco, T.M.; Garey, K.W. FKS mutant Candida glabrata: Risk factors and outcomes in patients with candidemia. Clin. Infect. Dis. 2014, 59, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Verweij, P.E.; Chowdhary, A.; Melchers, W.J.; Meis, J.F. Azole resistance in Aspergillus fumigatus: Can we retain the clinical use of mold-active antifungal azoles? Clin. Infect. Dis. 2016, 62, 362–368. [Google Scholar] [CrossRef]
- Burks, C.; Darby, A.; Gómez Londoño, L.; Momany, M.; Brewer, M.T. Azole-resistant Aspergillus fumigatus in the environment: Identifying key reservoirs and hotspots of antifungal resistance. PLoS Pathog. 2021, 17, e1009711. [Google Scholar] [CrossRef]
- Alvarez-Moreno, C.; Lavergne, R.-A.; Hagen, F.; Morio, F.; Meis, J.F.; Le Pape, P. Azole-resistant Aspergillus fumigatus harboring TR34/L98H, TR46/Y121F/T289A and TR53 mutations related to flower fields in Colombia. Sci. Rep. 2017, 7, 45631. [Google Scholar] [CrossRef]
- Ashton, P.; Thanh, L.; Trieu, P.; Van Anh, D.; Trinh, N.; Beardsley, J.; Kibengo, F.; Chierakul, W.; Dance, D.; Rattanavong, S. Three phylogenetic groups have driven the recent population expansion of Cryptococcus neoformans. Nat. Commun. 2019, 10, 2035. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, C.A.; Giamberardino, C.; Sykes, S.M.; Yu, C.-H.; Tenor, J.L.; Chen, Y.; Yang, T.; Jones, A.M.; Sun, S.; Haverkamp, M.R. Population genomics and the evolution of virulence in the fungal pathogen Cryptococcus neoformans. Genome Res. 2017, 27, 1207–1219. [Google Scholar] [CrossRef]
- Florent, M.; Noël, T.; Ruprich-Robert, G.; Da Silva, B.; Fitton-Ouhabi, V.; Chastin, C.; Papon, N.; Chapeland-Leclerc, F. Nonsense and missense mutations in FCY2 and FCY1 genes are responsible for flucytosine resistance and flucytosine-fluconazole cross-resistance in clinical isolates of Candida lusitaniae. Antimicrob. Agents Chemother. 2009, 53, 2982–2990. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Lamichhane, A.K.; Cai, H.; Walter, P.J.; Bennett, J.E.; Kwon-Chung, K.J. Moderate levels of 5-fluorocytosine cause the emergence of high frequency resistance in cryptococci. Nat. Commun. 2021, 12, 3418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Z.; Chen, Y.; Zhou, Z.; Yang, Q.; Fu, Y.; Zhao, F.; Li, X.; Chen, Q.; Fang, L. Antifungal susceptibility and molecular characteristics of Cryptococcus spp. based on whole-genome sequencing in Zhejiang Province, China. Front. Microbiol. 2022, 13, 991703. [Google Scholar] [CrossRef]
- Kaur, I.; Shaw, B.; Multani, A.; Pham, C.; Malhotra, S.; Smith, E.; Adachi, K.; Allyn, P.; Bango, Z.; Beaird, O.E. Real-world clinical impact of plasma cell-free DNA metagenomic next-generation sequencing assay. Infect. Control Hosp. Epidemiol. 2025, 46, 504–511. [Google Scholar] [CrossRef]
- Khalil, S.; Paras, M.L.; Eichenberger, E.; Sohail, M.R. The Next Step: Role of Metagenomic Next Generation Sequencing for Microbial Detection in Culture-negative Cardiovascular infections. Clin. Infect. Dis. 2025, ciaf361. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, R.; Zhu, Y.; Hu, L.; Xia, H.; Li, J.; Ye, Y. Metagenomic next-generation sequencing of plasma cell-free DNA improves the early diagnosis of suspected infections. BMC Infect. Dis. 2024, 24, 187. [Google Scholar] [CrossRef]
- Yang, H.; Xu, N.; Yan, M.; Yang, L.; Wen, S.; Wang, S.; Qu, C.; Xu, K.; Yang, X.; Wang, G. Comparison of metagenomic next-generation sequencing and conventional culture for the diagnostic performance in febrile patients with suspected infections. BMC Infect. Dis. 2024, 24, 350. [Google Scholar] [CrossRef]
- Zhou, Y.; Shi, W.; Wen, Y.; Mao, E.; Ni, T. Comparison of pathogen detection consistency between metagenomic next-generation sequencing and blood culture in patients with suspected bloodstream infection. Sci. Rep. 2023, 13, 9460. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, L.; Gao, D.; Wang, J.; Li, Y.; Sun, N. Comparison of metagenomic next-generation sequencing and blood culture for diagnosis of bloodstream infections. Front. Cell. Infect. Microbiol. 2024, 14, 1338861. [Google Scholar] [CrossRef]
- Eichenberger, E.M.; Degner, N.; Scott, E.R.; Ruffin, F.; Franzone, J.; Sharma-Kuinkel, B.; Shah, P.; Hong, D.; Dalai, S.C.; Blair, L. Microbial cell-free DNA identifies the causative pathogen in infective endocarditis and remains detectable longer than conventional blood culture in patients with prior antibiotic therapy. Clin. Infect. Dis. 2023, 76, e1492–e1500. [Google Scholar] [CrossRef] [PubMed]
- Derache, A.; Iwuji, C.C.; Danaviah, S.; Giandhari, J.; Marcelin, A.-G.; Calvez, V.; de Oliveira, T.; Dabis, F.; Pillay, D.; Gupta, R.K. Predicted antiviral activity of tenofovir versus abacavir in combination with a cytosine analogue and the integrase inhibitor dolutegravir in HIV-1-infected South African patients initiating or failing first-line ART. J. Antimicrob. Chemother. 2019, 74, 473–479. [Google Scholar] [CrossRef]
- Stower, H. Rapid lower respiratory tract infectious diagnosis. Nat. Med. 2019, 25, 1189. [Google Scholar] [CrossRef]
- Godsey, J.H.; Silvestro, A.; Barrett, J.C.; Bramlett, K.; Chudova, D.; Deras, I.; Dickey, J.; Hicks, J.; Johann, D.J.; Leary, R. Generic protocols for the analytical validation of next-generation sequencing-based ctDNA assays: A joint consensus recommendation of the BloodPAC’s Analytical Variables Working Group. Clin. Chem. 2020, 66, 1156–1166. [Google Scholar] [CrossRef]
- Febbo, P.G.; Martin, A.M.; Scher, H.I.; Barrett, J.C.; Beaver, J.A.; Beresford, P.J.; Blumenthal, G.M.; Bramlett, K.; Compton, C.; Dittamore, R. Minimum technical data elements for liquid biopsy data submitted to public databases. Clin. Pharmacol. Ther. 2020, 107, 730–734. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute (NCI) Biorepositories. NCI Biospecimen Evidence-Based Practices (BEBP) Cell-Free DNA: Biospecimen Collection and Processing; National Cancer Institute, National Institutes of Health: Bethesda, MD, USA, 2024. [Google Scholar]
- McArthur, A.G.; Waglechner, N.; Nizam, F.; Yan, A.; Azad, M.A.; Baylay, A.J.; Bhullar, K.; Canova, M.J.; De Pascale, G.; Ejim, L. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013, 57, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- AMA. CPT® Proprietary Laboratory Analyses (PLA) Codes: Long Descriptors. Available online: https://www.ama-assn.org/system/files/cpt-pla-codes-long.pdf (accessed on 15 September 2025).
- Anthem. Metagenomic Sequencing for Infectious Disease in the Outpatient Setting. Available online: https://www.anthem.com/medpolicies/abc/active/mp_pw_e002725.html (accessed on 15 September 2025).
- FDA. Letter to Kristen Kanack, Senior Vice President, Regulatory and Clinical Affairs, BioFire Diagnostics, LLC. Trade/Device Name: BioFire Blood Culture Identification 2 (BCID2) Panel; Regulation Number: 21 CFR 866.3365; Regulatory Class: Class II.; Product Code: PAM, PEO. 18 March 2020. Available online: https://www.fda.gov/media/146766/download (accessed on 11 November 2025).
- FDA. Letter to Karli Plenert, Sr. Director, Regulatory Affairs, BioFire Diagnostics, LLC. Trade/Device Name: BIOFIRE Blood Culture Identification 2 (BCID2) Panel (RFIT-ASY-0147, RFIT-ASY-0148); Regulation Number: 21 CFR 866.3365; Regulation Name: Multiplex Nucleic Acid Assay for Identification of Microorganisms and Resistance Markers from Positive Blood Cultures; Regulatory Class: Class II.; Product Code: PEN, PAM, PEO. 20 December 2024. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf24/K243544.pdf (accessed on 11 November 2025).
- FDA. 510(k) Premarket Notification: Device Classification Name: Gram-Negative Bacteria and Associated Resistance Markers; 510(k) Number: K193519; Device Name: BioFire Blood Culture Identification 2 (BCID2) Panel; Applicant: BioFire Diagnostics, LLC, 2019. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=K193519 (accessed on 11 November 2025).
- FDA. 510(k) Premarket Notification: Device Classification Name: Gram-Positive Bacteria and Their Resistance Markers; 510(k) Number: K181663; Device Name: ePlex Blood Culture Identification Panel—Gram Positive (BCID-GP) Panel; Applicant: GenMark Diagnostics, Inc., Carlsbad, CA, USA.; Regulation Number: 21 CFR 866.3365; Classification Product Code: PAM.; Subsequent Product Codes: PEN, PEO.; Decision: Substantially Equivalent (SESE). Decision Date: 20 December 2018. Available online: https://www.accessdata.fda.gov/cdrh_docs/reviews/K182619.pdf (accessed on 11 November 2025).
- Trivett, H.; Darby, A.C.; Oyebode, O. Academic and clinical perspectives of metagenome sequencing as a diagnostic tool for infectious disease: An interpretive phenomenological study. BMC Infect. Dis. 2025, 25, 448. [Google Scholar] [CrossRef]
- Yi, Q.; Zhang, G.; Wang, T.; Li, J.; Kang, W.; Zhang, J.; Liu, Y.; Xu, Y. Comparative Analysis of Metagenomic Next-Generation Sequencing, Sanger Sequencing, and Conventional Culture for Detecting Common Pathogens Causing Lower Respiratory Tract Infections in Clinical Samples. Microorganisms 2025, 13, 682. [Google Scholar] [CrossRef] [PubMed]
- Peri, A.M.; Chatfield, M.D.; Ling, W.; Furuya-Kanamori, L.; Harris, P.N.; Paterson, D.L. Rapid diagnostic tests and antimicrobial stewardship programs for the management of bloodstream infection: What is their relative contribution to improving clinical outcomes? A systematic review and network meta-analysis. Clin. Infect. Dis. 2024, 79, 502–515. [Google Scholar] [CrossRef]
- Liang, Q.; Bible, P.W.; Liu, Y.; Zou, B.; Wei, L. DeepMicrobes: Taxonomic classification for metagenomics with deep learning. NAR Genom. Bioinform. 2020, 2, lqaa009. [Google Scholar] [CrossRef]
- Peres da Silva, R.; Suphavilai, C.; Nagarajan, N. MetageNN: A memory-efficient neural network taxonomic classifier robust to sequencing errors and missing genomes. BMC Bioinform. 2024, 25, 153. [Google Scholar] [CrossRef] [PubMed]
- WHO. One Health Joint Plan of Action (2022–2026): Working Together for the Health of Humans, Animals, Plants and the Environment. Available online: https://www.who.int/publications/i/item/9789240059139 (accessed on 16 September 2025).
- Shanmugam, H.; Airen, L.; Rawat, S. Machine Learning and Deep Learning Models for Early Sepsis Prediction: A Scoping Review. Indian J. Crit. Care Med. Peer-Rev. Off. Publ. Indian Soc. Crit. Care Med. 2025, 29, 516. [Google Scholar]
- Boutin, S.; Welker, S.; Gerigk, M.; Miethke, T.; Heeg, K.; Nurjadi, D. Molecular surveillance of carbapenem-resistant Enterobacterales in two nearby tertiary hospitals to identify regional spread of high-risk clones in Germany, 2019–2020. J. Hosp. Infect. 2024, 149, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Oke, K.T.; Pitout, J.D.; Peirano, G.; Strydom, K.-A.; Kingsburgh, C.; Ehlers, M.M.; Ismail, A.; Takawira, F.T.; Kock, M.M. Molecular epidemiology of carbapenemase-producing Klebsiella pneumoniae in Gauteng South Africa. Sci. Rep. 2024, 14, 27337. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Bortolaia, V.; Tate, H.; Tyson, G.H.; Aarestrup, F.M.; McDermott, P.F. Using genomics to track global antimicrobial resistance. Front. Public Health 2019, 7, 242. [Google Scholar] [CrossRef]
| Task | Example Tools/Workflows | Primary Outputs | Key Notes | Key References |
|---|---|---|---|---|
| Human-read subtraction/alignment | Burrows Wheeler Aligner (BWA-MEM); Bowtie 2; minimap2 | Alignment of reads to large references; host-filtered BAM/FASTQ | BWA-MEM aligns short–long reads; Bowtie 2 fast gapped alignment; minimap2 supports long/short reads | [87,88,89] |
| Taxonomic classification | Kraken 2 with Bracken; Centrifuge | Taxonomic labels; abundance estimates | Kraken 2 high accuracy and low memory; Bracken re-estimates abundance; Centrifuge rapid classification | [80,90,91] |
| Background/contamination modeling | Negative control filtering; decontam (R) | Contaminant identification | Low biomass reagent contamination; decontam uses statistical models | [59,92] |
| AMR gene/variant detection | AMRFinderPlus; CARD/RGI; ResFinder; PointFinder | AMR gene and mutation detection | AMRFinderPlus detects genes & mutations; CARD curated families; ResFinder phenotype prediction; PointFinder mutation detection | [82,83,84,93] |
| Quality control summaries | FastQC; MultiQC | QC metrics; aggregated QC | FastQC QC checks; MultiQC aggregates | [94] |
| Index-swap mitigation | UDI | Reduced index hopping | UDI enables removal of swapped reads | [65,73] |
| Genome typing/outbreak relatedness | ONT positive BC; Guppy; Medaka; Flye | Rapid ID; assemblies; AMR detection | Rapid ID & AMR detection from BC | [24,37] |
| Viral variant calling | iVar; LoFreq | Consensus + variant calls | iVar amplicon framework; LoFreq low-frequency variant calling | [95,96] |
| Clinical reporting | Version-annotated reporting; STROBE-metagenomics | Structured reports | Improves transparency and reproducibility | [85] |
| Modality | Specimen and Minimum Volume | Typical TAT | Organism Scope | Quantitative Output | Best-Fit Clinical Scenarios | Key Strengths | Main Limitations and Pitfalls | Typical Stewardship Actions |
|---|---|---|---|---|---|---|---|---|
| Plasma mcfDNA | EDTA plasma, 5–10 mL; rapid separation; ≤1 freeze–thaw (e) | 24–48 h (a) | Bacteria (DNA), DNA viruses, fungi, parasites; around 1, 250 targets (a) | MPM (molecules/µL) | Culture negative or unobtainable; pretreated; immunocompromised; fastidious or occult pathogens; quantitative burden trending | One-test breadth; quantitative trending; minimally invasive | Nonlocalizing; no phenotypic MICs; residual or nonviable DNA; low-biomass contamination risk (f) | Protocolized escalation or deescalation based on pathogen and MPM trend; targeted imaging and source evaluation; ID stewardship |
| Direct mNGS plasma | EDTA plasma, 3–10 mL; hostDNA depletion recommended (e) | 24–72 h (b,c) | Broad, hypothesis-free | RPM or unique k-mers; relative abundance | Wide differential, including polymicrobial or unusual pathogens; culture negative or delayed | Unbiased detection; complements culture and panels | Low microbial biomass; contamination/background modeling required (f); thresholds needed | Narrow or expand therapy for high-confidence hits; order confirmatory tests; ID consult |
| Direct mNGS whole blood/cellular fraction | Whole blood 1–5 mL; optimized extraction for cellular fraction (e) | 24–72 h (c) | Broad; may enrich intracellular/pathogen DNA in cells | RPM or unique k-mers | When cellular fraction may add yield; complementary to plasma testing | Complementary to plasma; may capture different taxa | Higher host background; matrix-dependent performance; contamination and index controls (f) | As above; reconcile with plasma results and clinical context |
| Positive BC ONT | Positive BC broth (direct DNA) | 4–12 h (same-day) (d) | Primarily bacteria (from BC) | Depth/coverage; AMR gene presence | BC flagged positive; rapid ID/AMR; plasmid/resistance-context; rapid epidemiology | Same-day ID and genotypic AMR; plasmid context; supports rapid epidemiology | Requires culture positivity; genotype- phenotype for some pairs; thresholds must be validated | Targeted therapy within hours; infection-prevention notification; phenotypic confirmation per protocol |
| Targeted sequencing 16S rRNA | DNA from specimen or BC isolate | 24–48 h (g) | Bacteria (barcode) | Qualitative (ID call) | Culture negative; slow growing/fastidious; polymicrobial clarification | Broad bacterial ID; low input | Limited species-level resolution in some genera; copy number bias; database dependence | De-escalate; confirm unusual taxa; plan targeted cultures |
| Targeted sequencing ITS (fungi) | DNA from specimen or BC isolate | 24–48 h (g) | Fungi (barcode) | Qualitative (ID call) | Candidemia and other invasive mycoses; mixed fungal infections | Species-level calls that guide antifungal selection | Primer bias; molds/cryptic yeasts may need D1/D2, TEF1, β-tubulin (g) | Optimize antifungal choice and duration; epidemiologic linkage |
| Targeted AMR panels (bacterial and fungal) | DNA from specimen or positive BC | 6–24 h (g) | Focused AMR loci (blaESBL, carbapenemases; vanA/vanB; ERG11; FKS1/FKS2) | Gene/allele calls | Specific mechanisms suspected; need rapid resistance information | Fast and actionable; high depth over key loci | Panel limited; may miss off-panel mechanisms; genotype-phenotype gaps | Rapid escalation or de-escalation; isolation precautions for high-risk genes |
| Hybrid short + long read assemblies (outbreaks/plasmids) | DNA from isolate or positive BC | 1–3 days (h) | Bacterial genomes; plasmids | Closed/near closed assemblies | Outbreak resolution; plasmid and AMR-context mapping | Most reliable plasmid reconstruction; mobile-element context | More resources and time; specialized bioinformatics | Infection-prevention interventions; source tracing; stewardship and formulary updates |
| Pathogen Group (Example) | Clinical Question | Preferred Specimen | Sequencing Approach | Actionable Outputs | Typical TAT | Key References |
|---|---|---|---|---|---|---|
| MRSA | Outbreak investigation, lineage assignment, rapid identification and antimicrobial resistance | Positive BC broth | ONT from positive BC; WGS for final resolution | Species; MRSA lineage (for example ST22 or ST239); outbreak linkage | Hours for ONT; 24–72 h for WGS | [24,99] |
| VRE | vanA or vanB carriage and clustering | Positive BC | WGS | vanA or vanB; cgMLST clusters; transmission benchmarking | 24–72 h | [100] |
| K. pneumoniae (CG258/ST258) | Carbapenemase context and spread | Positive BC | WGS or hybrid (short plus long reads) | blaKPC, blaNDM, blaOXA-48-like; plasmid context; network spread | 48–72 h | [101,102] |
| P. aeruginosa | Importation versus transmission; resistance drivers | Positive BC | WGS | High risk clones; disinfectant tolerance; Verona integron encoded metallo-β-lactamase (VIM) and related determinants | 48–72 h | [108] |
| A. baumannii | Persistence and clonality | Positive BC | WGS | OXA carbapenemases; ward-persistence mapping | 48–72 h | [109] |
| HIV | Drug resistance at failure or baseline in selected settings | Plasma | Targeted NGS of pol (reverse transcriptase, protease, integrase) | Drug-resistance mutation profile informing regimen change | 2–5 days | [110,111,150] |
| HBV | Genotype and resistance assessment | Plasma | Deep sequencing of polymerase | RAS; genotype guiding therapy | 2–5 days | [113,115,116] |
| HCV (genotype 3) | Baseline NS5A RAS (for example Y93H) | Plasma | Targeted NGS of NS5A | RAS informing direct-acting antiviral selection | 2–5 days | [117,118] |
| CMV | Resistance during DNAemia | Plasma | Amplicon panel covering UL97, UL54, UL56, UL27 | Early detection of resistance informing switch of therapy | 2–4 days | [119,120,121] |
| SARS-CoV-2 | Prognosis based on RNAemia | Plasma | Quantitative PCR or sequencing based quantification | Risk stratification and monitoring | Same-day to 48 h | [86,123,125] |
| Candida auris | Clade assignment and azole or echinocandin resistance | Positive BC | WGS with or without targeted ERG11 and FKS1 | Clade identification; resistance markers guiding therapy and infection-prevention control | 48–96 h | [127,128,130] |
| Candida parapsilosis | Fluconazole resistance mechanism | Positive BC | Targeted ERG11 amplicon sequencing | Y132F detection guiding azole-sparing therapy | 24–72 h | [131,132] |
| Nakaseomyces glabratus | Echinocandin resistance mechanism | Positive BC | Targeted FKS1 and FKS2 sequencing | Hotspot mutations guiding change in therapy | 24–72 h | [133,134] |
| Aspergillus fumigatus | Azole resistance mechanism | Blood rarely; culture or bronchoalveolar lavage when available | Targeted cyp51A or WGS | TR34/L98H and TR46/Y121F/T289A informing therapy choice | 48–96 h | [135,136] |
| Cryptococcus species | Lineage assignment and 5-flucytosine resistance | Positive BC or CSF | WGS; targeted FUR1, FCY1, FCY2 | Lineage assignment; resistance mechanisms related to 5-flucytosine | 48–96 h | [139,140,142] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elbehiry, A.; Marzouk, E.; Edrees, H.M.; Abdelsalam, M.H.; Aljizani, F.; Alqarni, S.; Khateeb, E.; Alzaben, F.; Ibrahem, M.; Mousa, A.M.; et al. Next-Generation Sequencing for Bloodstream Infections: Shaping the Future of Rapid Diagnostics and Precision Medicine. Diagnostics 2025, 15, 2944. https://doi.org/10.3390/diagnostics15232944
Elbehiry A, Marzouk E, Edrees HM, Abdelsalam MH, Aljizani F, Alqarni S, Khateeb E, Alzaben F, Ibrahem M, Mousa AM, et al. Next-Generation Sequencing for Bloodstream Infections: Shaping the Future of Rapid Diagnostics and Precision Medicine. Diagnostics. 2025; 15(23):2944. https://doi.org/10.3390/diagnostics15232944
Chicago/Turabian StyleElbehiry, Ayman, Eman Marzouk, Husam M. Edrees, Moustafa H. Abdelsalam, Feras Aljizani, Saad Alqarni, Eyad Khateeb, Feras Alzaben, Mai Ibrahem, Ayman M. Mousa, and et al. 2025. "Next-Generation Sequencing for Bloodstream Infections: Shaping the Future of Rapid Diagnostics and Precision Medicine" Diagnostics 15, no. 23: 2944. https://doi.org/10.3390/diagnostics15232944
APA StyleElbehiry, A., Marzouk, E., Edrees, H. M., Abdelsalam, M. H., Aljizani, F., Alqarni, S., Khateeb, E., Alzaben, F., Ibrahem, M., Mousa, A. M., Huraysh, N., & Abu-Okail, A. (2025). Next-Generation Sequencing for Bloodstream Infections: Shaping the Future of Rapid Diagnostics and Precision Medicine. Diagnostics, 15(23), 2944. https://doi.org/10.3390/diagnostics15232944