Do LLMs Have ‘the Eye’ for MRI? Evaluating GPT-4o, Grok, and Gemini on Brain MRI Performance: First Evaluation of Grok in Medical Imaging and a Comparative Analysis
Abstract
1. Introduction
2. Materials and Methods
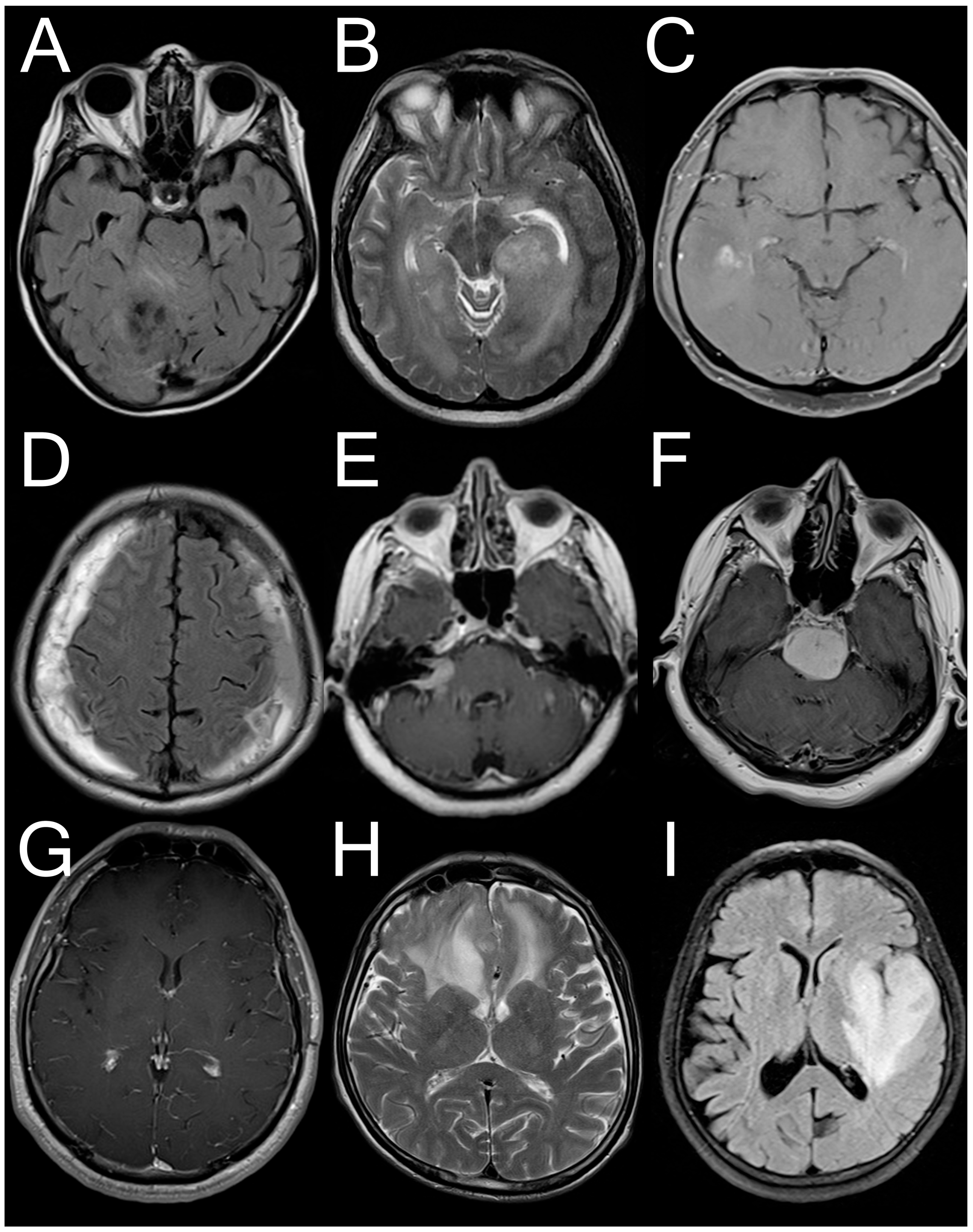
2.1. Dataset Used
2.2. Questioning LLMs
2.3. Statistical Analysis
3. Results
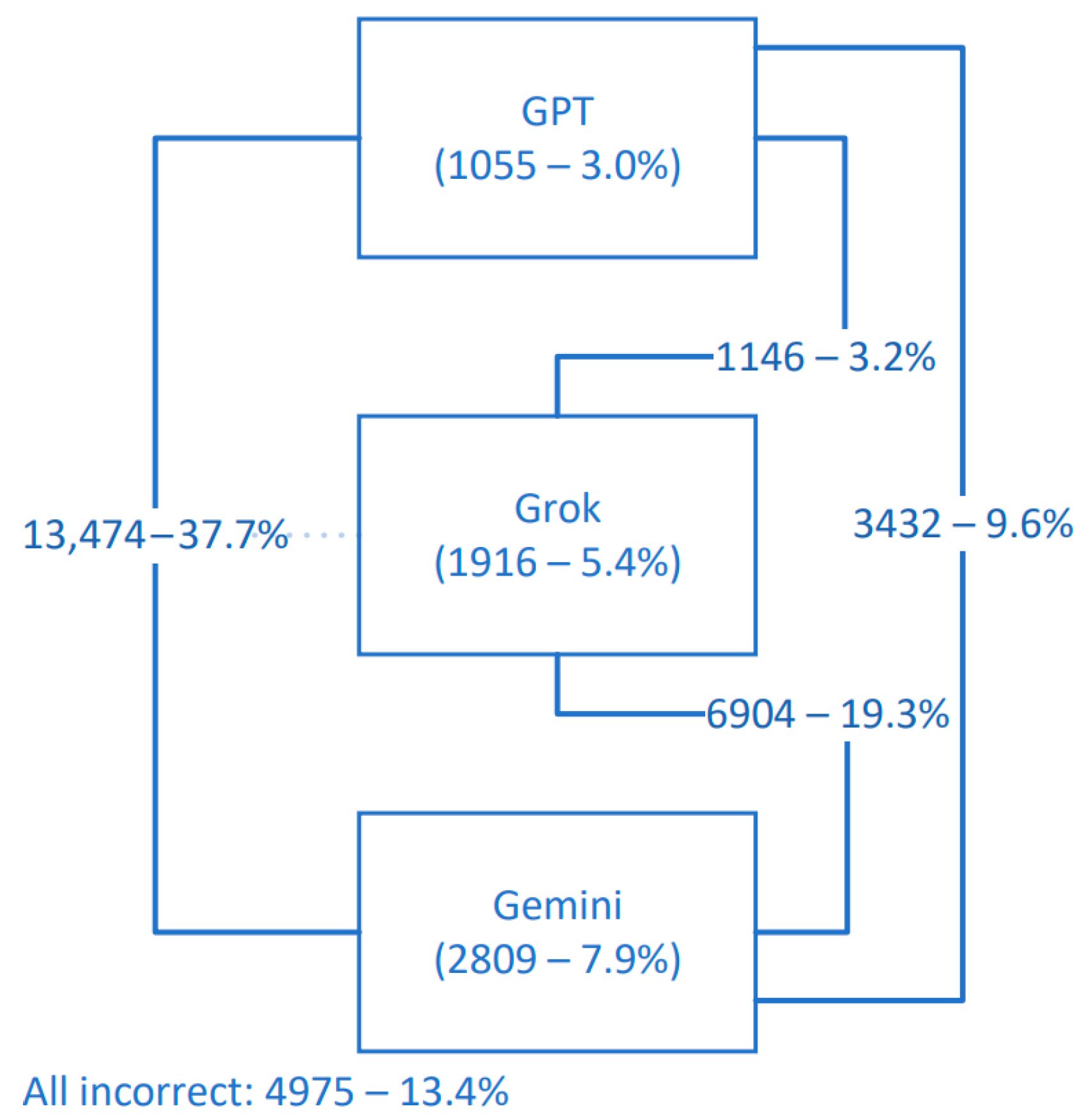
3.1. Dataset Descriptives and Answer Rates
3.2. Performance of GPT-4o
3.3. Performance of Grok
3.4. Performance of Gemini
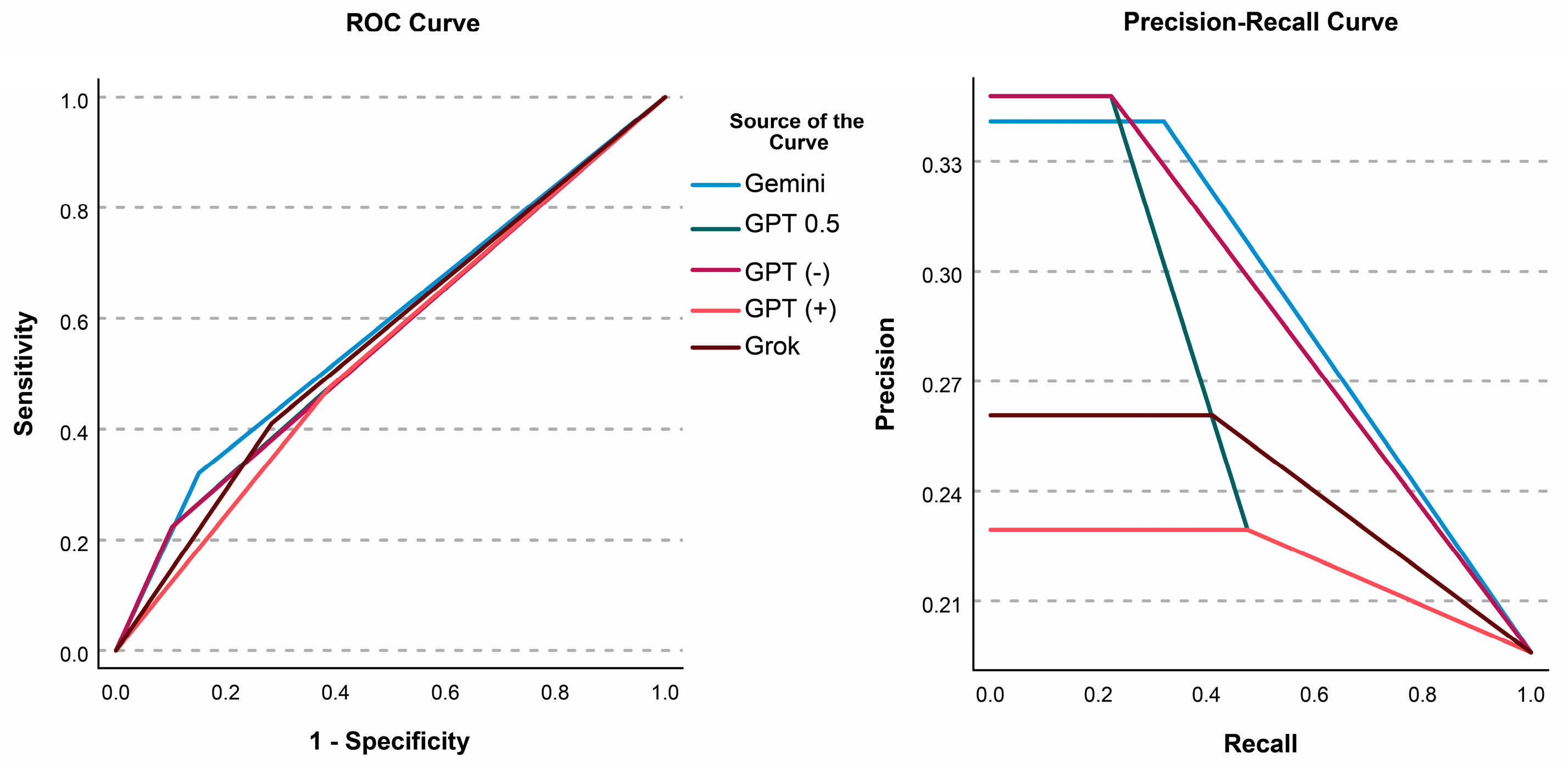
3.5. Comparing LLMs and ROC Analyses
3.6. Accuracy in Different Pathologies
3.7. Slice Difficulty
3.8. Sequence-Level Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LLM | Large Language Model |
| MRI | Magnetic Resonance Imaging/Image |
| AI | Artificial Intelligence |
| CNN | Convolutional Neural Network |
| ViT | Vision Transformer |
| FLAIR | Fluid-Attenuated Inversion Recovery |
| HGG | High Grade Glioma |
| LGG | Low Grade Glioma |
| SDK | Software Development Kit |
| OR | Odds Ratio |
| CI | Confidence Interval |
| PPV | Positive Predictive Value |
| NPV | Negative Predictive Value |
| ROC | Receiver Operating Characteristic |
| AUC | Area Under the Curve |
| ECG | Electrocardiogram |
| CT | Computerized Tomography |
Appendix A
- response = completions_with_backoff(
- # model = “gpt-4o”,
- # model = “grok-vision-beta”,
- # model = “gemini-1.5-pro”,
- messages= [
- {“role”: “system”, “content”: “You are a consultant neurosurgeon.”},
- {
- “role”: “user”,
- “content”: [
- {“type”: “text”, “text”: “You are given an axial MRI slice. Please evaluate whether there is any pathology. Your answer should be in the form of ‘true’ or ‘false,’ and you should also predict the MRI sequence of this slice. Format your answer as ‘‘True’ or ‘False’/MRI sequence’ without any additional descriptions. Possible MRI sequences for this task are ‘T1’, ‘T2’, ‘FLAIR’, ‘T1c’. You must provide an answer with the requested format.”},
- {
- “type”: “image_url”,
- “image_url”: {
- “url”: f”data:image/jpeg;base64,{base64_image}”,
- },
- },
- ],
- }
- ],
- max_tokens = 300,
- )
Appendix B
References
- Döhler, F.; Mormann, F.; Weber, B.; Elger, C.E.; Lehnertz, K. A Cellular Neural Network Based Method for Classification of Magnetic Resonance Images: Towards an Automated Detection of Hippocampal Sclerosis. J. Neurosci. Methods 2008, 170, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Ji, S.; Shen, D. Deep Convolutional Neural Networks for Multi-Modality Isointense Infant Brain Image Segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Aydoseli, A.; Unal, T.C.; Kardes, O.; Doguc, O.; Dolas, I.; Adiyaman, A.E.; Ortahisar, E.; Silahtaroglu, G.; Aras, Y.; Sabanci, P.A.; et al. An Early Warning System Using Machine Learning for the Detection of Intracranial Hematomas in the Emergency Trauma Setting. Turk. Neurosurg. 2022, 32, 459–465. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image Is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2021, arXiv:2010.11929v2. [Google Scholar]
- Hello GPT-4o. Available online: https://openai.com/index/hello-gpt-4o/ (accessed on 5 December 2024).
- xAI. Available online: https://x.ai/ (accessed on 5 December 2024).
- Introducing Gemini: Google’s Most Capable AI Model Yet. Available online: https://blog.google/technology/ai/google-gemini-ai/ (accessed on 5 December 2024).
- Sagiroglu, S.; Celtikci, E.; Demirezen, M.U.; Terzi, R.; Asan, A.; Celtikci, P.; Sozer, A.; Sahin, M.C.; Atici, B.M.; Ucar, M.; et al. Gazi Brains 2020 Dataset 2020. Available online: https://www.synapse.org/Synapse:syn25926092 (accessed on 2 May 2025).
- Demirezen, M.U.; Terzi, R.; Aşan, A.; Çeltikçi, E.; Sağiroğlu, Ş.; Börcek, A.Ö.; Atici, B.M. Beyin Görüntülerindeki Aykırılığın veya Anomalilerin Zeki Bir Sistem ile Otomatik Tespiti ve Bildirim Sistemi. Patient TR2021009196A1, 21 December 2022. [Google Scholar]
- OPENAPI Initative. Available online: https://learn.openapis.org/ (accessed on 5 December 2024).
- Base64—Base16, Base32, Base64, Base85 Data Encodings. Available online: https://docs.python.org/3/library/base64.html (accessed on 1 December 2024).
- Taha, A.A.; Hanbury, A. Metrics for Evaluating 3D Medical Image Segmentation: Analysis, Selection, and Tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Improving Model Safety Behavior with Rule-Based Rewards. Available online: https://openai.com/index/improving-model-safety-behavior-with-rule-based-rewards/ (accessed on 13 December 2024).
- Günay, S.; Öztürk, A.; Yiğit, Y. The Accuracy of Gemini, GPT-4, and GPT-4o in ECG Analysis: A Comparison with Cardiologists and Emergency Medicine Specialists. Am. J. Emerg. Med. 2024, 84, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Hindy, J.-R.; Souaid, T.; Kovacs, C.S. Capabilities of GPT-4o and Gemini 1.5 Pro in Gram Stain and Bacterial Shape Identification. Future Microbiol. 2024, 19, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Hamid, A.M.; Jhaveri, M.; Patel, N.; Suthar, P.P. Comparative Evaluation of AI Models Such as ChatGPT 3.5, ChatGPT 4.0, and Google Gemini in Neuroradiology Diagnostics. Cureus 2024, 16, e67766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Ma, Z.; Gong, R.; Lian, L.; Li, Y.; He, Z.; Han, Y.; Hui, J.; Huang, J.; Jiang, J.; et al. Using Natural Language Processing (GPT-4) for Computed Tomography Image Analysis of Cerebral Hemorrhages in Radiology: Retrospective Analysis. J. Med. Internet Res. 2024, 26, e58741. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Sun, W.; Mi, D.; Jin, S.; Wu, X.; Xin, B.; Zhang, H.; Wang, Y.; Sun, X.; He, X. Artificial Intelligence Diagnosing of Oral Lichen Planus: A Comparative Study. Bioengineering 2024, 11, 1159. [Google Scholar] [CrossRef] [PubMed]
- Brin, D.; Sorin, V.; Barash, Y.; Konen, E.; Glicksberg, B.S.; Nadkarni, G.N.; Klang, E. Assessing GPT-4 Multimodal Performance in Radiological Image Analysis. Eur. Radiol. 2025, 35, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Strotzer, Q.D.; Nieberle, F.; Kupke, L.S.; Napodano, G.; Muertz, A.K.; Meiler, S.; Einspieler, I.; Rennert, J.; Strotzer, M.; Wiesinger, I.; et al. Toward Foundation Models in Radiology? Quantitative Assessment of GPT-4V’s Multimodal and Multianatomic Region Capabilities. Radiology 2024, 313, e240955. [Google Scholar] [CrossRef] [PubMed]
- Asiri, A.A.; Shaf, A.; Ali, T.; Shakeel, U.; Irfan, M.; Mehdar, K.M.; Halawani, H.T.; Alghamdi, A.H.; Alshamrani, A.F.A.; Alqhtani, S.M. Exploring the Power of Deep Learning: Fine-Tuned Vision Transformer for Accurate and Efficient Brain Tumor Detection in MRI Scans. Diagnostics 2023, 13, 2094. [Google Scholar] [CrossRef] [PubMed]
- Ali Al-Hamza, K. ViT-BT: Improving MRI Brain Tumor Classification Using Vision Transformer with Transfer Learning. Int. J. Soft Comput. Eng. 2024, 14, 16–26. [Google Scholar] [CrossRef]
| Origin Patient | Sequence | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FLAIR (n = 9511, 26.63%) | T2 (n = 9347, 26.17%) | T1 (n = 9343, 26.16%) | T1c (n = 7510, 21.03%) | Total (n = 35,711) | ||||||
| Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | Positive | Negative | |
| Normal | 0 | 1854 (100%) | 0 | 1819 (100%) | 0 | 1819 (100%) | 0 | 429 (100%) | 0 | 5921 (100%) |
| HGG | 499 (40.54%) | 732 (59.46%) | 499 (40.94%) | 720 (59.06%) | 499 (41.%) | 718 (59.%) | 499 (41.07%) | 716 (58.93%) | 1996 (40.88%) | 2886 (59.12%) |
| LGG | 116 (20.21%) | 458 (79.79%) | 116 (20.42%) | 452 (79.58%) | 116 (20.42%) | 452 (79.58%) | 115 (20.32%) | 451 (79.68%) | 463 (20.34%) | 1813 (79.66%) |
| Meningioma | 577 (20.88%) | 2187 (79.12%) | 577 (21.27%) | 2136 (78.73%) | 577 (21.27%) | 2136 (78.73%) | 572 (21.72%) | 2061 (78.28%) | 2303 (21.28%) | 8520 (78.72%) |
| Schwannoma | 102 (14.55%) | 599 (85.45%) | 102 (15.04%) | 576 (84.96%) | 102 (14.96%) | 580 (85.04%) | 102 (14.96%) | 580 (85.04%) | 408 (14.87%) | 2335 (85.13%) |
| Cavernoma | 128 (12.46%) | 899 (87.54%) | 128 (12.62%) | 886 (87.38%) | 128 (12.67%) | 882 (87.33%) | 97 (12.29%) | 692 (87.71%) | 481 (12.53%) | 3359 (87.47%) |
| Metastatic Disease | 130 (34.39%) | 248 (65.61%) | 130 (34.57%) | 246 (65.43%) | 130 (34.76%) | 244 (65.24%) | 130 (34.67%) | 245 (65.33%) | 520 (34.6%) | 983 (65.4%) |
| Other | 84 (24.78%) | 255 (75.22%) | 84 (25.07%) | 251 (74.93%) | 84 (25.07%) | 251 (74.93%) | 71 (24.15%) | 223 (75.85%) | 323 (24.79%) | 980 (75.21%) |
| Post-op | 130 (20.22%) | 513 (79.78%) | 129 (20.64%) | 496 (79.36%) | 129 (20.64%) | 496 (79.36%) | 115 (21.82%) | 412 (78.18%) | 503 (20.79%) | 1917 (79.21%) |
| TOTAL | 1766 (18.57%) | 7745 (81.43%) | 1765 (18.88%) | 7582 (81.12%) | 1765 (18.89%) | 7578 (81.11%) | 1701 (22.65%) | 5809 (77.35%) | 6997 (19.59%) | 28,714 (80.41%) |
| TP | TN | Sensitivity | Specificity | LR+ | DOR | F1 (DICE) | Accuracy | Sequence Accuracy | |
|---|---|---|---|---|---|---|---|---|---|
| FP | FN | PPV | NPV | LR− | F0.5 | F2 | |||
| FLAIR | 643 (8.32%) | 4720 (61.11%) | 0.428 | 0.759 | 1.776 | 2.355 | 0.353 | 69.43% | 5652/7724 (73.17%) |
| 1501 (19.43%) | 860 (11.13%) | 0.300 | 0.846 | 0.754 | 0.319 | 0.394 | |||
| T2 | 354 (5.80%) | 4334 (70.98%) | 0.291 | 0.887 | 2.575 | 3.223 | 0.333 | 76.78% | 5469/6106 (89.57%) |
| 554 (9.07%) | 864 (14.15%) | 0.390 | 0.834 | 0.799 | 0.365 | 0.307 | |||
| T1 | 129 (1.92%) | 5189 (77.14%) | 0.100 | 0.955 | 2.222 | 2.359 | 0.155 | 79.05% | 6727/7285 (92.34%) 1 |
| 242 (3.60%) | 1167 (17.35%) | 0.348 | 0.816 | 0.942 | 0.233 | 0.117 | |||
| T1c | 435 (8.45%) | 3303 (64.16%) | 0.358 | 0.840 | 2.238 | 2.929 | 0.382 | 72.61% | 4733/5148 (91.94%) 1 |
| 629 (12.22%) | 781 (15.17%) | 0.409 | 0.809 | 0.764 | 0.398 | 0.367 | |||
| TOTAL | 1561 (6.07%) | 17,546 (68.26%) | 0.298 | 0.857 | 2.084 | 2.545 | 0.321 | 74.33% | 22,581/26,263 (85.98%) |
| 2926 (11.38%) | 3672 (14.29%) | 0.348 | 0.827 | 0.819 | 0.337 | 0.307 |
| TP | TN | Sensitivity | Specificity | LR+ | DOR | F1 (DICE) | Accuracy | Sequence Accuracy | |
|---|---|---|---|---|---|---|---|---|---|
| FP | FN | PPV | NPV | LR− | F0.5 | F2 | |||
| FLAIR | 847 (8.91%) | 5388 (56.65%) | 0.480 | 0.696 | 1.579 | 2.114 | 0.341 | 65.56% | 1610/9511 (16.93%) |
| 2357 (24.78%) | 919 (9.66%) | 0.264 | 0.854 | 0.747 | 0.290 | 0.413 | |||
| T2 | 762 (8.15%) | 5432 (58.11%) | 0.432 | 0.716 | 1.521 | 1.918 | 0.326 | 66.27% | 7314/9347 (78.25%) |
| 2150 (23.00%) | 1003 (10.73%) | 0.262 | 0.844 | 0.793 | 0.284 | 0.382 | |||
| T1 | 505 (5.41%) | 5852 (62.64%) | 0.286 | 0.772 | 1.254 | 1.356 | 0.252 | 68.04% | 8231/9343 (88.10%) 1 |
| 1726 (18.47%) | 1260 (13.49%) | 0.226 | 0.823 | 0.925 | 0.236 | 0.272 | |||
| T1c | 759 (10.11%) | 3895 (51.86%) | 0.446 | 0.671 | 1.356 | 1.642 | 0.347 | 61.97% | 6495/7510 (86.48%) 1 |
| 1914 (25.49%) | 942 (12.54%) | 0.284 | 0.805 | 0.826 | 0.306 | 0.400 | |||
| TOTAL | 2873 (8.05%) | 20,567 (57.59%) | 0.411 | 0.716 | 1.447 | 1.758 | 0.319 | 65.64% | 23,650/35,711 (66.23%) |
| 8147 (22.81%) | 4124 (11.55%) | 0.261 | 0.833 | 0.823 | 0.282 | 0.369 |
| TP | TN | Sensitivity | Specificity | LR+ | DOR | F1 (DICE) | Accuracy | Sequence Accuracy | |
|---|---|---|---|---|---|---|---|---|---|
| FP | FN | PPV | NPV | LR− | F0.5 | F2 | |||
| FLAIR | 606 (6.37%) | 6688 (70.32%) | 0.343 | 0.864 | 2.522 | 3.318 | 0.353 | 76.69% | 2135/9511 (22.45%) |
| 1057 (11.11%) | 1160 (12.20%) | 0.364 | 0.852 | 0.760 | 0.360 | 0.347 | |||
| T2 | 572 (6.12%) | 6658 (71.23%) | 0.324 | 0.878 | 2.656 | 3.449 | 0.351 | 77.35% | 1340/9347 (14.34%) |
| 924 (9.89%) | 1193 (12.76%) | 0.382 | 0.848 | 0.770 | 0.369 | 0.334 | |||
| T1 | 396 (4.24%) | 6557 (70.18%) | 0.224 | 0.865 | 1.659 | 1.849 | 0.248 | 74.42% | 7618/9343 (81.54%) 1 |
| 1021 (10.93%) | 1369 (14.65%) | 0.279 | 0.827 | 0.897 | 0.266 | 0.233 | |||
| T1c | 669 (8.91%) | 4473 (59.56%) | 0.393 | 0.770 | 1.709 | 2.169 | 0.361 | 68.47% | 5468/7510 (72.81%) 1 |
| 1336 (17.79%) | 1032 (13.74%) | 0.334 | 0.813 | 0.788 | 0.344 | 0.380 | |||
| TOTAL | 2243 (6.28%) | 24,376 (68.26%) | 0.321 | 0.849 | 2.126 | 2.658 | 0.331 | 74.54% | 16,561/35,711 (46.38%) |
| 4338 (12.15%) | 4754 (13.31%) | 0.341 | 0.837 | 0.800 | 0.337 | 0.325 |
| Cochran’s Q | GPT vs. Grok | GPT vs. Gemini | Grok vs. Gemini | LLM Order | |
|---|---|---|---|---|---|
| FLAIR | χ2(2) = 4863.681 p < 0.001 | χ2(1) = 3365.557, p < 0.001 | χ2(1) = 2566.381, p < 0.001 | χ2(1) = 122.088, p < 0.001 | GPT > Gemini > Grok |
| T2 | χ2(2) = 7041.433 p < 0.001 | χ2(1) = 1031.342, p < 0.001 | χ2(1) = 2924.384, p < 0.001 | χ2(1) = 5229.658, p < 0.001 | Grok > GPT > Gemini |
| T1 | χ2(2) = 1588.105 p < 0.001 | χ2(1) = 1358.836, p < 0.001 | χ2(1) = 628.697, p < 0.001 | χ2(1) = 186.248, p < 0.001 | Grok > Gemini > GPT |
| T1c | χ2(2) = 1156.113 p < 0.001 | χ2(1) = 1146.864, p < 0.001 | χ2(1) = 171.306, p < 0.001 | χ2(1) = 461.498, p < 0.001 | Grok > Gemini > GPT |
| TOTAL | χ2(2) = 196,901.084 p < 0.001 | χ2(1) = 189.132, p < 0.001 | χ2(1) = 1741.563, p < 0.001 | χ2(1) = 3759.616, p < 0.001 | Grok > GPT > Gemini |
| AUC (95%CI) | vs. Gemini | vs. Grok | vs. GPT 0.5 | vs. GPT (−) | |
|---|---|---|---|---|---|
| Gemini | 58.5% (57.9–59.1%) | ||||
| Grok | 56.3% (55.7–57.0%) | z = 6.462, p < 0.001 * | |||
| GPT 0.5 | 56.2% (55.5–56.9%) | z = 5.950, p < 0.001 * | z = 0.262, p = 0.793 | ||
| GPT (−) | 56.1% (55.5–56.6%) | z = 8.427, p < 0.001 * | z = 0.867, p = 0.386 | z = 0.733, p = 0.463 | |
| GPT (+) | 54.3% (53.7–55.0%) | z = 10.841, p < 0.001 * | z = 5.004, p < 0.001 * | z = 20.593, p < 0.001 * | z = 5.952, p < 0.001 * |
| Pathology | Number of Positive Slices | GPT 1 Accuracy | Grok Accuracy | Gemini Accuracy | GPT Accuracy 2 (n) |
|---|---|---|---|---|---|
| HGG | 1996 | 39.8% | 57.7% | 51.4% | 48.9% (1625) |
| LGG | 463 | 13.8% | 30.0% | 21.0% | 18.8% (341) |
| Meningioma | 2303 | 17.5% | 36.1% | 26.3% | 24.6% (1644) |
| Schwannoma | 408 | 10.8% | 25.7% | 15.0% | 14.4% (305) |
| Cavernoma | 481 | 12.9% | 29.1% | 16.2% | 17.4% (356) |
| Metastatic Disease | 520 | 20.0% | 42.1% | 31.5% | 26.8% (388) |
| Other | 323 | 8.7% | 34.7% | 27.9% | 13.5% (207) |
| Post-op | 503 | 11.9% | 34.8% | 24.1% | 16.4% (366) |
| TOTAL | 6997 | 22.3% | 41.1% | 32.1% | 29.8% (5232) |
| Subgroup | Number of Sequences | GPT Accuracy | Grok Accuracy | Gemini Accuracy |
|---|---|---|---|---|
| Normal | 299 | 42.5% | 0.3% | 31.1% |
| HGG | 236 | 92.8% | 100.0% | 97.9% |
| LGG | 112 | 69.6% | 92.9% | 86.6% |
| Meningioma | 548 | 70.6% | 98.9% | 85.6% |
| Schwannoma | 140 | 67.9% | 98.6% | 86.4% |
| Cavernoma | 193 | 70.5% | 99.0% | 77.7% |
| Metastatic Disease | 72 | 79.2% | 100.0% | 91.7% |
| Other | 66 | 74.2% | 98.5% | 87.9% |
| Post-op | 127 | 66.1% | 94.5% | 85.8% |
| TOTAL | 1793 | 68.7% | 81.9% | 77.7% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sozer, A.; Sahin, M.C.; Sozer, B.; Erol, G.; Tufek, O.Y.; Nernekli, K.; Demirtas, Z.; Celtikci, E. Do LLMs Have ‘the Eye’ for MRI? Evaluating GPT-4o, Grok, and Gemini on Brain MRI Performance: First Evaluation of Grok in Medical Imaging and a Comparative Analysis. Diagnostics 2025, 15, 1320. https://doi.org/10.3390/diagnostics15111320
Sozer A, Sahin MC, Sozer B, Erol G, Tufek OY, Nernekli K, Demirtas Z, Celtikci E. Do LLMs Have ‘the Eye’ for MRI? Evaluating GPT-4o, Grok, and Gemini on Brain MRI Performance: First Evaluation of Grok in Medical Imaging and a Comparative Analysis. Diagnostics. 2025; 15(11):1320. https://doi.org/10.3390/diagnostics15111320
Chicago/Turabian StyleSozer, Alperen, Mustafa Caglar Sahin, Batuhan Sozer, Gokberk Erol, Ozan Yavuz Tufek, Kerem Nernekli, Zuhal Demirtas, and Emrah Celtikci. 2025. "Do LLMs Have ‘the Eye’ for MRI? Evaluating GPT-4o, Grok, and Gemini on Brain MRI Performance: First Evaluation of Grok in Medical Imaging and a Comparative Analysis" Diagnostics 15, no. 11: 1320. https://doi.org/10.3390/diagnostics15111320
APA StyleSozer, A., Sahin, M. C., Sozer, B., Erol, G., Tufek, O. Y., Nernekli, K., Demirtas, Z., & Celtikci, E. (2025). Do LLMs Have ‘the Eye’ for MRI? Evaluating GPT-4o, Grok, and Gemini on Brain MRI Performance: First Evaluation of Grok in Medical Imaging and a Comparative Analysis. Diagnostics, 15(11), 1320. https://doi.org/10.3390/diagnostics15111320








