1. Introduction
Breast cancer is the most frequently diagnosed cancer and the second leading cause of cancer-related deaths among women globally [
1,
2]. Improving patient survival requires a multidisciplinary approach involving experts across pathology, medical, surgical, and radiation oncology [
3]. Accurate diagnosis, staging, and grading of the disease depend on tissue biopsies, initially processed with haematoxylin and eosin (H&E) staining. For further insights, immunohistochemistry (IHC) staining is employed to detect specific protein receptors, such as the Human Epidermal Growth Factor Receptor 2 (HER2) [
4]. HER2 overexpression, present in approximately
to
of breast cancer cases, is often associated with aggressive disease and a poorer prognosis [
5]. However, HER2-positive patients benefit from targeted therapies, underscoring the need for accurate HER2 status evaluation [
6,
7]. When HER2 status is equivocal (score 2+), further assessment using in situ hybridization (ISH) techniques is necessary to determine HER2 gene amplification.
While H&E and HER2 IHC provide essential insights, they do not confirm gene amplification. This assessment is conducted through ISH techniques, including fluorescence ISH (FISH), chromogenic ISH (CISH), and silver-enhanced ISH (SISH). HER2 protein overexpression is specifically evaluated by IHC [
8]. In clinical settings, such as in Korea, IHC serves as the primary test for HER2, with SISH used as a confirmatory test for equivocal cases due to its accuracy, efficiency, and consistency compared to FISH and CISH.
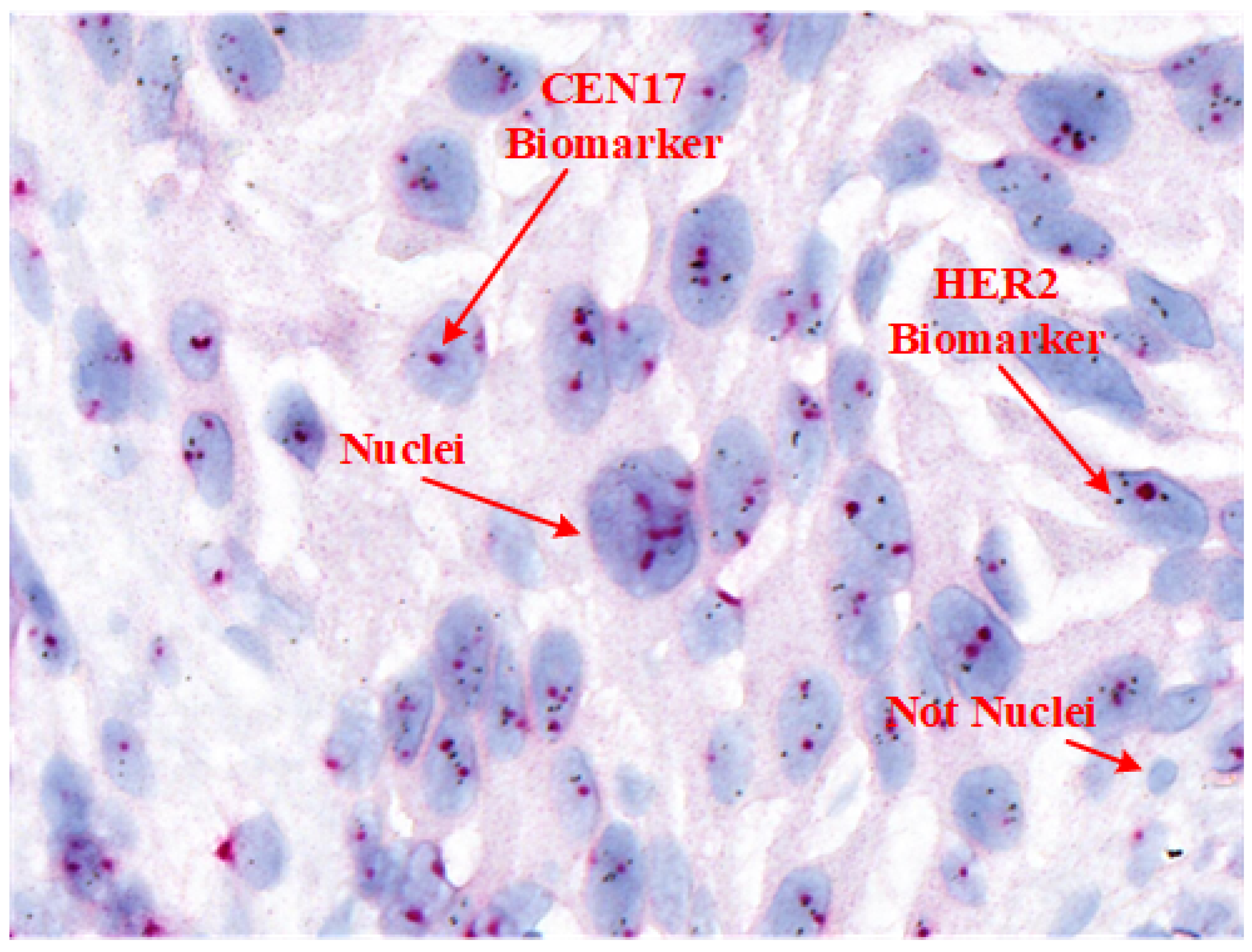
Table 1 provides a comparison of these techniques, and
Figure 1 shows an example SISH image highlighting its key features.
Advances in digital pathology, such as whole-slide imaging (WSI) and cloud-based data storage, have transformed diagnostic workflows [
9]. Integrating WSI data with telemedicine and big data analytics opens avenues for personalized healthcare and research-driven insights, incorporating data from epidemiological studies and genomic sequencing [
10]. Computer vision techniques are crucial in digital pathology, as they enable automated and standardized image analysis, reducing observer variability and enhancing diagnostic efficiency [
11,
12]. As a fully automated staining method, SISH enables pathologists to analyze tissue morphology under bright-field microscopy, which is vital for accurate interpretation [
13].
For accurate HER2 quantification in SISH, pathologists assess the ratio of HER2 signals (black spots) to centromere 17 (CEN17) signals (red dots) within selected areas containing 20 nuclei, following ASCO/CAP guidelines. However, manual nuclei segmentation in SISH WSIs is labor-intensive, as it involves variable nuclei shapes and distribution patterns essential for tissue and cancer characterization [
14].
Automated nuclei segmentation poses challenges due to overlapping nuclei, staining variability, and cytoplasmic morphology differences. Traditional methods, such as intensity thresholding, marker-based watershed segmentation, and active contour models, are limited by computational demands or lack robustness [
15,
16,
17]. Recent advances in machine learning, especially convolutional neural networks (CNNs), have demonstrated promise by learning features relevant to nuclei morphology and staining patterns [
16,
18].
Despite advancements, deep learning-based segmentation methods face limitations due to the lack of large, annotated datasets for training. To overcome these challenges, we propose a two-stage algorithm that combines a pretrained Stardist model with morphological preprocessing and fine-tuning on a novel, expert-annotated HER2-SISH dataset. This approach aims to improve nuclei detection accuracy in complex SISH images and streamline diagnostic workflows by offering automated tools for pathologists.
The primary contributions of this research include:
Developing a two-stage framework that employs morphological preprocessing, a pretrained Stardist 2D Fluo model, and fine-tuning for precise nuclei detection in HER2-SISH histopathology images;
Facilitating visualization and analysis of SISH-detected nuclei regions for enhanced clinical interpretation;
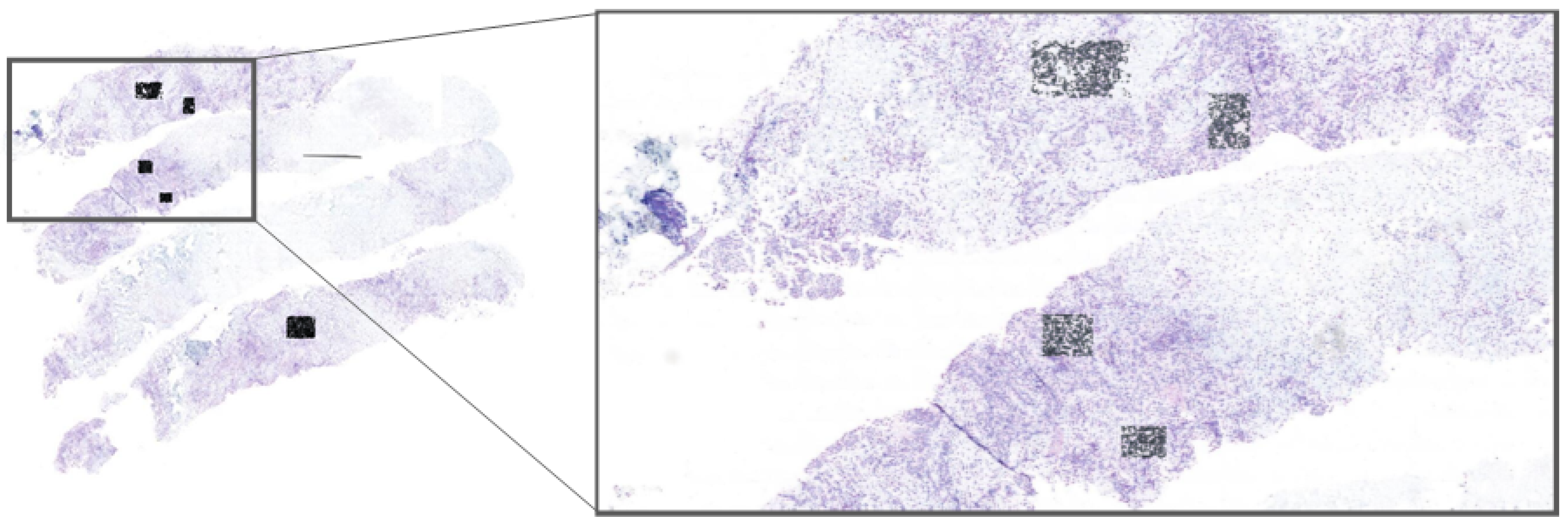
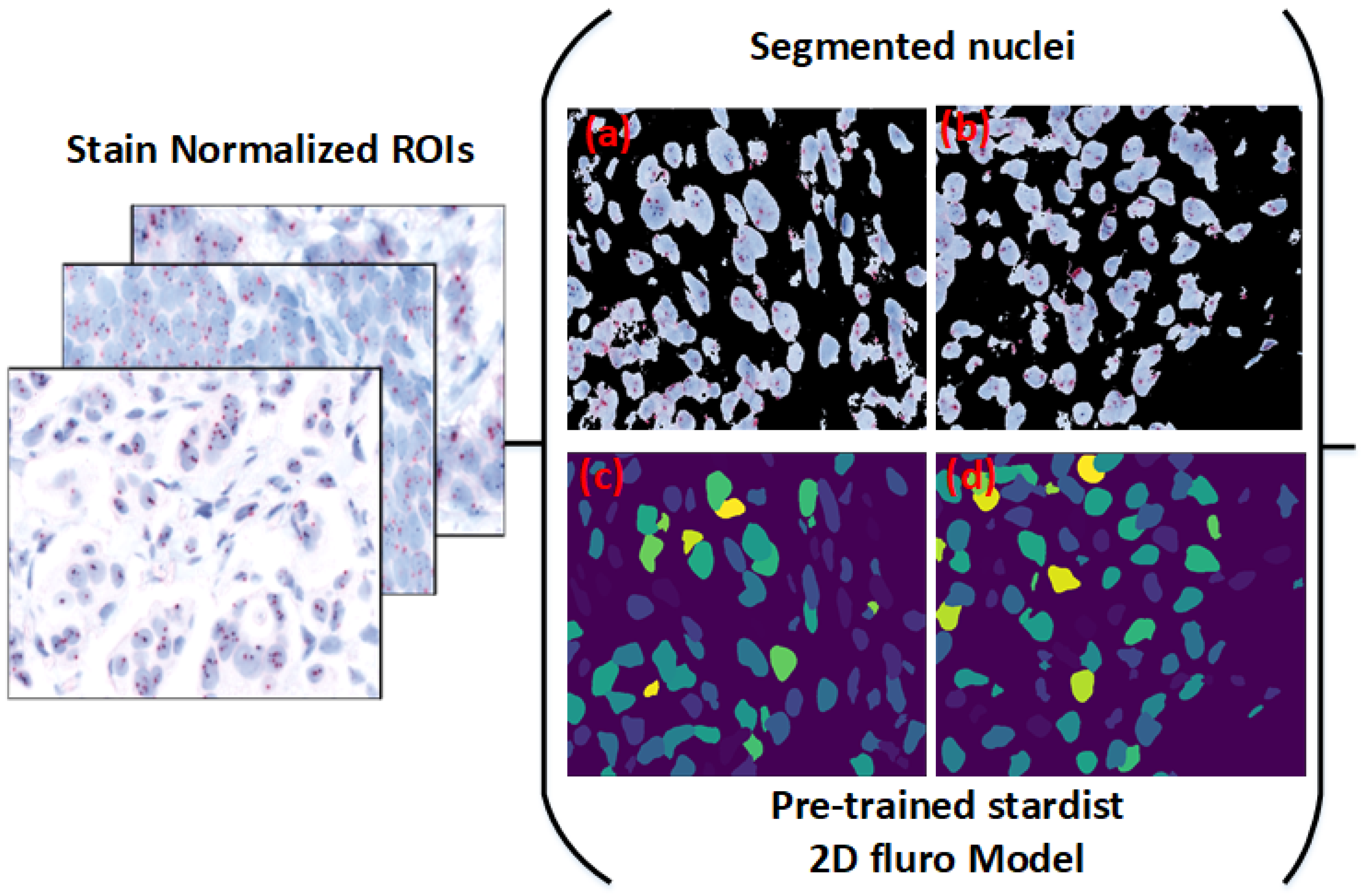
Implementing our model within the Cytomine platform, allowing experts to select regions of interest (ROIs) directly from whole-slide images (WSIs) for real-time nuclei segmentation; this application, shown in
Figure 2, enables pathologists to efficiently use our model for HER2 scoring, accelerating the evaluation process;
Demonstrating the model’s effectiveness for automatic nuclei detection in complex SISH images, thereby supporting pathologists in HER2 assessment and scoring.
The paper is structured as follows:
Section 2 reviews the literature on nuclei detection in histopathology.
Section 3 describes our methodology, including datasets and training processes.
Section 4 presents the experimental setup, and
Section 5 provides a detailed analysis and discussion of results. Finally, the conclusion and future directions of this research are outlined in
Section 7.
2. Related Work
In this section, we review the significance of nuclei regions in various histopathology-stained images, explore deep learning-based nuclei segmentation techniques for computational pathology, and discuss the role of artificial intelligence (AI) in addressing nuclei segmentation challenges. This review highlights the applications of deep learning and AI in digital pathology, especially as they pertain to automating complex and labor-intensive tasks in medical imaging.
2.1. Deep Learning-Based Nuclei Segmentation
Nuclei segmentation in histopathology images has been extensively studied, with numerous deep learning methods developed for this critical task. However, the inherent complexity of histopathology images continues to challenge the application of deep learning for nuclei segmentation.
Accurate nuclei segmentation is crucial for computer-assisted histopathology, as it underpins many downstream analyses essential for disease characterization and diagnosis. Yet, histopathology images present unique challenges: nuclei often form intricate structures such as clumps or nests, making precise annotation difficult. Furthermore, boundary delineation is complicated by the close proximity of epithelial cells and irregular mitotic patterns.
Addressing these challenges requires advanced deep learning techniques, innovative data augmentation strategies, and architectures capable of capturing the nuanced details of histopathology images. Collaborative data-sharing efforts within the research community are also essential to improve the performance and robustness of models for nuclei segmentation in histopathology. By overcoming these barriers, deep learning can enable more accurate and reliable computer-assisted pathology analyses, benefiting both clinicians and patients.
Several deep learning methods have been proposed for nuclei segmentation, often utilizing two-class probability maps followed by post-processing. For instance, ref. [
18] introduced CNN-3C, which reframes nuclei segmentation as a three-class problem, achieving promising results across multiple organ types. Cui et al. proposed a fully convolutional network-based method for nuclei segmentation [
19], while [
20] formulated the task as a regression problem by predicting nuclei distance maps. Ref. [
21] demonstrated that fusing networks led to better segmentation results compared to multi-task learning. Salvi et al. introduced a multi-scale adaptive nuclei segmentation (MANA) method, showing strong performance on multi-organ nuclei segmentation [
22]. Recent work such as Xu et al. [
23] explored transformer-based nuclei segmentation with improved generalization across staining variations. Similarly, Huang et al. [
24] proposed a Multiscale CNN Transformer approach, showing strong results across nuclei segmentation. Despite these advancements, these methods often remain limited by the availability of sufficiently large, annotated datasets and the diversity needed to generalize across tissue types.
Adaptability to multiple organ types, clinical data sources, patient samples, and disease conditions requires large and varied datasets. Simple CNN architectures frequently encounter difficulties with overlapping nuclei, necessitating complex post-processing techniques. Many previous studies have approached nuclei segmentation as a classification or combined classification–regression task. We propose an alternative approach by treating it as a self-learning process that uses pretrained predictions to refine segmented nuclei regions. This method provides greater context awareness and global consistency by optimizing the loss function across the entire image rather than just at the pixel level, enhancing robustness and adaptability for various histopathology applications.
2.2. Stardist for Medical Imaging Applications
Accurate detection and segmentation of cell nuclei in histopathology images are critical for numerous medical applications, including cancer diagnosis and treatment planning. Factors such as low signal-to-noise ratios and clustered cells often complicate the identification of nuclei using traditional intensity-based algorithms.
To address these issues, Stardist [
25,
26] leverages deep learning techniques to enhance segmentation performance. Stardist applies to both 2D and 3D data, representing objects as star-convex polygons, which facilitates effective segmentation of individual cells in dense tissue sections [
26].
A key innovation of Stardist is its use of a neural network to predict shape representations for nuclei in the form of star-convex polygons. This approach significantly improves segmentation accuracy. For each pixel within an object, the network predicts distances to the object boundary along multiple radial directions, effectively defining a star-convex polygon. This representation allows for accurate and reliable segmentation, even under challenging conditions, making Stardist a valuable tool for histopathology and other medical imaging tasks.
In addition to polygon prediction, Stardist incorporates object probability estimation to determine pixels belonging to nuclei, which contribute to shape estimation. To minimize redundant shape predictions, Stardist applies non-maximum suppression, eliminating overlapping shapes that likely represent the same object.
The approach outlined by [
26] lies at the intersection of object detection and instance segmentation. While Stardist may not achieve pixel-perfect precision, it provides high-fidelity segmentation suitable for cell nuclei, making it particularly effective in histopathology image analysis.
Although star-convex polygons have been previously investigated for object detection in natural images [
27], they were found to be less suitable for generic object classes like people or vehicles. In contrast, in histopathology, Stardist’s ability to segment cell nuclei has encouraged further research, particularly for HER2-SISH image analysis.
Recently, ref. [
28] assessed Stardist in segmenting nuclei in CellSearch images, specifically in peripheral blood and DLA samples. Their study highlighted significant improvements in segmenting CellSearch images, resulting in more accurate counts of circulating tumor cells (CTCs). These findings emphasize Stardist’s potential for nuclei localization using star-convex polygons, making it a promising tool for advanced histopathology analysis, including applications in HER2-SISH images.
3. Materials and Methods
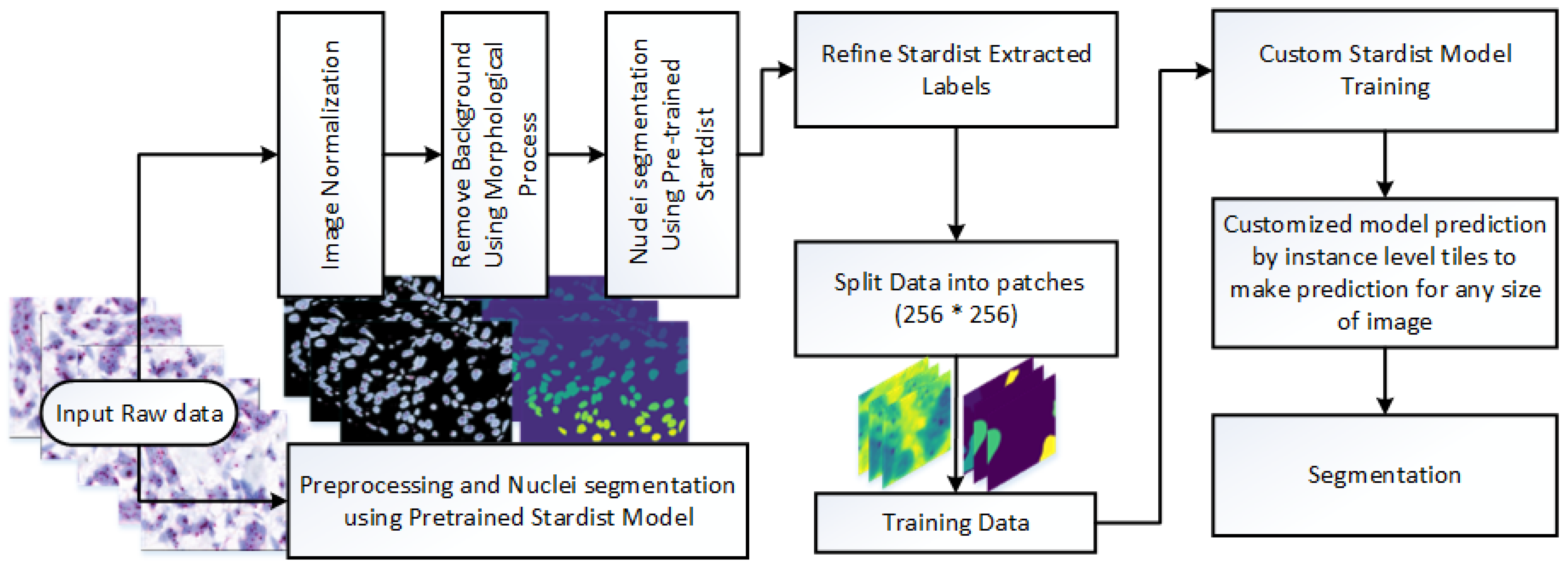
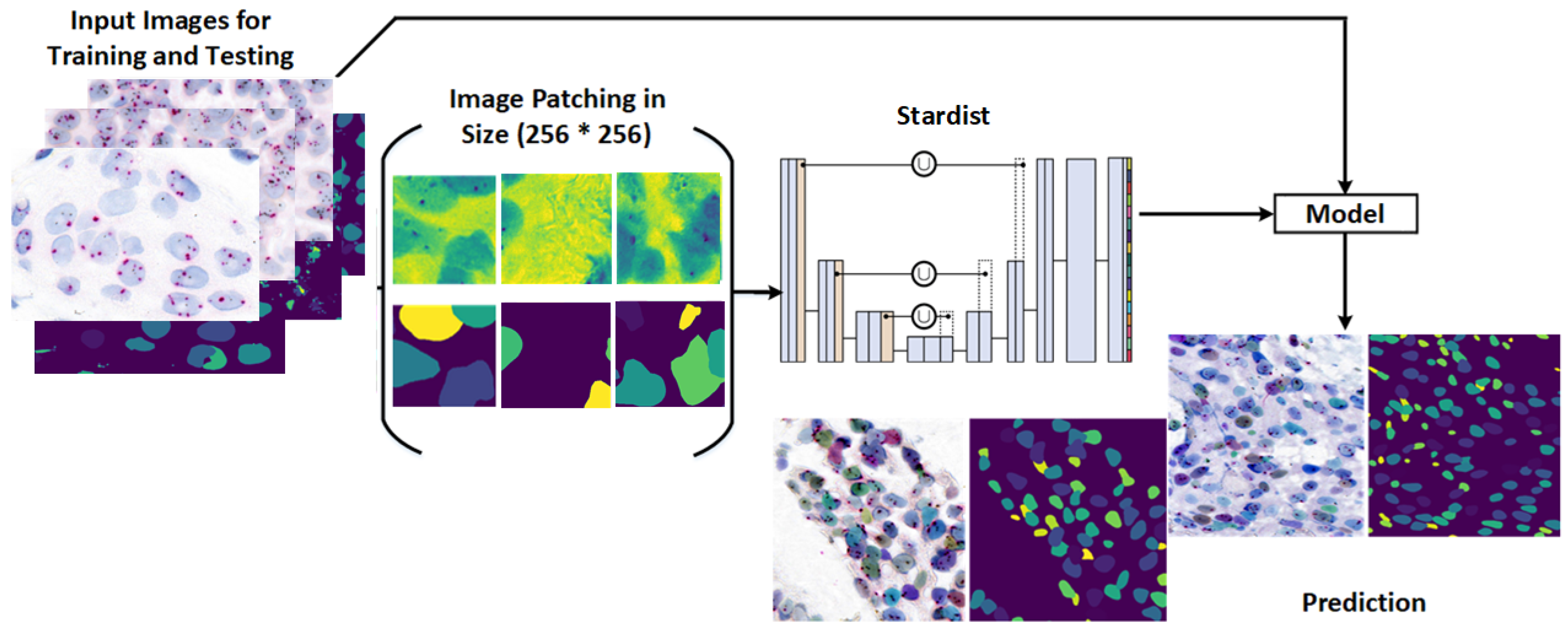
This study presents a novel framework for nuclei detection in HER2-SISH-stained whole slide images (WSIs) using a two-stage deep learning approach. Our methodology combines a pretrained Stardist model with morphological preprocessing and fine tuning on expert-annotated ROIs. The workflow is illustrated in
Figure 3, with each component detailed in the following subsections.
3.1. Dataset Description
A total of 20 HER2-SISH whole slide images (WSIs) were obtained from breast cancer biopsy cases at the University of Malaya Medical Center (UMMC), comprising 8 HER2-amplified and 12 non-amplified cases. The biopsy samples were stained using the SISH protocol and scanned using a 3DHistech Pannoramic DESK scanner, producing high-resolution WSIs with dimensions ranging from
pixels. From each WSI, expert pathologists selected five regions of interest (ROIs), resulting in 100 ROIs in total. The ROI dimensions varied from 859 × 755 to 5451 × 3136 pixels.
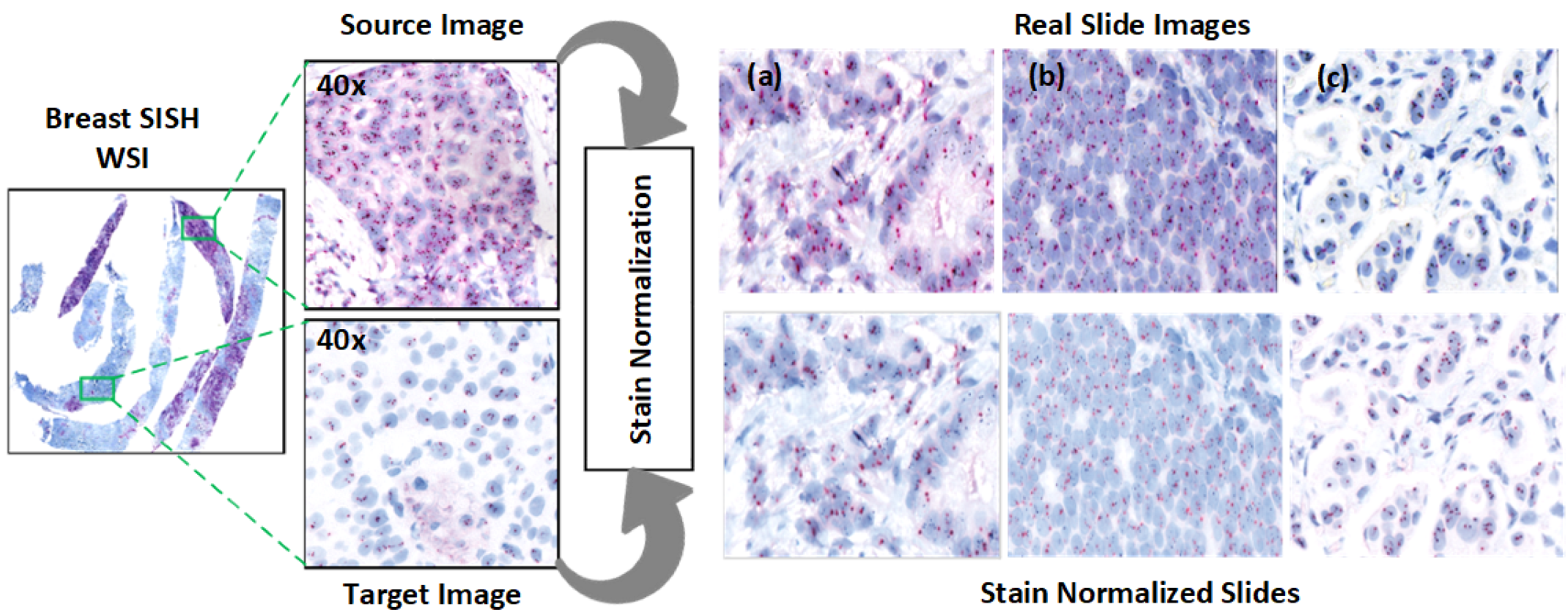
Figure 4 illustrates an example ROI following stain normalization, and
Table 2 summarizes the dataset characteristics.
The relatively small dataset size (20 WSIs with 100 ROIs) reflects the significant manual effort required for expert annotation and the high-resolution nature of the SISH images. Despite its size, this curated dataset enabled the model to generalize well, as evidenced by strong F1-scores across expert evaluations.
The dataset was split into an 80:20 ratio for training and testing. The training set consisted of 80 ROIs, used to create labels and fine tune a custom Stardist model. Initial annotations were generated using a pretrained Stardist model and refined through manual corrections. The remaining 20 ROIs were reserved for evaluation, both visually and using expert-marked nuclei as ground truth.
Inclusion criteria required archived HER2-SISH WSIs from pathologically confirmed breast cancer cases with complete HER2 scoring records. Exclusion criteria eliminated samples with poor tissue preservation, insufficient metadata, or image artifacts affecting nuclei visibility. ROIs were selected only if they showed clear nuclear morphology, good staining contrast, and minimal overlapping. Regions with ambiguous features or nuclei clustering were excluded.
Three pathologists participated in the ROI selection process, including a senior consultant with extensive experience in breast pathology. To mitigate inter-observer variability, ROIs were initially selected independently by two pathologists and later reviewed and finalized through consensus with the senior pathologist.
3.2. Cytomine Platform for ROI Selection and Real-Time Segmentation
To make our framework accessible for clinical application, we implemented it on the Cytomine platform [
29]. Cytomine allows pathologists to select ROIs directly from WSIs, enabling them to interactively apply our model for real-time nuclei segmentation (see
Figure 2). This integration with Cytomine streamlines the HER2 scoring process by allowing pathologists to focus on specific regions, thus enhancing diagnostic efficiency. Real-time feedback from pathologists also provides a mechanism to refine segmentation accuracy and ensure clinical relevance. ROIs were included only if they exhibited clear nuclear morphology with adequate staining contrast and no overlapping nuclei clusters. Excluded regions included artifacts, ambiguous staining, and overlapping nuclei unsuitable for HER2/CEN17 quantification. To reduce inter-observer variability, an initial set of candidate ROIs was independently selected by two junior pathologists and reviewed by a senior pathologist. Final ROIs were chosen through consensus agreement.
3.3. Ground Truth Preparation
To create reliable ground truth labels, we developed a preprocessing method that includes image normalization and nuclei region extraction. First, images were normalized to standardize contrast, and then background removal was applied to enhance nuclei visibility.
3.3.1. Stain Normalization
Normalization addresses the contrast variability in HER2-SISH images. We used the Macenko normalization algorithm [
30], which falls under unsupervised normalization techniques:
Stain Vector Estimation: Singular value decomposition (SVD) was applied to foreground pixels to estimate the SISH stain vectors.
Intensity Correction: Intensity variations, arising from staining and procedural factors, were corrected.
Projection to Reference Image: Images were projected to a reference image to maintain consistent color properties across normalized images.
The Macenko algorithm operates by converting RGB vectors to optical density (OD) values using
, allowing a linear combination of stains to yield OD values. An open-source Python 3.11 implementation of the Macenko method was utilized [
31].
Normalization effectiveness was confirmed through statistical testing, with results indicating that the Macenko method aligns image contrast with the reference image (
Table 3). Visual examples are shown in
Figure 4.
3.3.2. Extraction of Image Foreground
Following normalization, background removal was applied to improve nuclei visibility, facilitating accurate segmentation with Stardist. Histogram equalization and grayscale conversion were used to enhance contrast, followed by mask creation using thresholding and morphological operations.
Figure 5 shows the effect of these preprocessing steps.
The thresholding method involved both global and local adjustments to distinguish nuclei intensity. Morphological refinements were incorporated to improve boundary accuracy, resulting in clearer nuclei separation, as illustrated in
Figure 5.
3.4. Stardist Model for Nuclei Segmentation
Nuclei detection and segmentation in HER2-SISH images are crucial for HER2 scoring and analysis. Stardist [
25,
32] addresses segmentation challenges by predicting nuclei shapes as star-convex polygons, facilitating precise segmentation in 2D and 3D volumes.
Originally designed for cell nuclei segmentation, Stardist was adapted here for HER2-SISH nuclei with a fine-tuning process.
Figure 6 illustrates the workflow used for training and applying the Stardist model in our study.
Key features of the Stardist model include:
CNN Architecture: Built on a U-Net base, Stardist utilizes a contracting path for context capture and an expansive path for localization.
StarConvex Shape Prediction: Each image tile is divided into overlapping tiles, predicting object shapes and distance maps that refine boundary accuracy.
Fine-Grained Segmentation: Unlike bounding box predictions, Stardist achieves pixel-level boundary precision for accurate object shape capture.
Implementation: Our Stardist model was implemented in TensorFlow and fine tuned on the expert-annotated dataset.
3.5. Separating Overlapping Nuclei Using Pretrained Stardist
Traditional watershed methods produce excessive segmentation artifacts in clustered regions. To address this, we applied a pretrained Stardist model, specifically the Versatile fluorescent nuclei model, which performed effectively for HER2-SISH nuclei segmentation based on visual and expert evaluations.
A custom wrapper function was used to normalize the input, initialize Stardist, and generate segmentation masks.
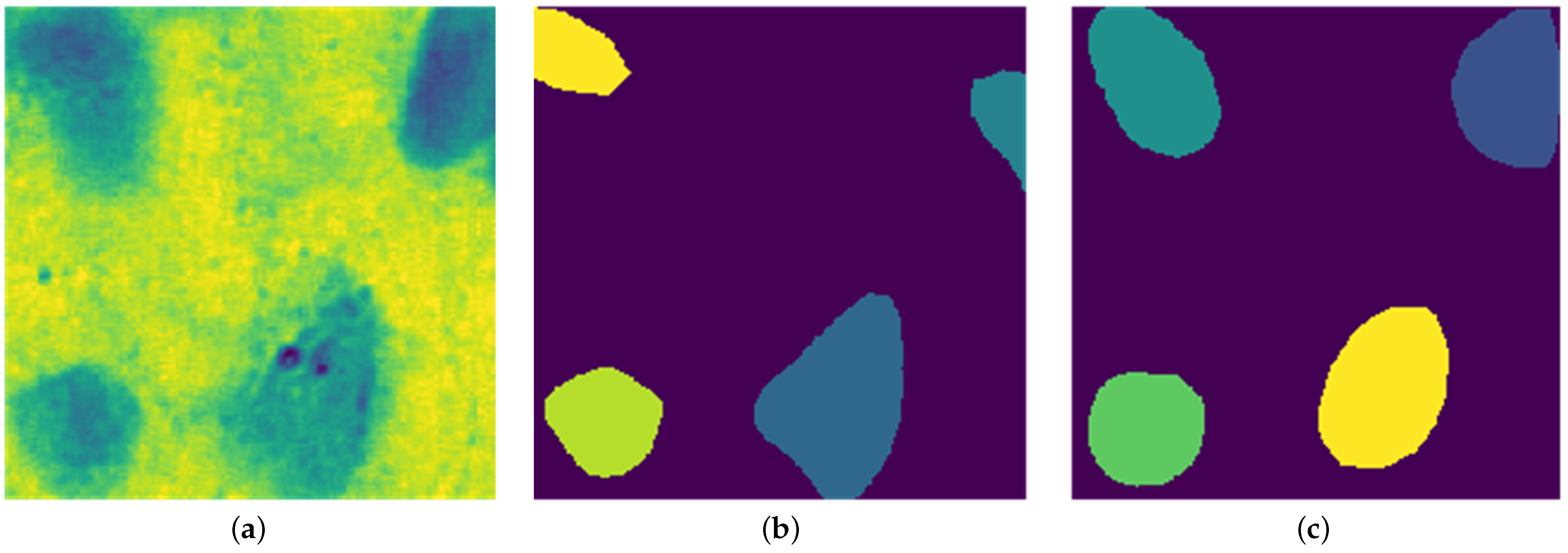
Figure 5 (bottom right) shows sample nuclei region predictions obtained from this model.
Post Processing
Post-processing refines Stardist predictions by removing extraneous small regions and dividing images into 256 × 256 patches, enhancing training efficiency and compatibility with Stardist’s settings.
Figure 7 shows the visual comparison between predicted nuclei and ground truth label patches after fine tuning.
5. Results and Discussion
This section presents the results of our proposed methodology for computational analysis and nuclei detection in HER2-SISH images. Given that this approach is novel, there is no direct comparison with existing methods; hence, we compare our results against expert-marked images provided by senior pathologists. A total of 20 images were marked by pathologists, following their established protocols for HER2 scoring from HER2-SISH stains. Our analysis, described in the following subsections, evaluates the results both visually and quantitatively, demonstrating that our proposed method effectively supports HER2-SISH image analysis.
Our framework is already integrated within the Cytomine platform, which is compatible with clinical digital pathology systems. Pathologists can select ROIs interactively, apply real-time segmentation, and incorporate the results into HER2 scoring workflows. This reduces diagnostic time and enhances reproducibility, making it feasible for deployment in clinical settings with digital infrastructure.
5.1. Objective Evaluation
Objective evaluation was conducted on
of the dataset, primarily due to the limited availability of expert-marked data and the large image sizes, ranging from 859 × 755 to approximately 5451 × 3136 pixels per ROI. Two evaluation approaches were used: one adhering to expert criteria and the other based on visual similarity to nuclei. The expert criteria included only nuclei regions with at least two red signals, non-overlapping, and non-border-touching, as singular nuclei are particularly suitable for HER2 quantification [
39]. The second approach considered any object resembling a nucleus, without additional constraints.
Table 4 presents visual nuclei detection counts based on visually identified regions, without strict expert constraints.
The evaluation metrics—recall, precision, and F1-score—had mean values of
,
, and
, respectively, for the test images (
Table 4). Individual metrics were computed for each test image, showing consistent model performance. For expert-marked, medically significant nuclei, recall, precision, and F1-score reached
,
, and
(
Table 5), indicating the model’s reliability for identifying critical nuclei. These results demonstrate a high concordance between model-detected nuclei and expert annotations, suggesting that our method is dependable for clinically relevant nuclei detection.
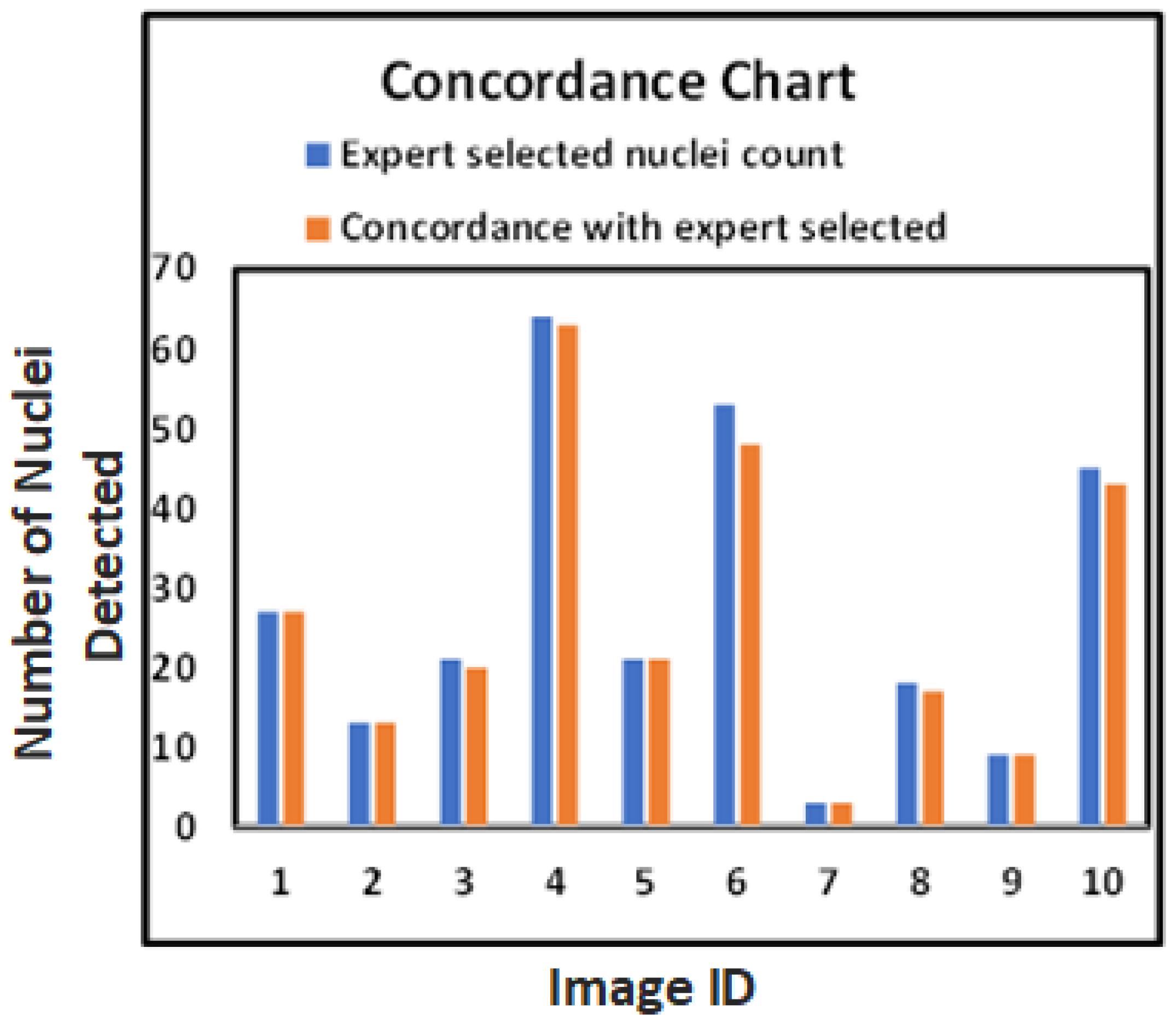
These results provide a foundation for reliable and reproducible HER2 quantification, as shown in
Table 4 and
Table 5, and the evaluation graph in
Figure 9, which highlights the close alignment of model-detected and expert-marked nuclei regions.
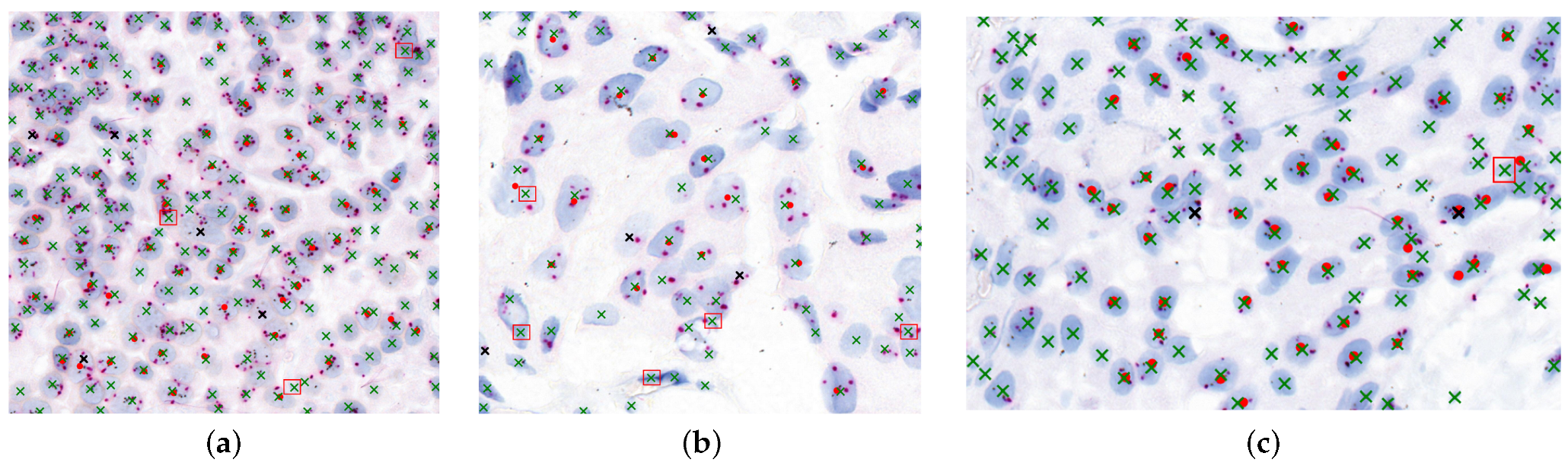
Figure 10 illustrates the agreement between model-detected nuclei and expert-marked regions essential for diagnosis. Red dots represent expert-targeted nuclei, green “x” marks are model-detected nuclei, black “x” marks indicate missed detections, and red boxes denote oversegmented regions.
The bar chart in
Figure 9 further confirms the close match between model-detected nuclei and expert annotations. Our method generalizes well across both visual and quantitative evaluations, even with limited annotated data, as demonstrated at the patch level in
Figure 7.
5.2. Comparison with Baseline Methods
To evaluate the effectiveness of our fine-tuned Stardist model, we compared it against two baseline approaches: (i) a conventional nuclei detection method based on marker-controlled watershed segmentation as proposed in our prior work [
40], and (ii) the pretrained Stardist model without fine tuning. Both baseline methods were applied to the same annotated dataset for consistency.
The fine-tuned Stardist model outperformed the other methods in terms of nuclei detection accuracy and F1-score, particularly in challenging regions with clustered or overlapping nuclei. This demonstrates the benefit of domain-specific fine tuning on expert-annotated SISH ROIs. The detailed quantitative results are summarized in
Table 6.
7. Conclusions
This study presents a robust and scalable framework for nuclei detection in HER2-SISH whole slide breast tissue images, leveraging expert-marked annotations for both HER2-amplified and non-amplified regions. By integrating a pretrained Stardist model with traditional image processing and fine tuning on a domain-specific dataset, the proposed approach addresses key challenges in SISH-stained nuclei segmentation, including morphological complexity, stain variability, and clustered nuclei.
Our method demonstrated superior performance over both the pretrained Stardist model and a conventional watershed segmentation approach from our previous work [
40]. Quantitative results confirm its effectiveness, achieving an average F1-score of
for visually assessed nuclei and
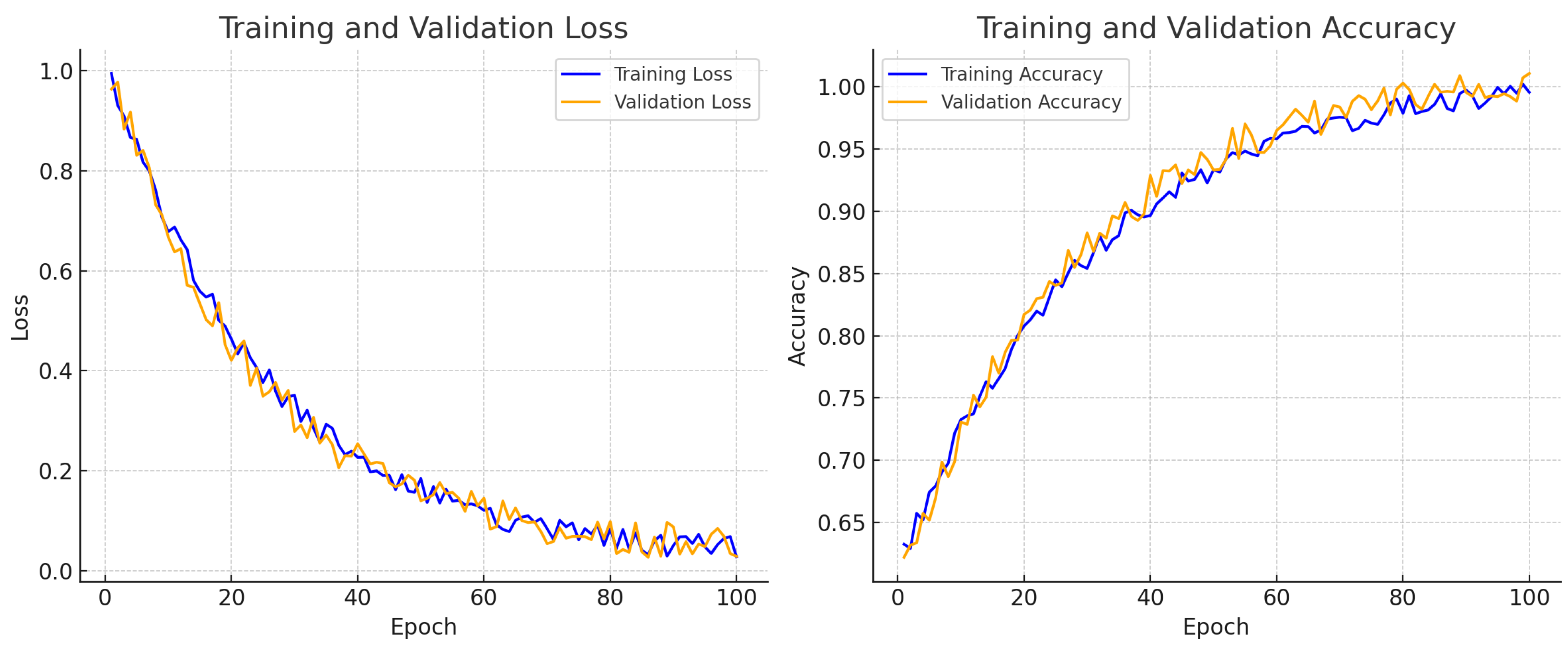
for expert-marked nuclei across 20 test images. These improvements were further supported by training–validation accuracy and loss curves, which illustrated stable convergence throughout the training process.
Currently in the testing phase, the framework has been integrated with the Cytomine platform to facilitate clinical deployment. This paves the way for real-time nuclei detection and HER2 scoring support within digital pathology workflows, offering greater consistency and efficiency for pathologists.
Future work will focus on expanding the dataset with more expert-annotated cases, enabling broader benchmarking and generalization. Additionally, we aim to extend the framework to include nuclei classification, signal quantification, and segmentation-based HER2 ratio calculation, contributing toward the automation of HER2 diagnostic scoring protocols.