The Value of Circulating Tumor HPV DNA in Head and Neck Squamous Cell Cancer: A Review †
Abstract
1. Introduction
2. Cell Free DNA as a Biomarker in HNSCC
3. ctHPV-DNA Detection as a Biomarker for HPV-Related OPSCC
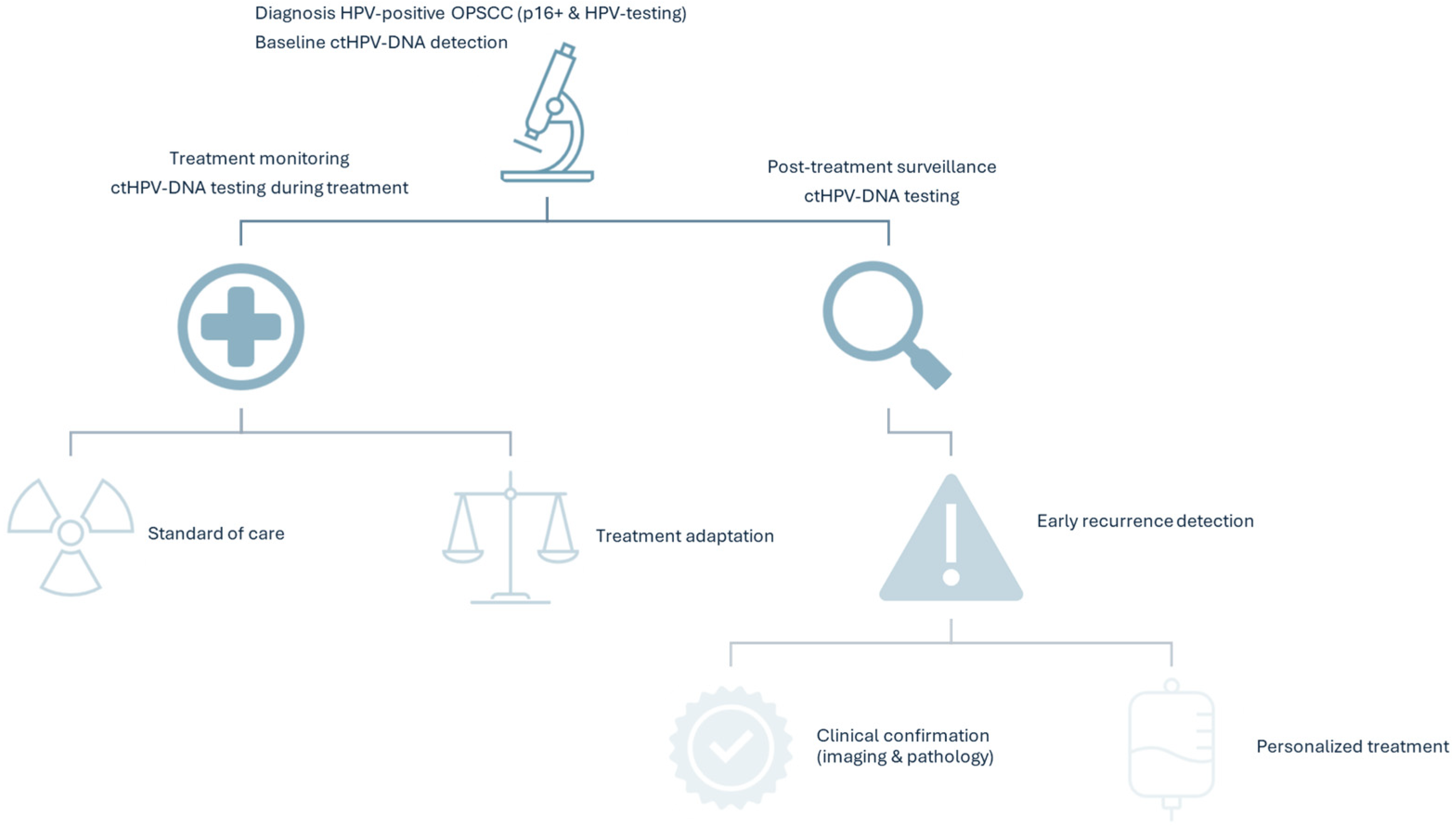
4. Role of ctHPV-DNA in Detection, Treatment Monitoring, and Surveillance
4.1. ctHPV-DNA Detection
4.2. Treatment Monitoring and Surveillance
5. Future Perspectives
6. Economic Considerations and Implementation Barriers
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Wu, Y.; Li, X.; Lin, Y.; Chen, Y.; Yang, H.; Shen, Y. Clinical and molecular characterizations of HNSCC patients with occult lymph node metastasis. Sci. Rep. 2025, 15, 25263. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef] [PubMed]
- Malagón, T.; Franco, E.L.; Tejada, R.; Vaccarella, S. Epidemiology of HPV-associated cancers past, present and future: Towards prevention and elimination. Nat. Rev. Clin. Oncol. 2024, 21, 522–538. [Google Scholar] [CrossRef]
- Kang, J.J.; Yu, Y.; Chen, L.; Zakeri, K.; Gelblum, D.Y.; McBride, S.M.; Riaz, N.; Tsai, C.J.; Kriplani, A.; Hung, T.K.W.; et al. Consensuses, controversies, and future directions in treatment deintensification for human papillomavirus-associated oropharyngeal cancer. CA Cancer J. Clin. 2023, 73, 164–197. [Google Scholar] [CrossRef]
- Mehanna, H.; Rischin, D.; Wong, S.J.; Gregoire, V.; Ferris, R.; Waldron, J.; Le, Q.-T.; Forster, M.; Gillison, M.; Laskar, S.; et al. De-Escalation After DE-ESCALATE and RTOG 1016: A Head and Neck Cancer InterGroup Framework for Future De-Escalation Studies. J. Clin. Oncol. 2020, 38, 2552–2557. [Google Scholar] [CrossRef]
- Golusinski, P.; Corry, J.; Poorten, V.V.; Simo, R.; Sjögren, E.; Mäkitie, A.; Kowalski, L.P.; Langendijk, J.; Braakhuis, B.J.M.; Takes, R.P.; et al. De-escalation studies in HPV-positive oropharyngeal cancer: How should we proceed? Oral Oncol. 2021, 123, 105620. [Google Scholar] [CrossRef]
- McLean, T.; Fitzgerald, C.; Boyle, J.O. Therapeutic strategies: Surgery for human papillomavirus-associated oropharyngeal carcinoma. J. Surg. Oncol. 2021, 124, 935–944. [Google Scholar] [CrossRef]
- Sabatini, M.E.; Chiocca, S. Human papillomavirus as a driver of head and neck cancers. Br. J. Cancer 2020, 122, 306–314. [Google Scholar] [CrossRef]
- Brandt, A.; Thiele, B.; Schultheiss, C.; Daetwyler, E.; Binder, M. Circulating Tumor DNA in Head and Neck Squamous Cell Carcinoma. Cancers 2023, 15, 2051. [Google Scholar] [CrossRef] [PubMed]
- Bruhm, D.C.; Vulpescu, N.A.; Foda, Z.H.; Phallen, J.; Scharpf, R.B.; Velculescu, V.E. Genomic and fragmentomic landscapes of cell-free DNA for early cancer detection. Nat. Rev. Cancer 2025, 25, 341–358. [Google Scholar] [CrossRef]
- Parisi, F.M.; Lentini, M.; Chiesa-Estomba, C.M.; Mayo-Yanez, M.; Leichen, J.R.; White, M.; Giurdanella, G.; Cocuzza, S.; Bianco, M.R.; Fakhry, N.; et al. Liquid Biopsy in HPV-Associated Head and Neck Cancer: A Comprehensive Review. Cancers 2025, 17, 977. [Google Scholar] [CrossRef]
- Chennareddy, S.; Chen, S.; Levinson, C.; Genden, E.M.; Posner, M.R.; Roof, S.A. Circulating tumor DNA in human papillomavirus-associated oropharyngeal cancer management: A systematic review. Oral Oncol. 2025, 164, 107262. [Google Scholar] [CrossRef]
- Yang, R.; Li, T.; Zhang, S.; Shui, C.; Ma, H.; Li, C. The effect of circulating tumor DNA on the prognosis of patients with head and neck squamous cell carcinoma: A systematic review and meta-analysis. BMC Cancer 2024, 24, 1434. [Google Scholar] [CrossRef]
- Chatfield-Reed, K.; Roche, V.P.; Pan, Q. cfDNA detection for HPV+ squamous cell carcinomas. Oral Oncol. 2021, 115, 104958. [Google Scholar] [CrossRef]
- Sivars, L.; Palsdottir, K.; Crona Guterstam, Y.; Falconer, H.; Hellman, K.; Tham, E. The current status of cell-free human papillomavirus DNA as a biomarker in cervical cancer and other HPV-associated tumors: A review. Int. J. Cancer 2023, 152, 2232–2242. [Google Scholar] [CrossRef]
- Wotman, M.; Xiao, W.; Du, R.; Jiang, B.; Liu, S.; Gillison, M.L. Development and validation of an assay to quantify plasma cell-free human papillomavirus DNA for 13 high-risk types that cause 98% of HPV-positive cancers. J. Clin. Oncol. 2024, 42 (Suppl. S16), 6065. [Google Scholar] [CrossRef]
- Cillo, A.R.; Kürten, C.H.L.; Tabib, T.; Qi, Z.; Onkar, S.; Wang, T.; Liu, A.; Duvvuri, U.; Kim, S.; Soose, R.J.; et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020, 52, 183–199.E9. [Google Scholar] [CrossRef]
- Diao, P.; Dai, Y.; Wang, A.; Bu, X.; Wang, Z.; Li, J.; Wu, Y.; Jiang, H.; Wang, Y.; Cheng, J. Integrative Multiomics Analyses Identify Molecular Subtypes of Head and Neck Squamous Cell Carcinoma with Distinct Therapeutic Vulnerabilities. Cancer Res. 2024, 84, 3101–3117. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Fenton, T.R. The Genomics, Epigenomics, and Transcriptomics of HPV-Associated Oropharyngeal Cancer--Understanding the Basis of a Rapidly Evolving Disease. Adv. Genet. 2016, 93, 1–56. [Google Scholar] [CrossRef]
- Serafini, M.S.; Lopez-Perez, L.; Fico, G.; Licitra, L.; De Cecco, L.; Resteghini, C. Transcriptomics and Epigenomics in head and neck cancer: Available repositories and molecular signatures. Cancers Head Neck 2020, 5, 2. [Google Scholar] [CrossRef]
- Bhambhani, C.; Kang, Q.; Hovelson, D.H.; Sandford, E.; Olesnavich, M.; Dermody, S.M.; Wolfgang, J.; Tuck, K.L.; Brummel, C.; Bhangale, A.D.; et al. ctDNA transiting into urine is ultrashort and facilitates noninvasive liquid biopsy of HPV+ oropharyngeal cancer. JCI Insight 2024, 9, e177759. [Google Scholar] [CrossRef]
- Ahn, S.M.; Chan, J.Y.; Zhang, Z.; Wang, H.; Khan, Z.; Bishop, J.A.; Westra, W.; Koch, W.M.; Califano, J.A. Saliva and plasma quantitative polymerase chain reaction-based detection and surveillance of human papillomavirus-related head and neck cancer. JAMA Otolaryngol. Head Neck Surg. 2014, 140, 846–854. [Google Scholar] [CrossRef]
- Berger, B.M.; Hanna, G.J.; Posner, M.R.; Genden, E.M.; Lautersztain, J.; Naber, S.P.; Del Vecchio Fitz, C.; Kuperwasser, C. Detection of Occult Recurrence Using Circulating Tumor Tissue Modified Viral HPV DNA among Patients Treated for HPV-Driven Oropharyngeal Carcinoma. Clin. Cancer Res. 2022, 28, 4292–4301. [Google Scholar] [CrossRef] [PubMed]
- Bhide, S. Circulating Human Papillomavirus DNA as a Marker of Minimally Residual Disease After Surgery for Oropharyngeal Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 539–541. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Beaty, B.T.; Marron, D.; Jefferys, S.; Green, R.; Goldman, E.C.; Amdur, R.; Sheets, N.; Dagan, R.; et al. Rapid Clearance Profile of Plasma Circulating Tumor HPV Type 16 DNA during Chemoradiotherapy Correlates with Disease Control in HPV-Associated Oropharyngeal Cancer. Clin. Cancer Res. 2019, 25, 4682–4690. [Google Scholar] [CrossRef]
- Ferrandino, R.M.; Chen, S.; Kappauf, C.; Barlow, J.; Gold, B.S.; Berger, M.H.; Westra, W.H.; Teng, M.S.; Khan, M.N.; Posner, M.R.; et al. Performance of Liquid Biopsy for Diagnosis and Surveillance of Human Papillomavirus–Associated Oropharyngeal Cancer. JAMA Otolaryngol. Head Neck Surg. 2023, 149, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Hanna, G.J.; Chang, S.S.; Siddiqui, F.; Bain, P.A.; Takiar, V.; Ward, M.C.; Shukla, M.E.; Hu, K.S.; Robbins, J.; Witek, M.E.; et al. Imaging and Biomarker Surveillance for Head and Neck Squamous Cell Carcinoma: A Systematic Review and American Radium Society Appropriate Use Criteria Statement. Int. J. Radiat. Oncol. Biol. Phys. 2024, 119, 786–802. [Google Scholar] [CrossRef]
- Hanna, G.J.; Roof, S.A.; Jabalee, J.; Rettig, E.M.; Ferrandino, R.; Chen, S.; Posner, M.R.; Misiukiewicz, K.J.; Genden, E.M.; Chai, R.L.; et al. Negative Predictive Value of Circulating Tumor Tissue Modified Viral (TTMV)-HPV DNA for HPV-driven Oropharyngeal Cancer Surveillance. Clin. Cancer Res. 2023, 29, 4306–4313. [Google Scholar] [CrossRef]
- Hanna, G.J.; Supplee, J.G.; Kuang, Y.; Mahmood, U.; Lau, C.J.; Haddad, R.I.; Jänne, P.A.; Paweletz, C.P. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann. Oncol. 2018, 29, 1980–1986. [Google Scholar] [CrossRef]
- Lang Kuhs, K.A.; Brenner, J.C.; Holsinger, F.C.; Rettig, E.M. Circulating Tumor HPV DNA for Surveillance of HPV-Positive Oropharyngeal Squamous Cell Carcinoma: A Narrative Review. JAMA Oncol. 2023, 9, 1716–1724. [Google Scholar] [CrossRef]
- Rettig, E.M.; Faden, D.L.; Sandhu, S.; Wong, K.; Faquin, W.C.; Warinner, C.; Stephens, P.; Kumar, S.; Kuperwasser, C.; Richmon, J.D.; et al. Detection of circulating tumor human papillomavirus DNA before diagnosis of HPV-positive head and neck cancer. Int. J. Cancer 2022, 151, 1081–1085. [Google Scholar] [CrossRef]
- Siravegna, G.; O’Boyle, C.J.; Varmeh, S.; Queenan, N.; Michel, A.; Stein, J.; Thierauf, J.; Sadow, P.M.; Faquin, W.C.; Perry, S.K.; et al. Cell-Free HPV DNA Provides an Accurate and Rapid Diagnosis of HPV-Associated Head and Neck Cancer. Clin. Cancer Res. 2022, 28, 719–727. [Google Scholar] [CrossRef]
- Chera, B.S.; Kumar, S.; Shen, C.; Amdur, R.; Dagan, R.; Green, R.; Goldman, E.; Weiss, J.; Grilley-Olson, J.; Patel, S.; et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J. Clin. Oncol. 2020, 38, 1050–1058. [Google Scholar] [CrossRef]
- Chikuie, N.; Urabe, Y.; Ueda, T.; Hamamoto, T.; Taruya, T.; Kono, T.; Yumii, K.; Takeno, S. Utility of plasma circulating tumor DNA and tumor DNA profiles in head and neck squamous cell carcinoma. Sci. Rep. 2022, 12, 9316. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Chen, C.; Hu, Y.; Zhang, W.; Yang, X.; Qi, Y.; Zhu, C.; Chen, X.; Shen, X.; Ji, W. Clinical application of molecular residual disease detection by circulation tumor DNA in solid cancers and a comparison of technologies: Review article. Cancer Biol. Ther. 2023, 24, 2274123. [Google Scholar] [CrossRef] [PubMed]
- Hanna, G.J.; Dennis, M.J.; Scarfo, N.; Mullin, M.S.; Sethi, R.K.V.; Sehgal, K.; Annino, D.J., Jr.; Goguen, L.A.; Haddad, R.I.; Tishler, R.B.; et al. Personalized ctDNA for Monitoring Disease Status in Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res. 2024, 30, 3329–3336. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.X.; D’Silva, N.J. HPV-associated oropharyngeal cancer: In search of surrogate biomarkers for early lesions. Oncogene 2024, 43, 543–554. [Google Scholar] [CrossRef]
- Mattox, A.K.; D’Souza, G.; Khan, Z.; Allen, H.; Henson, S.; Seiwert, T.Y.; Koch, W.; Pardoll, D.M.; Fakhry, C. Comparison of next generation sequencing, droplet digital PCR, and quantitative real-time PCR for the earlier detection and quantification of HPV in HPV-positive oropharyngeal cancer. Oral Oncol. 2022, 128, 105805. [Google Scholar] [CrossRef]
- Naegele, S.; Ruiz-Torres, D.A.; Zhao, Y.; Goss, D.; Faden, D.L. Comparing the Diagnostic Performance of Quantitative PCR, Digital Droplet PCR, and Next-Generation Sequencing Liquid Biopsies for Human Papillomavirus-Associated Cancers. J. Mol. Diagn. 2024, 26, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.; Breeze, C.E.; O’Mahony, J.F.; Masterson, L. Early detection of HPV-associated oropharyngeal cancer. Lancet 2019, 393, 2123. [Google Scholar] [CrossRef] [PubMed]
- Haring, C.T.; Brummel, C.; Bhambhani, C.; Jewell, B.; Neal, M.H.; Bhangale, A.; Casper, K.; Malloy, K.; McLean, S.; Shuman, A.; et al. Implementation of human papillomavirus circulating tumor DNA to identify recurrence during treatment de-escalation. Oral Oncol. 2021, 121, 105332. [Google Scholar] [CrossRef]
- Hirayama, S.; Al-Inaya, Y.; Aye, L.; Bryan, M.E.; Das, D.; Mendel, J.; Naegele, S.; Faquin, W.C.; Sadow, P.; Fisch, A.S.; et al. Prospective validation of ctHPVDNA for detection of minimal residual disease and prediction of recurrence in patients with HPV-associated head and neck cancer treated with surgery. J. Clin. Oncol. 2024, 42 (Suppl. S16), 6010. [Google Scholar] [CrossRef]
- Kabarriti, R.; Lloyd, S.; Jabalee, J.; Del Vecchio Fitz, C.; Tao, R.; Slater, T.; Jacobs, C.; Inocencio, S.; Rutenberg, M.; Matthiesen, C.; et al. Evaluating Tumor Tissue Modified Viral (TTMV)-HPV DNA for the Early Detection of Anal Squamous Cell Carcinoma Recurrence. Cancers 2025, 17, 174. [Google Scholar] [CrossRef]
- Zhong, L.-P.; Zhou, Z.-H.; Huang, Y.-Y.; Zhang, Y.-Y.; Lu, Y.; Li, B.; Wang, J.; Zhao, T.-C.; Ju, W.-T.; Zhu, D.-W. Associations of ctDNA clearance and pathological response after neoadjuvant treatment in patients with locally advanced oral cancer. J. Clin. Oncol. 2024, 42 (Suppl. S16), 6043. [Google Scholar] [CrossRef]
- Nocon, C.C.; Kennedy, A.; Jaffe, J.; Pruitt, J.; Kuchta, K.; Bhayani, M.K. Costs Associated With Imaging Surveillance After Treatment for Head and Neck Cancer. JAMA Otolaryngol. Head Neck Surg. 2021, 147, 632–637. [Google Scholar] [CrossRef]
- Agarwal, A.; Bhatt, A.; Patel, S.; Bathla, G.; Murray, J.; Rhyner, P. Preliminary Results from Retrospective Correlation of Circulating Tumor DNA (ct-DNA) with Imaging for HPV-Positive Oropharyngeal Squamous Cell Carcinoma. AJNR Am. J. Neuroradiol. 2024, 45, 1135–1140. [Google Scholar] [CrossRef]
- Almerén, A.O.; Waenerlund, M.; Landström, F.; von Beckerath, M.; Qvick, A.; Carlsson, J.; Helenius, G. Circulating Tumour DNA as a Complementary Tool for Treatment Evaluation in HPV-Associated Head and Neck Squamous Cell Carcinoma: An Observational Cohort Study. Clin. Otolaryngol. 2025, 50, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Lele, S.J.; Adilbay, D.; Lewis, E.; Pang, J.; Asarkar, A.A.; Nathan, C.O. ctDNA as an Adjunct to Posttreatment PET for Head and Neck Cancer Recurrence Risk Assessment. Otolaryngol. Head Neck Surg. 2024, 171, 439–444. [Google Scholar] [CrossRef]
- Gunning, A.; Kumar, S.; Williams, C.K.; Berger, B.M.; Naber, S.P.; Gupta, P.B.; Del Vecchio Fitz, C.; Kuperwasser, C. Analytical Validation of NavDx, a cfDNA-Based Fragmentomic Profiling Assay for HPV-Driven Cancers. Diagnostics 2023, 13, 725. [Google Scholar] [CrossRef] [PubMed]
- Campo, F.; Iocca, O.; Paolini, F.; Manciocco, V.; Moretto, S.; De Virgilio, A.; Moretti, C.; Vidiri, A.; Venuti, A.; Bossi, P.; et al. The landscape of circulating tumor HPV DNA and TTMV-HPVDNA for surveillance of HPV-oropharyngeal carcinoma: Systematic review and meta-analysis. J. Exp. Clin. Cancer Res. 2024, 43, 215. [Google Scholar] [CrossRef]
- Chen, L.; Cohen, M.; Hatzoglou, V.; Zhang, Z.; Riaz, N.; Wong, R.J.; McBride, S.; Gelblum, D.; Boyle, J.O.; Ganly, I.; et al. Early Disease Recurrence Following Post-operative HPV ctDNA Directed Active Surveillance in Oropharyngeal Carcinoma—Outcomes of a Prospective Pilot Study. Int. J. Radiat. Oncol. Biol. Phys. 2024, 118, e91. [Google Scholar] [CrossRef]
- Adrian, G.; Forslund, O.; Pedersen, L.; Sjövall, J.; Gebre-Medhin, M. Circulating tumour HPV16 DNA quantification—A prognostic tool for progression-free survival in patients with HPV-related oropharyngeal carcinoma receiving curative chemoradiotherapy. Radiother. Oncol. 2023, 186, 109773. [Google Scholar] [CrossRef]
- Cao, Y.; Haring, C.T.; Brummel, C.; Bhambhani, C.; Aryal, M.; Lee, C.; Heft Neal, M.; Bhangale, A.; Gu, W.; Casper, K.; et al. Early HPV ctDNA Kinetics and Imaging Biomarkers Predict Therapeutic Response in p16+ Oropharyngeal Squamous Cell Carcinoma. Clin. Cancer Res. 2022, 28, 350–359. [Google Scholar] [CrossRef]
- Rosenberg, A.J.; Izumchenko, E.; Juloori, A.; Katipally, R.; Cursio, J.; Choudhury, N.; Gooi, Z.; Blair, E.; Chin, J.; Hasina, R.; et al. Early Dynamics of Circulating Tumor HPV-DNA with Neoadjuvant Chemotherapy and Response-Adapted De-escalation in Human Papillomavirus-Associated Oropharyngeal Cancer. Clin. Cancer Res. 2025, 31, 3150–3159. [Google Scholar] [CrossRef] [PubMed]
- Routman, D.M.; Van Abel, K.M.; Price, K.A.; Moore, E.J.; Patel, S.H.; Hinni, M.L.; Fruth, B.; Foster, N.R.; Yin, L.X.; Neben-Wittich, M.; et al. ctDNA and Recurrence Risk for Adjuvant De-Escalation in HPV-Positive Oropharyngeal Carcinoma: A Secondary Analysis of the DART Phase 3 Randomized Clinical Trial. JAMA Otolaryngol. Head Neck Surg. 2025, 151, 665–672. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, Q.; Yuan, J.; Dai, L.; Luo, R.; Huang, T.; Wu, Y. Evaluation of ctDNA-guided adjuvant therapy de-escalation in head and neck squamous cell carcinoma: A comparative cohort study. Front. Immunol. 2025, 16, 1576042. [Google Scholar] [CrossRef] [PubMed]
- Mesher, D.; Panwar, K.; Thomas, S.L.; Edmundson, C.; Choi, Y.H.; Beddows, S.; Soldan, K. The Impact of the National HPV Vaccination Program in England Using the Bivalent HPV Vaccine: Surveillance of Type-Specific HPV in Young Females, 2010–2016. J. Infect. Dis. 2018, 218, 911–921. [Google Scholar] [CrossRef]
- Prue, G.; Baker, P.; Graham, D.; Nutting, C.; Greenhouse, P.; Lawler, M. It is time for universal HPV vaccination. Lancet 2018, 392, 913–914. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Bastien, A.J.; Ng, J.; Cong, I.; Garcia, J.; Walgama, E.S.; Luu, M.; Jang, J.K.; Mita, A.C.; Scher, K.S.; Moyers, J.T.; et al. Patient perceptions underlying ctDNA molecular surveillance for HPV(+) oropharyngeal squamous cell carcinoma. Oral Oncol. 2024, 156, 106894. [Google Scholar] [CrossRef]
- Chan, A.T.C.; Hui, E.P.; Ngan, R.K.C.; Tung, S.Y.; Cheng, A.C.K.; Ng, W.T.; Lee, V.H.F.; Ma, B.B.Y.; Cheng, H.C.; Wong, F.C.S.; et al. Analysis of Plasma Epstein-Barr Virus DNA in Nasopharyngeal Cancer After Chemoradiation to Identify High-Risk Patients for Adjuvant Chemotherapy: A Randomized Controlled Trial. J. Clin. Oncol. 2018, 36, 3091–3100. [Google Scholar] [CrossRef]
- Pearson, A.T.; Seiwert, T.Y.; Cohen, R.B.; Saba, N.F.; Kaczmar, J.M.; Fidler, M.J.; Wade, J.L.; Castellucci, E.; Karrison, T.; Katipally, R.R.; et al. A randomized, double-blind, placebo-controlled phase II study of adjuvant pembrolizumab versus placebo in patients with head and neck squamous cell cancers at high risk for recurrence: The PATHWay study. J. Clin. Oncol. 2024, 42 (Suppl. S16), 6008. [Google Scholar] [CrossRef]
- Han, K.; Zou, J.; Zhao, Z.; Baskurt, Z.; Zheng, Y.; Barnes, E.; Croke, J.; Ferguson, S.E.; Fyles, A.; Gien, L.; et al. Clinical Validation of Human Papilloma Virus Circulating Tumor DNA for Early Detection of Residual Disease After Chemoradiation in Cervical Cancer. J. Clin. Oncol. 2023, 42, 431–440. [Google Scholar] [CrossRef]
- Honore, N.; Laliotis, G.; Aushev, V.; Velichko, S.; Palsuledesai, C.; Dahou, H.; van Marcke, C.; Galot, R.; Liu, M.; Machiels, J.H. Tumor-informed ctDNA assay to predict recurrence in locally advanced squamous-cell carcinoma of the head and neck (SCCHN). ESMO Open 2025, 10, 104534. [Google Scholar] [CrossRef]
- Aye, L.; Bryan, M.E.; Das, D.; Hirayama, S.; Al-Inaya, Y.; Mendel, J.; Naegele, S.; Fisch, A.S.; Faquin, W.C.; Sadow, P.; et al. Multi-feature next-generation liquid biopsy for diagnosis and prognosis in HPV-associated head and neck cancer. J. Clin. Oncol. 2024, 42 (Suppl. S16), 6064. [Google Scholar] [CrossRef]
- Diplas, B.; Brown, D.N.; Pei, X.; Chen, L.; Graybill, C.; Korn, W.M.; Westheimer, E.; Novaj, A.; Humm, J.; Shukla-Dave, A.; et al. The combination of patient-specific tumor and HPV sequencing to enable high-sensitivity detection of ctDNA in patients with HPV-associated oropharyngeal carcinoma. J. Clin. Oncol. 2024, 42 (Suppl. S16), 6058. [Google Scholar] [CrossRef]
- Sanz-Garcia, E.; Zou, J.; Avery, L.; Spreafico, A.; Waldron, J.; Goldstein, D.; Hansen, A.; Cho, B.C.J.; de Almeida, J.; Hope, A.; et al. Multimodal detection of molecular residual disease in high-risk locally advanced squamous cell carcinoma of the head and neck. Cell Death Differ. 2024, 31, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Sangphukieo, A.; Noisagul, P.; Thongkumkoon, P.; Chaiyawat, P. Ultra-low coverage fragmentomic model of cell-free DNA for cancer detection based on whole-exome regions. eLife 2024, 13, RP95320. [Google Scholar] [CrossRef]
- Neumann, M.H.D.; Bender, S.; Krahn, T.; Schlange, T. ctDNA and CTCs in Liquid Biopsy—Current Status and Where We Need to Progress. Comput. Struct. Biotechnol. J. 2018, 16, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Kowalchuk, R.O.; Kamdem Talom, B.C.; Van Abel, K.M.; Ma, D.M.; Waddle, M.R.; Routman, D.M. Estimated Cost of Circulating Tumor DNA for Posttreatment Surveillance of Human Papillomavirus-Associated Oropharyngeal Cancer. JAMA Netw. Open 2022, 5, e2144783. [Google Scholar] [CrossRef]
- Febbo, P.G.; Allo, M.; Alme, E.B.; Cuyun Carter, G.; Dumanois, R.; Essig, A.; Kiernan, E.; Kubler, C.B.; Martin, N.; Popescu, M.C.; et al. Recommendations for the Equitable and Widespread Implementation of Liquid Biopsy for Cancer Care. JCO Precis. Oncol. 2024, 8, e2300382. [Google Scholar] [CrossRef]
- Nguyen Hoang, V.-A.; Nguyen, N.; Le, D.N.; Nguyen, V.S.; Nguyen Thi, H.G.; Vu, H.T.; Ho, H.H.; Le, H.M.; Nguyen, T.D.; Vo, H.N.; et al. Real-World Utilization and Performance of Circulating Tumor DNA Monitoring to Predict Recurrence in Solid Tumors. JCO Oncol. Adv. 2025, 2, e2400084. [Google Scholar] [CrossRef]
- Sánchez-Herrero, E.; Serna-Blasco, R.; Robado de Lope, L.; González-Rumayor, V.; Romero, A.; Provencio, M. Circulating Tumor DNA as a Cancer Biomarker: An Overview of Biological Features and Factors That may Impact on ctDNA Analysis. Front. Oncol. 2022, 12, 943253. [Google Scholar] [CrossRef] [PubMed]
- Bonstingl, L.; Skofler, C.; Ulz, C.; Zinnegger, M.; Sallinger, K.; Schönberger, J.; Schuch, K.; Pankratz, K.; Borrás-Cherrier, A.; Somodi, V.; et al. Clinical Application of ISO and CEN/TS Standards for Liquid Biopsies—Information Everybody Wants but Nobody Wants to Pay For. Clin. Chem. 2024, 70, 1140–1150. [Google Scholar] [CrossRef]
| Method | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|
| qPCR | 0.66 (0.58–0.74) | 0.94 (0.59–0.99) |
| dPCR | 0.89 (0.78–0.94) | 0.97 (0.94–0.99) |
| NGS | 0.91 (0.81–0.96) | 0.97 (0.90–0.99) |
| NCT Number | Study Title | Recruitment Status | Condition | Sponsor | Study Type |
|---|---|---|---|---|---|
| NCT05814549 | A Study Using Human Papillomavirus (HPV) DNA Testing to Detect HPV-Related Oropharyngeal Cancer (OPC) (NavDx®) | Active, not recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Memorial Sloan Kettering Cancer Center/New York, NY, USA | Observational |
| NCT05539638 | The Role of Circulating Tumor DNA in Head and Neck Cancer | Recruiting | Head and Neck Cancer | University of Edinburgh/Edinburgh, UK | Observational |
| NCT05904327 | Circulating Biomarkers in Oropharyngeal Cancers | Active, not recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Region Örebro County/Sweden | Observational |
| NL-OMON56995 | Personalized follow-up in HPV related oropharyngeal cancer patients using liquid biopsies | Recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Universitair Medisch Centrum Utrecht/Utrecht, The Netherlands | Observational |
| NCT05307939 | A Study on Using Cell-Free Tumor DNA (ctDNA) Testing to Decide When to Start Routine Treatment in People With Human Papilloma Virus (HPV)- Associated Oropharynx Cancer (OPC) | Recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Memorial Sloan Kettering Cancer Center/New York, NY, USA | Interventional |
| NCT05268614 | Risk Adapted De-Intensification of Radio-Chemotherapy for Oropharyngeal Squamous Cell Carcinoma (Phase 2) | Recruiting | Oropharyngeal Squamous Cell carcinoma | University of Florida/Gainesville, FL, USA | Interventional |
| NCT05541016 | De-Escalated Adjuvant and Definitive Radiation Therapy Informed by DART 2.0 ctHPV-DNA | Recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Mayo Clinic/Rochester, MN, USA | Interventional |
| NCT06821243 | Patients with Human Papillomavirus-associated Head and Neck Cancer for the Discovery of Predictive Biomarkers to Guide Clinical Intervention | Recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Regina Elena Cancer Institute/ Rome, Italy | Observational |
| NCT05582122 | SURVEILLE-HPV: Evaluation of HPV16 Circulating DNA as Biomarker to Detect the Recurrence, in Order to Improve Post Therapeutic Surveillance of HPV16-driven Oropharyngeal Cancers | Recruiting | Oropharyngeal Squamous Cell Carcinoma | UNICANCER/Paris, France | Interventional: Randomized |
| NCT04900623 | Risk-adapted Therapy in HPV+ Oropharyngeal Cancer Using Circulating Tumor (ct)HPV DNA Profile—The ReACT Study | Recruiting | Oropharyngeal Squamous Cell Carcinoma | Dana-Farber Cancer Institute/Boston, MA, USA | Interventional |
| NCT04965792 | Post-treatment Surveillance in HPV+ Oropharyngeal SCC | Active, not recruiting | Oropharyngeal Human Papillomavirus-Positive Squamous Cell Carcinoma | Dana-Farber Cancer Institute/Boston, MA, USA | Observational |
| NCT Number | Study Title | Recruitment Status | Condition | Sponsor/Location | Study Type |
|---|---|---|---|---|---|
| NCT06036563 | Prospective Screening and Differentiating Common Cancers Using Peripheral Blood Cell-Free DNA Sequencing | Recruiting | Pancancer | Air Force Military Medical University/Xi’an, China | Observational |
| NCT05366881 | cfDNA Assay Prospective Observational Validation for Early Cancer Detection and Minimal Residual Disease (CAMPERR) | Recruiting | Pancancer | Adela Inc./Foster City, CA, USA | Observational |
| NCT07035587 | Diagnosis of Multiple Cancer and Monitoring of Minimal Residual Tumors After Treatment Using Blood and High-Sensitivity Genetic Analysis Techniques | Recruiting | Pancancer | Yonsei University/
Seoul, Republic of Korea | Observational |
| NCT05685524 | Clinical Study of Pan-cancer DNA Methylation Test in Plasma | Unknown | Pancancer | Wuhan Ammunition Life-tech Co. Ltd./
Wuhan, China | Observational |
| NCT03926468 | Liquid Biopsy in Head and Neck Cancer | Unknown | Head and Neck Cancer | Turku University Hospital/
Turku, Finland | Observational |
| NCT03942380 | Cell-free Tumor DNA in Head and Neck Cancer Patients | Unknown | Head and Neck Cancer | Rigshospitalet/
København, Denmark | Interventional |
| NCT06356272 | Oropharynx (OPX) Biomarker Trial | Recruiting | Oropharyngeal Squamous Cell carcinoma | Mayo Clinic/Rochester, MN,
USA | Observational |
| NCT04599309 | Real-time Detection of ctDNA and/or HPV DNA in High-risk Locally advanced Head and Neck Squamous Cell Carcinoma | Active, not recruiting | Locally Advanced Head and Neck Carcinoma | University Health Network Toronto/
Toronto, ON, Canada | Observational |
| NCT05710679 | Prediction of Residual Disease by Circulating DNA Detection After Potentiated Radiotherapy for Locally Advanced Head and Neck Cancer (NeckTAR) | Recruiting | Locally Advanced Head and Neck Carcinoma | Centre Jean Perrin/
Clermont-Ferrand, France | Interventional |
| NCT02245100 | Circulating Tumor DNA in Predicting Outcomes in Patients with Stage IV Head and Neck Cancer or Stage III-IV Non-small Cell Lung Cancer | Completed | Head and Neck Cancer and Non-small Cell Lung Cancer | Sidney Kimmel Cancer Center at Thomas Jefferson University/Philadelphia, PA, USA | Observational |
| NCT04606940 | Study of Circulating Tumor DNA (ctDNA) Kinetics in Immuno-oncology (IO-KIN) | Completed | Recurrent of metastatic or advanced Head and Neck Cancer | University Health Network Toronto/
Toronto, ON, Canada | Observational |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dok, R.; Nuyts, S.; Lopez, F.; Bradford, C.; Forastiere, A.A.; Strojan, P.; Agaimy, A.; Stenman, G.; Mariano, F.V.; Leivo, I.; et al. The Value of Circulating Tumor HPV DNA in Head and Neck Squamous Cell Cancer: A Review. Diagnostics 2025, 15, 2708. https://doi.org/10.3390/diagnostics15212708
Dok R, Nuyts S, Lopez F, Bradford C, Forastiere AA, Strojan P, Agaimy A, Stenman G, Mariano FV, Leivo I, et al. The Value of Circulating Tumor HPV DNA in Head and Neck Squamous Cell Cancer: A Review. Diagnostics. 2025; 15(21):2708. https://doi.org/10.3390/diagnostics15212708
Chicago/Turabian StyleDok, Rüveyda, Sandra Nuyts, Fernando Lopez, Carol Bradford, Arlene A. Forastiere, Primož Strojan, Abbas Agaimy, Göran Stenman, Fernanda V. Mariano, Ilmo Leivo, and et al. 2025. "The Value of Circulating Tumor HPV DNA in Head and Neck Squamous Cell Cancer: A Review" Diagnostics 15, no. 21: 2708. https://doi.org/10.3390/diagnostics15212708
APA StyleDok, R., Nuyts, S., Lopez, F., Bradford, C., Forastiere, A. A., Strojan, P., Agaimy, A., Stenman, G., Mariano, F. V., Leivo, I., Rao, K. N., Williams, M., Eisbruch, A., Saba, N. F., & Ferlito, A. (2025). The Value of Circulating Tumor HPV DNA in Head and Neck Squamous Cell Cancer: A Review. Diagnostics, 15(21), 2708. https://doi.org/10.3390/diagnostics15212708











