Beyond the Urogenital Tract, the Role of Ureaplasma parvum in Invasive Infection in Adults: A Case Series and Literature Review
Abstract
1. Introduction
2. Methods
2.1. Patients Selection and Data Collection
2.2. Specimen Collection and Next-Generation Sequencing
2.3. Treatment and Outcome Assessment
2.4. Statistical Analysis
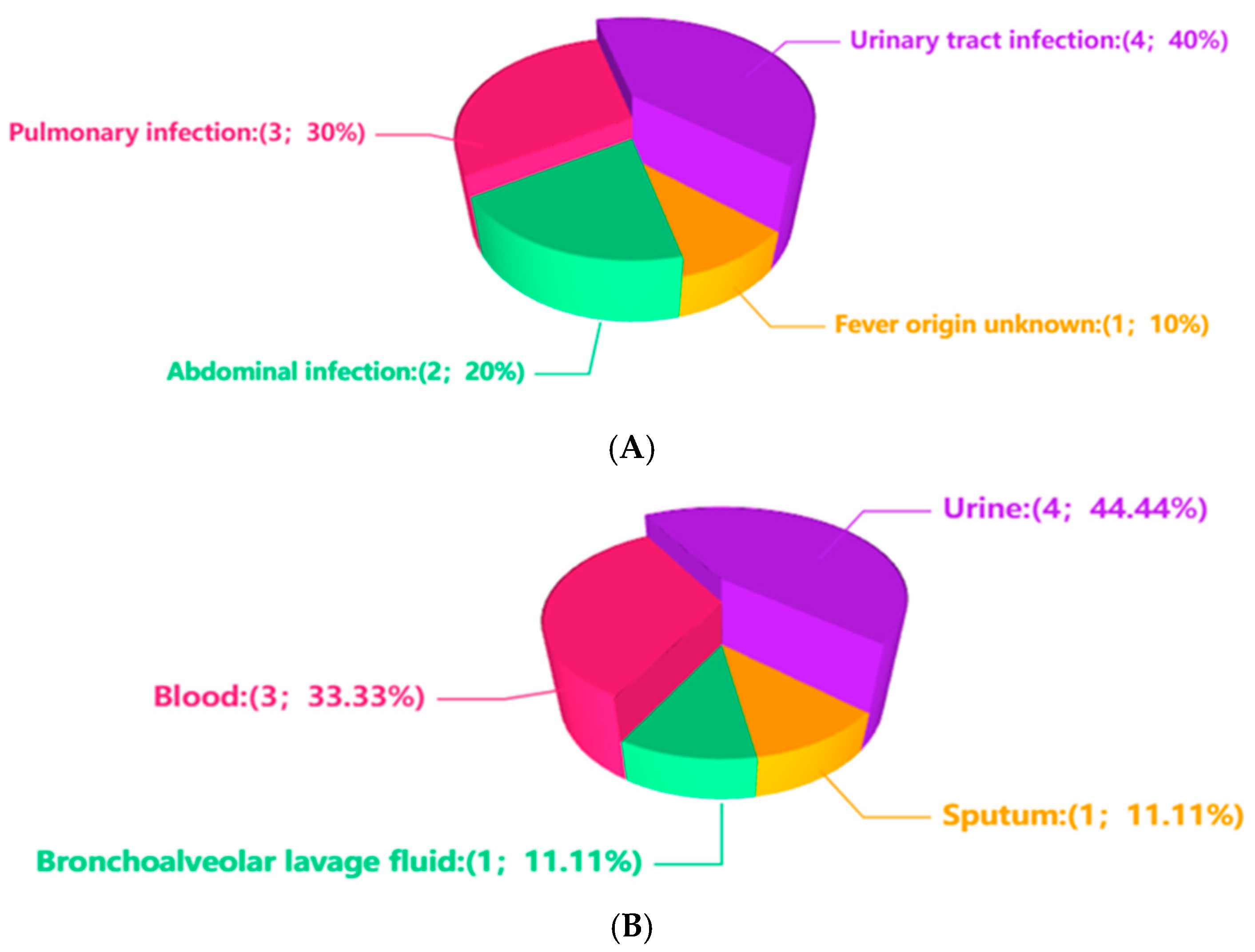
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Up | Ureaplasma parvum |
| NGS | next-generation sequencing |
| mNGS | metagenomic next-generation sequencing |
| IRB | Institutional Review Board |
| BALF | bronchoalveolar lavage fluid |
| CMV | Cytomegalovirus |
| NE | neutrophils |
| WBC | white blood cells |
| PA | Pseudomonas aeruginosa |
References
- Kotrotsiou, T.; Exindari, M.; Diza, E.; Gioula, G.; Melidou, A.; Kaplanis, K.; Malisiovas, N. Prevalence and antimicrobial susceptibility of Ureaplasma urealyticum in asymptomatic women in Northern Greece. Hippokratia 2013, 17, 319–321. [Google Scholar]
- Bartkeviciene, D.; Opolskiene, G.; Bartkeviciute, A.; Arlauskiene, A.; Lauzikiene, D.; Zakareviciene, J.; Ramasauskaite, D. The impact of Ureaplasma infections on pregnancy complications. Libyan J. Med. 2020, 15, 1812821. [Google Scholar] [CrossRef] [PubMed]
- Cassell, G.H.; Waites, K.B.; Watson, H.L.; Crouse, D.T.; Harasawa, R. Ureaplasma urealyticum intrauterine infection: Role in prematurity and disease in newborns. Clin. Microbiol. Rev. 1993, 6, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Qin, L.; Li, Y.H.; Cao, X.J.; Wang, X.J.; Mao, R.P.; Yang, H.Y.; Li, L. Clinical metagenomic sequencing for rapid diagnosis of neonatal meningitis caused by Ureaplasma parvum: A case report. Medicine 2022, 101, e28662. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, P.J.; Regan, J.A. Vertical transmission of Ureaplasma urealyticum in full term infants. Pediatr. Infect. Dis. J. 1987, 6, 825–828. [Google Scholar] [CrossRef] [PubMed]
- Gray, D.J.; Robinson, H.B.; Malone, J.; Thomson, R.B., Jr. Adverse outcome in pregnancy following amniotic fluid isolation of Ureaplasma urealyticum. Prenat. Diagn. 1992, 12, 111–117. [Google Scholar] [CrossRef]
- Perni, S.C.; Vardhana, S.; Korneeva, I.; Tuttle, S.L.; Paraskevas, L.R.; Chasen, S.T.; Kalish, R.B.; Witkin, S.S. Mycoplasma hominis and Ureaplasma urealyticum in midtrimester amniotic fluid: Association with amniotic fluid cytokine levels and pregnancy outcome. Am. J. Obstet. Gynecol. 2004, 191, 1382–1386. [Google Scholar] [CrossRef]
- Deguchi, T.; Shimada, Y.; Horie, K.; Mizutani, K.; Seike, K.; Tsuchiya, T.; Yokoi, S.; Yasuda, M.; Ito, S. Bacterial loads of Ureaplasma parvum contribute to the development of inflammatory responses in the male urethra. Int. J. STD AIDS 2015, 26, 1035–1039. [Google Scholar] [CrossRef]
- Ito, K.; Akai, K.; Nishiumi, F.; Nakura, Y.; Ning Wu, H.; Kurata, T.; Onodera, A.; Kawai, Y.; Kajiyama, S.; Yanagihara, I. Ability of Ureaplasma parvum to invade mouse sperm, fertilize eggs through infected sperm, and impair mouse sperm function and embryo development. F S Sci. 2021, 2, 13–23. [Google Scholar] [CrossRef]
- Pan, X.; Xu, J.; Pan, L.; Wang, C.; Qiu, J.; Huang, X.; Yan, C.; Mao, M. Hyperammonemia in a septic patient with Ureaplasma parvum arthritis: A case report. BMC Infect. Dis. 2022, 22, 958. [Google Scholar] [CrossRef]
- Pailhoriès, H.; Chenouard, R.; Eveillard, M.; Kempf, M.; Pereyre, S.; Bébéar, C.; Lemarié, C. A case of Ureaplasma parvum meningitis in an adult after transphenoidal ablation of craniopharyngioma. Int. J. Infect. Dis. 2019, 84, 5–7. [Google Scholar] [CrossRef]
- Tadera, K.; Kitagawa, H.; Kitano, H.; Hara, T.; Kashiyama, S.; Nomura, T.; Omori, K.; Shigemoto, N.; Yokozaki, M.; Ohge, H. Prevalence of Mycoplasma hominis, Ureaplasma urealyticum, and Ureaplasma parvum detection in urine and respiratory tract samples in Hiroshima, Japan. Heliyon 2023, 9, e14543. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Nie, Z.; Zhao, Y.; Bao, B. Peritonitis-associated with peritoneal dialysis following Ureaplasma parvum infection: A case report and literature review. Indian J. Med. Microbiol. 2023, 45, 100410. [Google Scholar] [CrossRef]
- Liu, D.; Zhou, H.; Xu, T.; Yang, Q.; Mo, X.; Shi, D.; Ai, J.; Zhang, J.; Tao, Y.; Wen, D.; et al. Multicenter assessment of shotgun metagenomics for pathogen detection. eBioMedicine 2021, 74, 103649. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Zhang, J.; Zhang, D.; Zhou, Y.; Wu, P.; Ding, W.; Wang, J.; Ouyang, C.; Yang, Q. Background Filtering of Clinical Metagenomic Sequencing with a Library Concentration-Normalized Model. Microbiol. Spectr. 2022, 10, e0177922. [Google Scholar] [CrossRef] [PubMed]
- Noma, I.H.Y.; Shinobu-Mesquita, C.S.; Suehiro, T.T.; Morelli, F.; De Souza, M.V.F.; Damke, E.; Da Silva, V.R.S.; Consolaro, M.E.L. Association of Righ-Risk Human Papillomavirus and Ureaplasma parvum Co-Infections with Increased Risk of Low-Grade Squamous Intraepithelial Cervical Lesions. Asian Pac. J. Cancer Prev. 2021, 22, 1239–1246. [Google Scholar] [CrossRef]
- Galdiero, M.; Trotta, C.; Schettino, M.T.; Cirillo, L.; Sasso, F.P.; Petrillo, F.; Petrillo, A. Normospermic Patients Infected with Ureaplasma parvum: Role of Dysregulated miR-122-5p, miR-34c-5, and miR-141-3p. Pathog. Immun. 2023, 8, 16–36. [Google Scholar] [CrossRef]
- Wu, J.; Hu, Y. A late-onset hyperammonemia syndrome caused by Ureaplasma parvum infection after kidney transplantation. Heliyon 2024, 10, e32134. [Google Scholar] [CrossRef]
- Asif, A.A.; Roy, M.; Ahmad, S. Rare case of Ureaplasma parvum septic arthritis in an immunocompetent patient. BMJ Case Rep. 2020, 13, e236396. [Google Scholar] [CrossRef]
- Suknuntha, K.; Lepak, A.J.; Rehrauer, W.M.; Chen, D.J. Identification of Ureaplasma parvum as a Cause of Culture-Negative Septic Monoarthitis Using 16S rRNA Gene Sequencing. Infect. Dis. Clin. Pract. 2019, 27, e12–e14. [Google Scholar] [CrossRef]
- Siles-Guerrero, V.; Cardona-Benavides, I.; Liébana-Martos, C.; Vázquez-Alonso, F.; Expósito-Ruiz, M.; Navarro-Marí, J.M.; Gutiérrez-Fernández, J. Recent clinical relevance of mono-genital colonization/infection by Ureaplasma parvum. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1899–1905. [Google Scholar] [CrossRef] [PubMed]
- Rehman, S.U.; Day, J.; Afshar, B.; Rowlands, R.S.; Billam, H.; Joseph, A.; Guiver, M.; Maddocks, S.E.; Chalker, V.J.; Beeton, M.L. Molecular exploration for Mycoplasma amphoriforme, Mycoplasma fermentans and Ureaplasma spp. in patient samples previously investigated for Mycoplasma pneumoniae infection. Clin. Microbiol. Infect. 2021, 27, e1691–e1697. [Google Scholar] [CrossRef] [PubMed]
- Triantafilou, M.; De Glanville, B.; Aboklaish, A.F.; Spiller, O.B.; Kotecha, S.; Triantafilou, K. Synergic activation of toll-like receptor (TLR) 2/6 and 9 in response to Ureaplasma parvum & urealyticum in human amniotic epithelial cells. PLoS ONE 2013, 8, e61199. [Google Scholar] [CrossRef]
- Biernat-Sudolska, M.; Rojek-Zakrzewska, D.; Drożdż, K.; Bilska-Wilkosz, A. Antimicrobial Activity of N,N-Diethyldithiocarbamate against Ureaplasma parvum and Ureaplasma urealyticum. Int. J. Mol. Sci. 2023, 25, 40. [Google Scholar] [CrossRef]
- Madlener, M.; Breuninger, M.; Meißner, A.; Stetefeld, H.; Telentschak, S.; Wille, T.; van Eimeren, T.; Jung, N. Brain abscess with Ureaplasma parvum in a patient with granulomatosis with polyangiitis. Infection 2023, 51, 779–782. [Google Scholar] [CrossRef] [PubMed]
| Case | Genda | Age | Infection Sites | Specimen Types | Relative Abundance of Up% | Sequence Number of Up | Target Therapy | Prognosis |
|---|---|---|---|---|---|---|---|---|
| 1 | female | 70–80 | Pulmonary infection | blood | <1 | ≤1 | No | Unresolved infection |
| 2 | male | 20–30 | Fever origin unknown | blood | <1 | ≤1 | No | Unclear |
| 3 | male | 40–50 | Pulmonary infection | BALF | <1 | 1–100 | No | Unclear |
| 4 | female | 40–50 | Urinary tract infection | urine | 1–10 | 100–1000 | Minocycline | Relieved |
| 5 | female | 30–40 | Abdominal infection | blood | 1–10 | ≤1 | Tigecycline | Relieved |
| 6 | male | 20–30 | Urinary tract infection | urine | >10 | 2000–3000 | Moxifloxacin | Relieved |
| 7 | female | 40–50 | Urinary tract infection | urine | 1–10 | 1000–2000 | Minocycline | Relieved |
| 8 | male | 50–60 | Pulmonary infection, Abdominal infection | Sputum | <1 | 1–100 | No | Relieved |
| 9 | female | 20–30 | Urinary tract infection | urine | <1 | 100–1000 | Minocycline | Relieved |
| Case | Immunosuppressed individuals | Other microbial species | Relative Abundance of other microbial species(%) | Sequence number of other microbial species | ||||
| 1 | Yes | None | None | None | ||||
| 2 | No | None | None | None | ||||
| 3 | Yes | Mycobacterium tuberculosis complex | 1–10 | 2000–3000 | ||||
| 4 | No | Lactobacillus crispatus | >10 | 70,000–80,000 | ||||
| 5 | Yes | Cytomegalovirus | 1–10 | ≤1 | ||||
| 6 | Yes | None | None | None | ||||
| 7 | Yes | Acinetobacter junii | <1 | 1–100 | ||||
| 8 | Yes | PA | >10 | 90,000–100,000 | ||||
| 9 | No | Enterococcusfaecalis | 1–10 | 1000–2000 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Li, X.; Liu, D.; Yao, J.; Li, X.; Wang, Y. Beyond the Urogenital Tract, the Role of Ureaplasma parvum in Invasive Infection in Adults: A Case Series and Literature Review. Diagnostics 2025, 15, 2242. https://doi.org/10.3390/diagnostics15172242
Hu L, Li X, Liu D, Yao J, Li X, Wang Y. Beyond the Urogenital Tract, the Role of Ureaplasma parvum in Invasive Infection in Adults: A Case Series and Literature Review. Diagnostics. 2025; 15(17):2242. https://doi.org/10.3390/diagnostics15172242
Chicago/Turabian StyleHu, Linhui, Xiangyan Li, Dan Liu, Jie Yao, Xueying Li, and Yan Wang. 2025. "Beyond the Urogenital Tract, the Role of Ureaplasma parvum in Invasive Infection in Adults: A Case Series and Literature Review" Diagnostics 15, no. 17: 2242. https://doi.org/10.3390/diagnostics15172242
APA StyleHu, L., Li, X., Liu, D., Yao, J., Li, X., & Wang, Y. (2025). Beyond the Urogenital Tract, the Role of Ureaplasma parvum in Invasive Infection in Adults: A Case Series and Literature Review. Diagnostics, 15(17), 2242. https://doi.org/10.3390/diagnostics15172242





