Exploring IL-10 and NOS3 Genetic Variants as a Risk Factor for Neonatal Respiratory Distress Syndrome and Its Outcome
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Consideration
2.2. Study Population
2.3. Sample Collection and DNA Extraction
2.4. Selection of Variants
2.5. Genotyping
2.6. Molecular Analysis
2.7. Evaluation of Data
2.7.1. Overview Statistics
2.7.2. Statistical Analysis
3. Results
3.1. Patient Demographics and Clinical Parameters
3.2. Analysis of Genotypic and Allelic Distributions of IL-10 and NOS3 Variants in Relation to RDS Risk
3.3. Haplotype and Linkage Disequilibrium Analysis of IL-10 and NOS3 Variants in Neonates with RDS
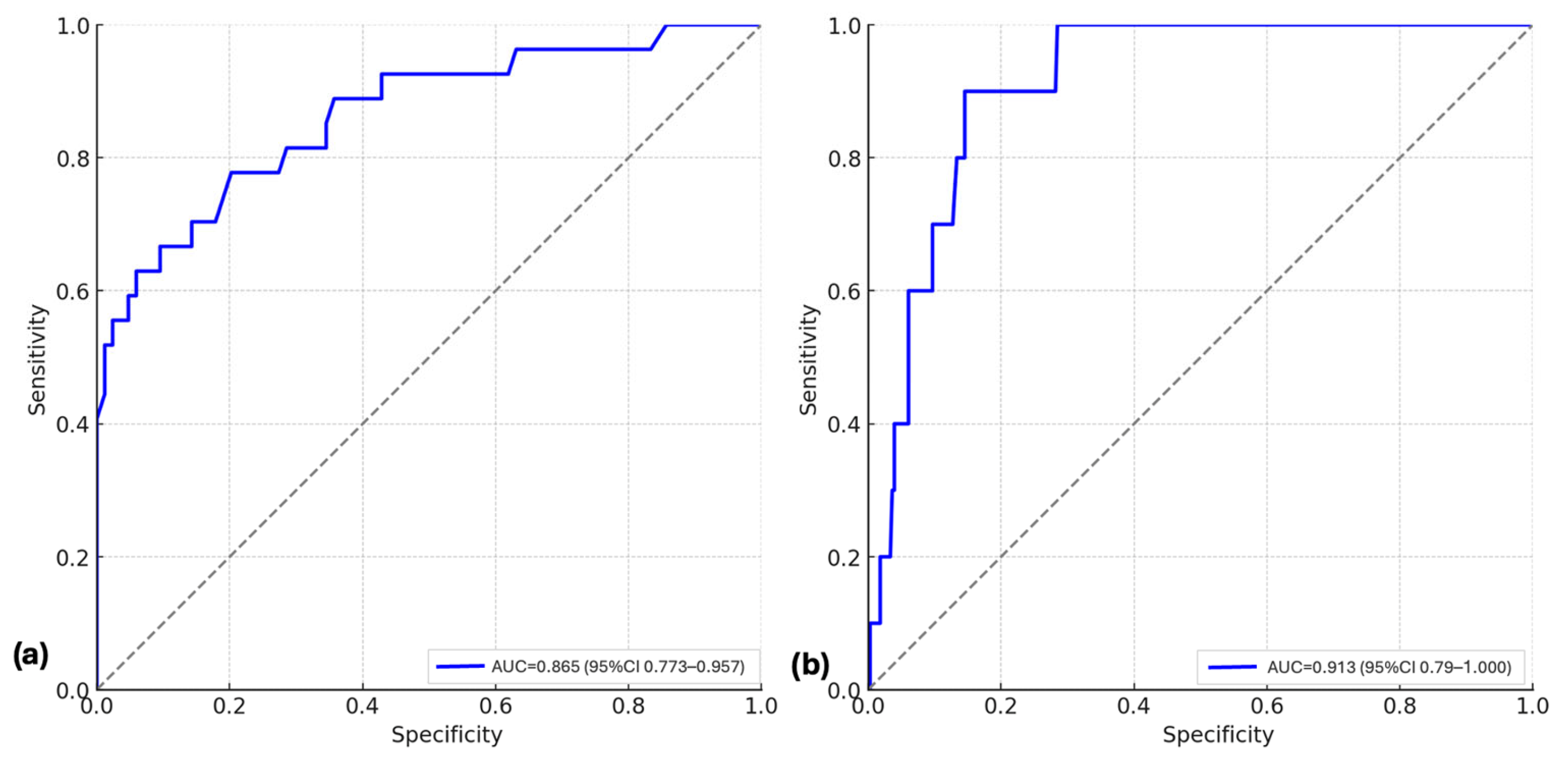
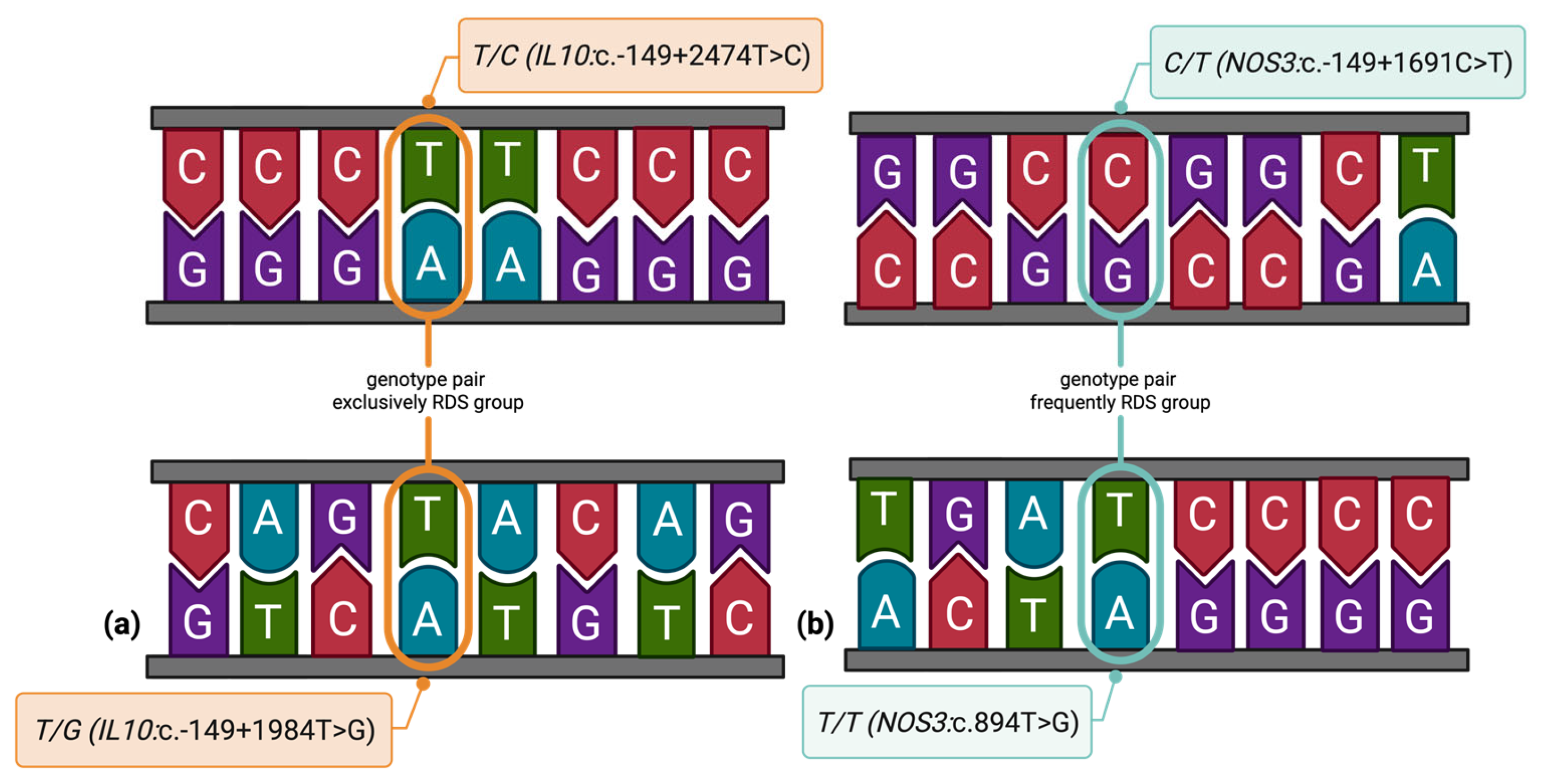
3.4. Epistatic and Environmental Modifiers of Genetic Risk in Neonatal RDS
3.5. Genetic Associations with Neonatal Complications
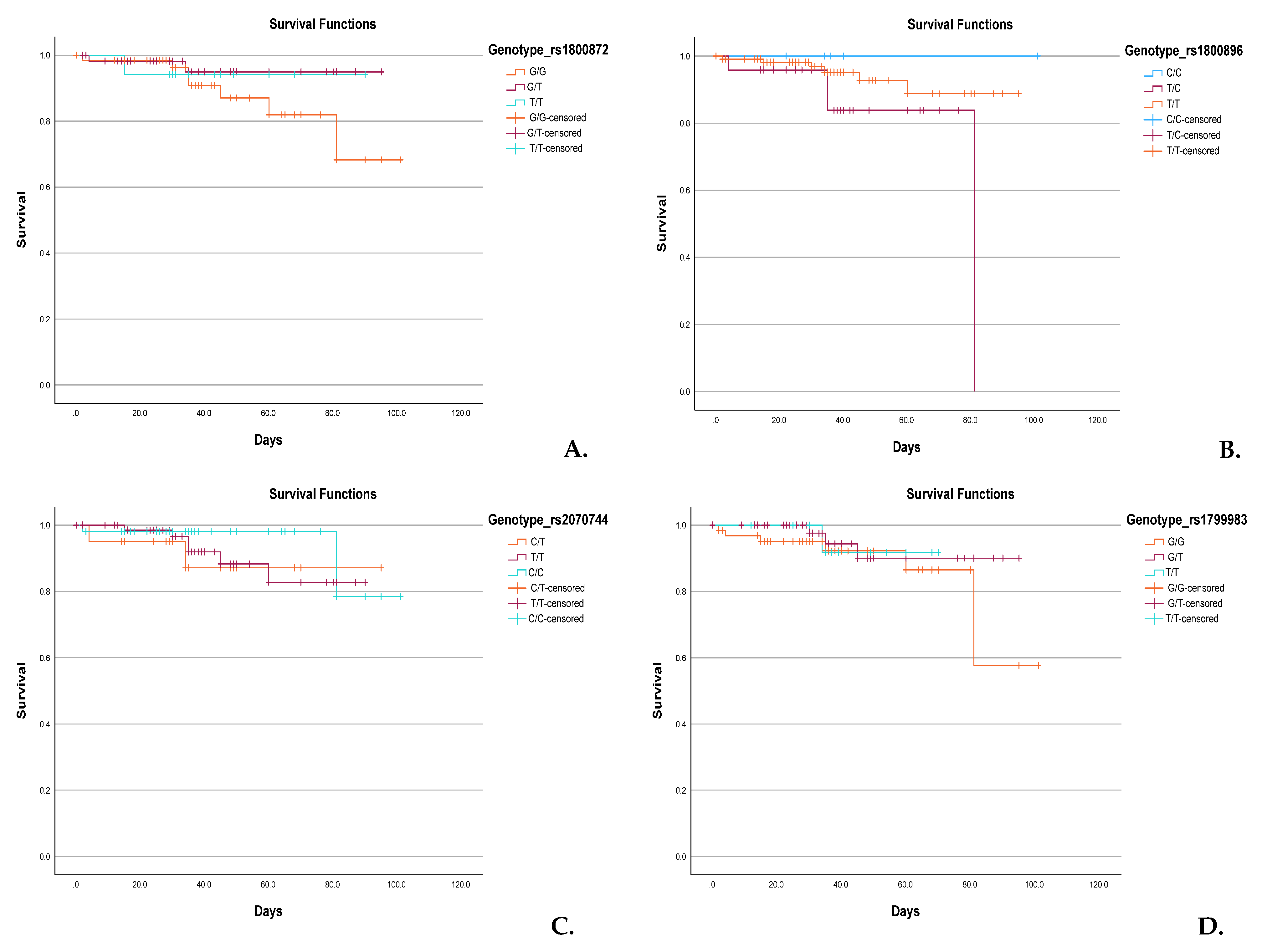
3.6. Genotype-Based Kaplan–Meier Survival Analysis
4. Discussion
4.1. IL-10 Variants and Their Role in RDS Susceptibility
4.2. NOS3 Gene Variants and Neonatal Pulmonary Outcomes
4.3. Variant-to-Variant Interactions Between IL-10 and NOS3 Genes
4.4. In Silico Analysis of the Investigated Variants
4.5. Population-Specific Context
4.6. Clinical Implications and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| NOS3 | endothelial nitric oxide synthase |
| IL-10 | Interleukin-10 |
| RDS | Respiratory distress syndrome |
| BPD | bronchopulmonary dysplasia |
| PDA | patent ductus arteriosus |
| CSIF | cytokine synthesis inhibitory factor |
| Th1 | T-helper 1 cell |
| Th2 | T-helper 2 cell |
| SNP | single-nucleotide polymorphism |
| NO | nitric oxide |
| FiO2 | fraction of inspired oxygen |
| DNA | deoxyribonucleic acid |
| RNA | ribonucleic acid |
| PCR | polymerase chain reaction |
| NCBI | National Center for Biotechnology Information |
| MAF | minor allele frequency |
| qPCR | real-time polymerase chain reaction |
| PROMs | premature rupture of membranes |
| PIH | pregnancy-induced hypertension |
| IVF | in vitro fertilization |
| GDM | gestational diabetes mellitus |
| LISA | less invasive surfactant administration |
| INSURE | intubation–surfactant administration-extubation |
| MV | mechanical ventilation |
| IVH | intraventricular hemorrhage |
| NICU | neonatal intensive care unit |
| ROP | retinopathy of prematurity |
| HIE | hypoxic–ischemic encephalopathy |
| PPHN | persistent pulmonary hypertension of the newborn |
| OR | odds ratio |
| 95%CI | 95% confidence interval |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under the Curve |
| NPVs | negative predictive values |
| PPVs | positive predictive values |
References
- Moore, K.W.; O’Garra, A.; de Waal Malefyt, R.; Vieira, P.; Mosmann, T.R. Interleukin-10. Annu. Rev. Immunol. 1993, 11, 165–190. [Google Scholar] [CrossRef] [PubMed]
- Antoniv, T.T.; Park-Min, K.H.; Ivashkiv, L.B. Kinetics of IL-10-induced gene expression in human macrophages. Immunobiology 2005, 210, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.T.; Oldstone, M.B. Infected CD8α-dendritic cells are the predominant source of IL-10 during establishment of persistent viral infection. Proc. Natl. Acad. Sci. USA 2012, 109, 14116–14121. [Google Scholar] [CrossRef]
- Cheng, S.B.; Sharma, S. Interleukin-10: A pleiotropic regulator in pregnancy. Am. J. Reprod. Immunol. 2015, 73, 487–500. [Google Scholar] [CrossRef]
- Schumacher, A.; Wafula, P.O.; Bertoja, A.Z.; Sollwedel, A.; Thuere, C.; Wollenberg, I.; Yagita, H.; Volk, H.-D.; Zenclussen, A.C. Mechanisms of action of regulatory T cells specific for paternal antigens during pregnancy. Obstet. Gynecol. 2007, 110, 1137–1145. [Google Scholar] [CrossRef]
- Na, Q.; Chudnovets, A.; Liu, J.; Lee, J.Y.; Dong, J.; Shin, N.; Elsayed, N.; Lei, L.; Burd, I. Placental macrophages demonstrate sex-specific response to intrauterine inflammation and may serve as a marker of perinatal neuroinflammation. J. Reprod. Immunol. 2021, 147, 103360. [Google Scholar] [CrossRef]
- Talebi, H.; Dastgheib, S.A.; Vafapour, M.; Bahrami, R.; Golshan-Tafti, M.; Danaei, M.; Azizi, S.; Shahbazi, A.; Pourkazemi, M.; Yeganegi, M.; et al. Advancements in biomarkers and machine learning for predicting of bronchopulmonary dysplasia and neonatal respiratory distress syndrome in preterm infants. Front. Pediatr. 2025, 13, 1521668. [Google Scholar] [CrossRef]
- Kadivnik, M.; Plečko, D.; Kralik, K.; Arvaj, N.; Wagner, J. Role of IL-6, IL-10 and TNFα Gene Variants in Preterm Birth. J. Clin. Med. 2024, 13, 2429. [Google Scholar] [CrossRef]
- Blanco-Quirós, A.; Arranz, E.; Solis, G.; Garrote, J.A.; Mayo, A. High cord blood IL-10 levels in preterm newborns with respiratory distress syndrome. Allergol. Immunopathol. 2004, 32, 189–196. [Google Scholar] [CrossRef]
- Mohany, K.M.; Sayed, A.A.; El-Asheer, O.M.; Raheem, Y.A.; Abbas, A.M.; Fawzy, A.M.; El-Baz, M.A.E.-H.H. The association of LPCAT1-rs9728 polymorphism with cord blood IL-10, MIF, and VEGF levels in neonatal respiratory distress syndrome: A case–control study. Egypt. J. Bronchol. 2024, 18, 25. [Google Scholar] [CrossRef]
- Matyas, M.; Hasmasanu, M.; Silaghi, C.N.; Samasca, G.; Lupan, I.; Orsolya, K.; Zaharie, G. Early Preeclampsia Effect on Preterm Newborns Outcome. J. Clin. Med. 2022, 11, 452. [Google Scholar] [CrossRef]
- Liu, C.; He, Y.; Ai, Q.; Shi, Y. A pilot study of plasma interleukin-6 and interleukin-27 in differential diagnosis of acute respiratory distress syndrome and neonatal respiratory distress syndrome in preterm infants. Zhongguo Dang Dai Er Ke Za Zhi 2022, 24, 428–432. (In Chinese) [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Chen, T.; Xue, F. The association of IL-10-1082G/A gene polymorphism with the risk of acute lung injury/respiratory distress syndrome (ALI/RDS): A meta-analysis. Heart Lung 2023, 58, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Capasso, M.; Avvisati, R.A.; Piscopo, C.; Laforgia, N.; Raimondi, F.; de Angelis, F.; Iolascon, A. Cytokine gene polymorphisms in Italian preterm infants: Association between interleukin-10-1082 G/A polymorphism and respiratory distress syndrome. Pediatr. Res. 2007, 61, 313–317. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Afolayan, A.J.; Eis, A.; Alexander, M.; Michalkiewicz, T.; Teng, R.J.; Lakshminrusimha, S.; Konduri, G.G. Decreased endothelial nitric oxide synthase expression and function contribute to impaired mitochondrial biogenesis and oxidative stress in fetal lambs with persistent pulmonary hypertension. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2016, 310, L40–L49. [Google Scholar] [CrossRef]
- Poggi, C.; Giusti, B.; Gozzini, E.; Sereni, A.; Romagnuolo, I.; Kura, A.; Pasquini, E.; Abbate, R.; Dani, C. Genetic Contributions to the Development of Complications in Preterm Newborns. PLoS ONE 2015, 10, e0131741. [Google Scholar] [CrossRef]
- Huang, H.C.; Yang, M.Y.; Huang, C.B.; Yang, K.D. Profiles of inflammatory cytokines in bronchoalveolar lavage fluid from premature infants with respiratory distress disease. J. Microbiol. Immunol. Infect. 2000, 33, 9–24. [Google Scholar]
- Yanamandra, K.; Boggs, P.; Loggins, J.; Baier, R.J. Interleukin-10-1082 G/A polymorphism and risk of death or bronchopulmonary dysplasia in ventilated very low birth weight infants. Pediatr. Pulmonol. 2005, 39, 426–432. [Google Scholar] [CrossRef]
- Al-shaer, O.S.; Behiry, E.G.; Elsadek, A.E.; Salama, S.A. Association between interleukin-10 genetic variant (-1082G>A) with detection and severity of respiratory distress syndrome in preterm neonates. Int. J. Immunogenet. 2019, 47, 50–56. [Google Scholar] [CrossRef]
- Demirçubuk, A.G.; Coşkun, M.Y.; Demiryürek, Ş.; Dokuyucu, R.; Öztuzcu, S.; Taviloğlu, Z.Ş.; Arslan, A.; Sivaslı, E. Endothelial NOS gene Glu298Asp polymorphism in preterm neonates with respiratory distress syndrome. Pediatr. Pulmonol. 2013, 48, 976–980. [Google Scholar] [CrossRef]
- Sivasli, E.; Babaoğlu, M.; Yaşar, U.; Yurdakök, M.; Bozkurt, A.; Korkmaz, A.; Yiğit, S.; Tekinalp, G. Association between the Glu298Asp and T(−786)C polymorphisms of the endothelial nitric oxide synthase gene and respiratory distress in preterm neonates. Turk. J. Pediatr. 2010, 52, 145–149. [Google Scholar] [PubMed]
- Sweet, D.G.; Carnielli, V.P.; Greisen, G.; Hallman, M.; Klebermass-Schrehof, K.; Ozek, E.; Te Pas, A.; Plavka, R.; Roehr, C.C.; Saugstad, O.D.; et al. European Consensus Guidelines on the Management of Respiratory Distress Syndrome: 2022 Update. Neonatology 2023, 120, 3–23. [Google Scholar] [CrossRef] [PubMed]
- De Luca, D. Respiratory distress syndrome in preterm neonates in the era of precision medicine: A modern critical care-based approach. Pediatr. Neonatol. 2021, 62, S3–S9. [Google Scholar] [CrossRef] [PubMed]
- Panthee, K.; Laxman, P.; Sumi, M. Use of Silverman Andersen Respiratory Severity Score in predicting need of respiratory support and outcome of neonates in Tertiary care center. Devdaha Med. J. 2023, 4, 1–5. [Google Scholar]
- Hedstrom, A.B.; Gove, N.E.; Mayock, D.E.; Batra, M. Performance of the Silverman Andersen Respiratory Severity Score in predicting PCO2 and respiratory support in newborns: A prospective cohort study. J. Perinatol. 2018, 38, 505–511. [Google Scholar] [CrossRef]
- Faul, F.; Erdfelder, E.; Lang, A.-G.; Buchner, A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav. Res. Methods 2007, 39, 175–191. [Google Scholar] [CrossRef]
- Jacob, C. Statistical Power Analysis for the Behavioral Sciences, 2nd ed.; Lawrence Erlbaum Associates: New York, NY, USA, 1988. [Google Scholar] [CrossRef]
- Ensembl. rs1800872 Variant in Homo Sapiens. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Explore?r=1:206772562-206773562;v=rs1800872;vdb=variation;vf=122471 (accessed on 29 July 2025).
- Ensembl. rs1800896 Variant in Homo Sapiens. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Explore?r=1:206773052-206774052;v=rs1800896;vdb=variation;vf=122482 (accessed on 29 July 2025).
- Ensembl. rs2070744 Variant in Homo Sapiens. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Explore?r=7:150992491-150993491;v=rs2070744;vdb=variation;vf=480762918 (accessed on 29 July 2025).
- Ensembl. rs1799983 Variant in Homo Sapiens. Available online: https://www.ensembl.org/Homo_sapiens/Variation/Explore?r=7:150998523-150999523;v=rs1799983;vdb=variation;vf=480750083 (accessed on 29 July 2025).
- ELMeneza, S.; Agaba, N.; Fawaz, R.A.E.S.; Abd Elgawad, S.S. Review of Precision Medicine and Diagnosis of Neonatal Illness. Diagnostics 2025, 15, 478. [Google Scholar] [CrossRef]
- Jo, H.S. Genetic risk factors associated with respiratory distress syndrome. Korean J. Pediatr. 2014, 57, 157–163. [Google Scholar] [CrossRef]
- Costa, F.; Titolo, A.; Ferrocino, M.; Biagi, E.; Dell’Orto, V.; Perrone, S.; Esposito, S. Lung Ultrasound in Neonatal Respiratory Distress Syndrome: A Narrative Review of the Last 10 Years. Diagnostics 2024, 14, 2793. [Google Scholar] [CrossRef]
- Popa, A.E.; Popescu, S.D.; Tecuci, A.; Bot, M.; Vladareanu, S. Current Trends in the Imaging Diagnosis of Neonatal Respiratory Distress Syndrome (NRDS): Chest X-ray Versus Lung Ultrasound. Cureus 2024, 16, e69787. [Google Scholar] [CrossRef]
- Balasubramaniam, V.; Maxey, A.M.; Morgan, D.B.; Markham, N.E.; Abman, S.H. Inhaled NO restores lung structure in eNOS-deficient mice recovering from neonatal hypoxia. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2006, 291, L119–L127. [Google Scholar] [CrossRef]
- Garingo, A.; Tesoriero, L.; Cayabyab, R.; Durand, M.; Blahnik, M.; Sardesai, S.; Ramanathan, R.; Jones, C.; Kwong, K.; Li, C.; et al. Constitutive IL-10 Expression by Lung Inflammatory Cells and Risk for Bronchopulmonary Dysplasia. Pediatr. Res. 2007, 61, 197–202. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mobini, M.; Mortazavi, M.; Nadi, S.; Zare-Bidaki, M.; Pourtalebi, S.; Arababadi, M.K. Significant roles played by interleukin-10 in outcome of pregnancy. Iran. J. Basic Med. Sci. 2016, 19, 119–124. [Google Scholar] [PubMed][Green Version]
- Hajeer, A.H.; Lazarus, M.; Turner, D.; Mageed, R.A.; Vencovsky, J.; Sinnott, P.; Hutchinson, I.V.; Ollier, W.E. IL-10 gene promoter polymorphisms in rheumatoid arthritis. Scand. J. Rheumatol. 1998, 27, 142–145. [Google Scholar] [CrossRef]
- Been, J.V.; Rours, I.G.; Kornelisse, R.F.; Lima Passos, V.; Kramer, B.W.; Schneider, T.A.; de Krijger, R.R.; Zimmermann, L.J. Histologic chorioamnionitis, fetal involvement, and antenatal steroids: Effects on neonatal outcome in preterm infants. Am. J. Obs. Gynecol. 2009, 201, 587.e1–587.e8. [Google Scholar] [CrossRef]
- Zheng, L.Y.; Sun, P.C. Increased Expression of IL-23 and IL-17 in Serum of Patients with Neonatal Respiratory Distress Syndrome and its Clinical Significance. Clin. Lab. 2020, 66, 1569. [Google Scholar] [CrossRef]
- Wang, H.; Oei, J.; Lui, K.; Henry, R. Decreased interleukin-10 in tracheal aspirates from preterm infants developing chronic lung disease. Acta Paediatr. 2002, 91, 1194–1199. [Google Scholar]
- Hobbs, K.; Negri, J.; Klinnert, M.; Rosenwasser, L.J.; Borish, L. Interleukin-10 and transforming growth factor-β promoter polymorphisms in allergies and asthma. Am. J. Respir. Crit. Care Med. 1998, 158, 1958–1962. [Google Scholar] [CrossRef]
- Vera-Lozada, G.; Minnicelli, C.; Segges, P.; Stefanoff, G.; Kristcevic, F.; Ezpeleta, J.; Tapia, E.; Niedobitek, G.; Barros, M.H.M.; Hassan, R. Interleukin 10 (IL10) proximal promoter polymorphisms beyond clinical response in classical Hodgkin lymphoma: Exploring the basis for the genetic control of the tumor microenvironment. Oncoimmunology 2018, 7, e1389821. [Google Scholar] [CrossRef]
- Steinke, J.W.; Barekzi, E.; Hagman, J.; Borish, L. Functional analysis of −571 IL-10 promoter polymorphism reveals a repressor element controlled by Sp1. J. Immunol. 2004, 173, 3215–3222. [Google Scholar] [CrossRef]
- Suarez, A.; Castro, P.; Alonso, R.; Mozo, L.; Gutierrez, C. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms. Transplantation 2003, 75, 711–717. [Google Scholar] [CrossRef] [PubMed]
- Reuss, E.; Fimmers, R.; Kruger, A.; Becker, C.; Rittner, C.; Höhler, T. Differential regulation of interleukin-10 production by genetic and environmental factors—A twin study. Genes. Immun. 2002, 3, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Beresford, M.W.; Shaw, N.J. Detectable IL-8 and IL-10 in bronchoalveolar lavage fluid from preterm infants ventilated for respiratory distress syndrome. Pediatr. Res. 2002, 52, 973–978. [Google Scholar] [CrossRef] [PubMed]
- van der Poll, T.; Marchant, A.; Buurman, W.A.; Berman, L.; Keogh, C.V.; Lazarus, D.D.; Nguyen, L.; Goldman, M.; Moldawer, L.L.; Lowry, S.F. Endogenous IL-10 protects mice from death during septic peritonitis. J. Immunol. 1995, 155, 5397–5401. [Google Scholar] [CrossRef]
- Nikkari, S.T.; Määttä, K.M.; Kunnas, T.A. Functional inducible nitric oxide synthase gene variants associate with hypertension: A case-control study in a Finnish population—The TAMRISK study. Medicine 2015, 94, e1958. [Google Scholar] [CrossRef]
- Wei, L.; An, Y.; Wang, J. Association between functional polymorphisms in the nitric oxide synthase 3 gene and pediatric acute respiratory distress syndrome. Genet. Mol. Res. 2016, 15, 1–7. [Google Scholar] [CrossRef]
- Szpecht, D.; Gadzinowski, J.; Seremak-Mrozikiewicz, A.; Kurzawińska, G.; Szymankiewicz, M. Role of endothelial nitric oxide synthase and endothelin-1 polymorphism genes with the pathogenesis of intraventricular hemorrhage in preterm infants. Sci. Rep. 2017, 7, 42541. [Google Scholar] [CrossRef]
- Choręziak-Michalak, A.; Szpecht, D.; Woźniak, T.; Chmielarz-Czarnocińska, A.; Gazińska, P.; Gotz-Więckowska, A.; Strauss, E. Association of endothelial nitric oxide synthase (NOS3) rs2070744 variant with advanced retinopathy of prematurity: A case-control study and meta-analysis. Sci. Rep. 2025, 15, 329. [Google Scholar] [CrossRef]
- Kuzmanić Samija, R.; Primorac, D.; Resić, B.; Lozić, B.; Krzelj, V.; Tomasović, M.; Stoini, E.; Samanović, L.; Benzon, B.; Pehlić, M.; et al. Association of NOS3 tag polymorphisms with hypoxic-ischemic encephalopathy. Croat. Med. J. 2011, 52, 396–402. [Google Scholar] [CrossRef]
- Han, R.N.; Babaei, S.; Robb, M.; Lee, T.; Ridsdale, R.; Ackerley, C.; Post, M.; Duncan, J. Defective lung vascular development and fatal respiratory distress in endothelial NO synthase-deficient mice: A model of alveolar capillary dysplasia? Circ. Res. 2004, 94, 1115–1123. [Google Scholar] [CrossRef]
- Shen, W.; Du, J.; Wang, B.; Zeng, Q. Analysis of nitric oxide synthase gene polymorphisms in neonatal respiratory distress syndrome among the Chinese Han population. Ital. J. Pediatr. 2014, 40, 27. [Google Scholar] [CrossRef]
- Steinhorn, R. Neonatal pulmonary hypertension. Pediatr. Crit. Care Med. 2010, 11, S79–S84. [Google Scholar] [CrossRef]
- Keszler, M. Guidelines for rational and cost-effective use of iNO therapy in term and preterm infants. J. Clin. Neonatol. 2012, 1, 59–63. [Google Scholar] [CrossRef]
- Chandrasekharan, P.; Lakshminrusimha, S.; Abman, S.H. When to say no to inhaled nitric oxide in neonates? Semin. Fetal Neonatal Med. 2021, 26, 101200. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, X.; Xu, X.; Cui, Q.; Wu, F. Efficacy of inhaled nitric oxide in preterm infants ≤ 34 weeks: A systematic review and meta—Analysis of randomized controlled trials. Front. Pharmacol. 2024, 14, 1268795. [Google Scholar] [CrossRef]
| Gene | NCBI dbSNP ID | Variant Localization | MAF * |
|---|---|---|---|
| IL-10 | rs1800872 | c.-149+1984T>G | 0.49 |
| IL-10 | rs1800896 | c.-149+2474T>C | 0.27 |
| NOS3 | rs2070744 | c.-149+1691C>T | 0.40 |
| NOS3 | rs1799983 | c.894T>G (p.Glu298Asp) | 0.24 |
| Variables | RDS Patients n = 113 | Controls (Without RDS) n = 227 | p-Values | OR; 95% IC |
|---|---|---|---|---|
| Gestational age (weeks): mean ± SD | 31 ± 1.46 | 31 ± 1.57 | 1.0 * | 0.66; 0.13–3.34 |
| Gender: n (%) | 0.8016 ‡ | 0.91; 0.56–1.46 | ||
| Female | 39 (34.51) | 83 (36.56) | ||
| Male | 74 (65.48) | 144 (63.43) | ||
| Birth weight (g): mean ± SD | 1476.44 ± 467.48 | 1504.24 ± 261.25 | 0.5574 * | 1.12; 0.71–1.76 |
| Preterm labor, n (%) | 63 (55.75) | 115 (50.66) | 0.4412 ‡ | 1.23; 0.78–1.93 |
| Singleton pregnancy, n (%) | 86 (76.10) | 197 (86.78) | 0.0199 ‡ | 0.49; 0.27–0.86 |
| Antenatal care, n (%) | 41 (36.28) | 134 (59.03) | 0.0001 ‡ | 0.40; 0.25–0.63 |
| Mother’s age, mean ± SD | 29.7 ± 6.43 | 30.5 ± 4.78 | 0.2431 * | 1.23; 0.69–2.19 |
| Delivery mode, n (%) | 0.0113 ‡ | 0.54; 0.34–0.85 | ||
| Spontaneous | 45 (39.82) | 125 (55.06) | ||
| C-section | 68 (60.17) | 102 (44.93) | ||
| Delivery in a tertiary center, n (%) | 87 (76.99) | 188 (82.81) | 0.2539 ‡ | 0.69; 0.40–1.21 |
| Apgar Score, mean | ||||
| 1 min | 6 | 7 | <0.0001 * | 2.21; 1.37–3.56 |
| 5 min | 7 | 8 | <0.0001 * | 2.20; 1.39–3.50 |
| Surfactant use, n (%) | 62 (54.86) | - | ||
| LISA | 36 (31.85) | - | ||
| INSURE | 7 (6.19) | - | ||
| Standard | 19 (16.81) | - | ||
| Need for subsequent surfactant doses, n (%) | 11 (9.73) | - | ||
| Duration of non-invasive ventilation, hours (mean) | 297.4 ± 138.9 | - | ||
| Duration of MV, hours (mean) | 169.3 ± 35.8 | - | ||
| Chronic lung disease, n (%) | 21 (18.58) | - | ||
| PDA: n (%) | 16 (14.15) | 6 (2.64) | 0.0001 † | 6.08; 2.31–15.99 |
| Pulmonary hemorrhage, n (%) | 6 (5.30) | - | ||
| NICU days, mean ± SD | 19.5 ± 6.3 | - | ||
| Deaths, n (%) | 10 (8.84) | - |
| Variants | Gene Models | Genotypes | Controls (%) | RDS Cases (%) | OR (% 95) | p Value | OR (% 95) * | p Value * |
|---|---|---|---|---|---|---|---|---|
| IL-10:c.-149+1984T>G | Codominant | TT | 37 (16) | 16 (14) | - | - | ||
| TG | 91 (40) | 47 (42) | 1.19 (0.60–2.36) | 0.73 | 1.63 (0.31–8.38) | 0.55 | ||
| GG | 99 (44) | 50 (44) | 1.16 (0.59–2.30) | 0.72 | 1.95 (0.18–2.70) | 0.57 | ||
| Dominant | TT | 37 (16) | 16 (14) | - | - | - | - | |
| TG + GG | 190 (84) | 97 (86) | 1.18 (0.62–2.22) | 0.63 | 0.66 (0.07–5.94) | 0.71 | ||
| Recessive | TT + TG | 128 (56) | 63 (56) | - | - | - | - | |
| GG | 99 (44) | 50 (44) | 1.02 (0.65–1.61) | 1.00 | 0.58 (0.12–2.74) | 0.49 | ||
| Overdominant | TT + GG | 136 (60) | 66 (58) | - | - | - | - | |
| TG | 91 (40) | 47 (42) | 1.06 (0.67–1.68) | 0.81 | 1.37 (0.30–6.23) | 0.41 | ||
| IL-10:c.-149+2474T>C | Codominant | TT | 181 (80) | 86 (76) | - | - | - | - |
| TC | 43 (19) | 22 (20) | 1.07 (0.60–1.91) | 0.88 | 0.88 (0.29–12.3) | 0.95 | ||
| CC | 3 (1) | 5 (5) | 3.50 (0.81–15.02) | 0.12 | 0.52 (0.34–13.4) | 0.75 | ||
| Dominant | TT | 181 (80) | 86 (76) | - | - | - | - | |
| TC + CC | 46 (20) | 27 (25) | 1.23(0.71–2.12) | 0.48 | 1.71 (0.28–3.22) | 0.55 | ||
| Recessive | TT + TC | 224 (99) | 108 (96) | - | - | - | - | |
| CC | 3 (1) | 5 (5) | 3.45 (0.81–14.73) | 0.12 | 2.84 (0.64–10.24) | 0.55 | ||
| Overdominant | TT + CC | 184 (81) | 91 (81) | - | - | - | - | |
| TC | 43 (19) | 22 (20) | 1.03 (0.58–1.83) | 1.00 | 1.62 (0.24–3.94) | 0.49 | ||
| NOS3:c.-149+1691C>T | Codominant | CC | 44 (19) | 17 (15) | - | - | - | - |
| CT | 86 (38) | 57 (50) | 1.71 (0.89–3.29) | 0.11 | 3.70 (0.44–6.54) | 0.22 | ||
| TT | 97 (43) | 39 (35) | 1.04 (0.53–1.03) | 1.00 | 2.57 (0.28–5.42) | 0.40 | ||
| Dominant | CC | 44 (19) | 17 (15) | - | - | - | - | |
| CT + TT | 183 (81) | 96 (85) | 1.35 (0.73–2.50) | 0.37 | 3.18 (0.43–6.34) | 0.25 | ||
| Recessive | CC + CT | 130 (57) | 74 (65) | - | - | - | - | |
| TT | 97 (43) | 39 (35) | 0.70 (0.44–1.12) | 0.15 | 3.18 (0.43–7.23) | 0.25 | ||
| Overdominant | CC + TT | 141 (62) | 56 (50) | - | - | - | - | |
| CT | 86 (38) | 57 (50) | 1.66 (1.05–2.64) | 0.03 | 1.95 (0.42–6.98) | 0.38 | ||
| NOS3:c.894T>G | Codominant | TT | 27 (12) | 15 (14) | - | - | - | - |
| TG | 95 (42) | 49 (43) | 0.92 (0.45–1.90) | 0.85 | 0.96 (0.25–3.22) | 0.96 | ||
| GG | 104 (46) | 49 (43) | 0.84 (0.41–1.73) | 0.71 | 0.41 (0.23–3.58) | 0.42 | ||
| Dominant | TT | 27 (12) | 15 (14) | - | - | - | - | |
| TG + GG | 199 (88) | 98 (86) | 0.88 (0.45–1.74) | 0.72 | 1.38 (2.64–8.31) | 0.39 | ||
| Recessive | TT + TG | 122 (54) | 64 (57) | - | - | - | - | |
| GG | 104 (46) | 49 (43) | 0.89 (0.56–1.41) | 0.72 | 1.39 (0.32–1.72) | 0.39 | ||
| Overdominant | TT + GG | 131 (58) | 64 (86) | - | - | - | - | |
| TG | 95 (42) | 49 (43) | 1.05 (0.66–1.66) | 0.81 | 1.22 (0.36–2.32) | 0.79 |
| Variants | Variant Alleles | Overall Variant Allele Frequency/European Variant Allele Frequency/East Asian Allele Frequency (gnomAD v4.1.0) | Frequency of Variant Allele in Control Group (%) | Frequency of Variant Allele in RDS Group (%) | OR (% 95 CI) * | p Value |
|---|---|---|---|---|---|---|
| IL-10:c.-149+1984T>G | G | 68/76/31 | 289 (64) | 147 (65) | 1.06 (0.76–1.48) | 0.73 |
| IL-10:c.-149+2474T>C | C | 40/48/5 | 49 (11) | 32 (14) | 1.36 (0.84–2.19) | 0.21 |
| NOS3:c.-149+1691C>T | T | 70/61/88 | 280 (62) | 135 (60) | 0.93 (0.67–1.29) | 0.67 |
| NOS3:c.894T>G | G | 69/66/90 | 303 (67) | 147 (65) | 0.91 (0.65–1.28) | 0.60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anciuc-Crauciuc, M.; Crauciuc, G.-A.; Tripon, F.; Simon, M.; Cucerea, M.C.; Bănescu, C.V. Exploring IL-10 and NOS3 Genetic Variants as a Risk Factor for Neonatal Respiratory Distress Syndrome and Its Outcome. Diagnostics 2025, 15, 2259. https://doi.org/10.3390/diagnostics15172259
Anciuc-Crauciuc M, Crauciuc G-A, Tripon F, Simon M, Cucerea MC, Bănescu CV. Exploring IL-10 and NOS3 Genetic Variants as a Risk Factor for Neonatal Respiratory Distress Syndrome and Its Outcome. Diagnostics. 2025; 15(17):2259. https://doi.org/10.3390/diagnostics15172259
Chicago/Turabian StyleAnciuc-Crauciuc, Mădălina, George-Andrei Crauciuc, Florin Tripon, Marta Simon, Manuela Camelia Cucerea, and Claudia Violeta Bănescu. 2025. "Exploring IL-10 and NOS3 Genetic Variants as a Risk Factor for Neonatal Respiratory Distress Syndrome and Its Outcome" Diagnostics 15, no. 17: 2259. https://doi.org/10.3390/diagnostics15172259
APA StyleAnciuc-Crauciuc, M., Crauciuc, G.-A., Tripon, F., Simon, M., Cucerea, M. C., & Bănescu, C. V. (2025). Exploring IL-10 and NOS3 Genetic Variants as a Risk Factor for Neonatal Respiratory Distress Syndrome and Its Outcome. Diagnostics, 15(17), 2259. https://doi.org/10.3390/diagnostics15172259






