Feasible and Rapid Screening of IDH1/2 and FLT3-TKD2 Mutations by High-Resolution Melting for Patients with Acute Myeloid Leukemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Samples
2.2. DNA Extraction
2.3. PCR-HRM
2.4. Analytical Performance of PCR-HRM
2.5. Sanger Sequencing
2.6. Next Generation Sequencing (NGS)
2.7. Turnaround Times
3. Results
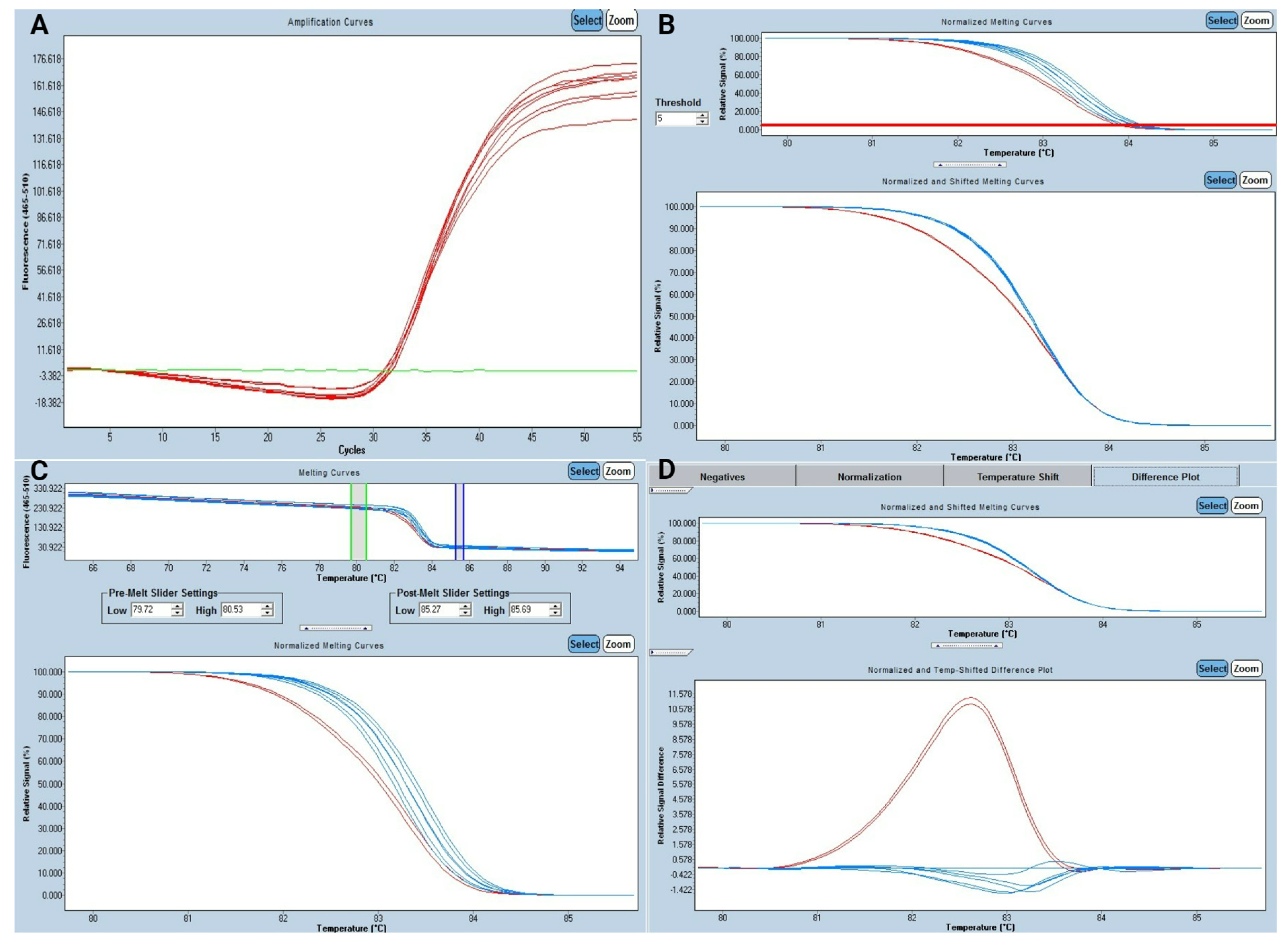
3.1. Study of High-Resolution Melting
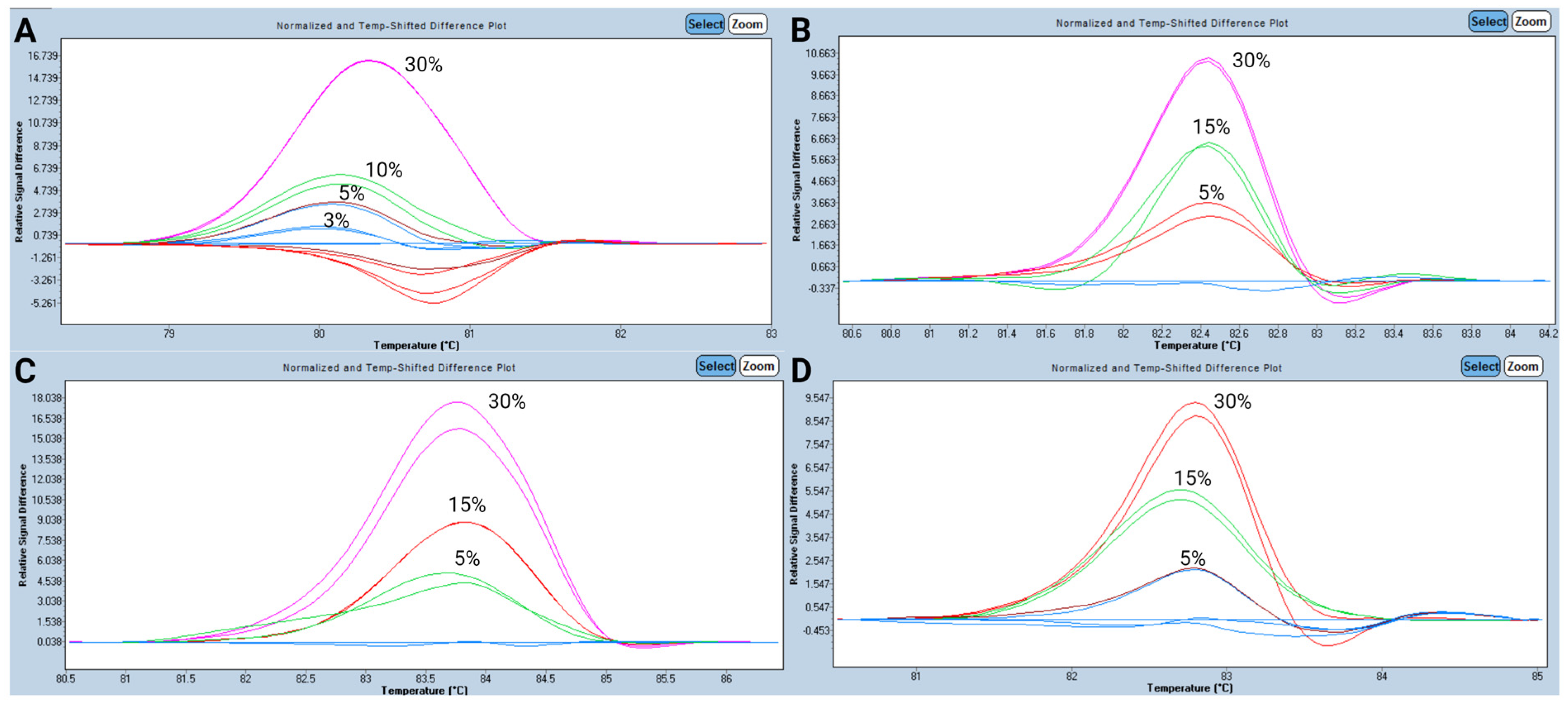
3.2. Analytical Performance for HRM Analysis
3.3. Comparison of HRM with Sequencing Studies
3.4. Turnaround Times
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Short, N.J.; Konopleva, M.; Kadia, T.M.; Borthakur, G.; Ravandi, F.; DiNardo, C.D.; Daver, N. Advances in the Treatment of Acute Myeloid Leukemia: New Drugs and New Challenges. Cancer Discov. 2020, 10, 506–525. [Google Scholar] [CrossRef] [PubMed]
- Döhner, H.; Wei, A.H.; Appelbaum, F.R.; Craddock, C.; DiNardo, C.D.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Godley, L.A.; Hasserjian, R.P.; et al. Diagnosis and management of AML in adults: 2022 Recommendations from an international expert panel on behalf of the ELN. Blood 2022, 140, 1345–1377. [Google Scholar] [CrossRef]
- Yu, J.; Jiang, P.Y.Z.; Sun, H.; Zhang, X.; Jiang, Z.; Li, Y.; Song, Y. Advances in targeted therapy for acute myeloid leukemia. Biomark. Res. 2020, 8, 1–11. [Google Scholar] [CrossRef]
- Bewersdorf, J.P.; Shallis, R.M.; Derkach, A.; Goldberg, A.D.; Stein, A.; Stein, E.M.; Marcucci, G.; Zeidan, A.M.; Shimony, S.; DeAngelo, D.J.; et al. Efficacy of FLT3 and IDH1/2 inhibitors in patients with acute myeloid leukemia previously treated with venetoclax. Leuk. Res. 2022, 122, 106942. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Liu, Y.; Cai, S.J.; Qian, M.; Ding, J.; Larion, M.; Gilbert, M.R.; Yang, C. IDH mutation in glioma: Molecular mechanisms and potential therapeutic targets. Br. J. Cancer 2020, 122, 1580–1589. [Google Scholar] [CrossRef]
- Bill, M.; Jentzsch, M.; Bischof, L.; Kohlschmidt, J.; Grimm, J.; Schmalbrock, L.K.; Backhaus, D.; Brauer, D.; Goldmann, K.; Franke, G.-N.; et al. Impact ofIDH1andIDH2mutation detection at diagnosis and in remission in patients with AML receiving allogeneic transplantation. Blood Adv. 2022, 7, 436–444. [Google Scholar] [CrossRef]
- Libura, M.; Bialopiotrowicz, E.; Giebel, S.; Wierzbowska, A.; Roboz, G.J.; Piatkowska-Jakubas, B.; Pawelczyk, M.; Gorniak, P.; Borg, K.; Wojtas, M.; et al. IDH2 mutations in patients with normal karyotype AML predict favorable responses to daunorubicin, cytarabine and cladribine regimen. Sci. Rep. 2021, 11, 10017. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Xu, R.; Yang, Y. Acute myeloid leukemia treatment outcomes with isocitrate dehydrogenase mutations: A systematic review and meta-analysis. Medicine 2024, 103, e40565. [Google Scholar] [CrossRef]
- Tomuleasa, C.; Tigu, A.-B.; Munteanu, R.; Moldovan, C.-S.; Kegyes, D.; Onaciu, A.; Gulei, D.; Ghiaur, G.; Einsele, H.; Croce, C.M. Therapeutic advances of targeting receptor tyrosine kinases in cancer. Signal Transduct. Target. Ther. 2024, 9, 201. [Google Scholar] [CrossRef]
- Chen, Y.; Zou, Z.; Găman, M.-A.; Xu, L.; Li, J. NADPH oxidase mediated oxidative stress signaling in FLT3-ITD acute myeloid leukemia. Cell Death Discov. 2023, 9, 208. [Google Scholar] [CrossRef]
- Marensi, V.; Keeshan, K.R.; MacEwan, D.J. Pharmacological impact of FLT3 mutations on receptor activity and responsiveness to tyrosine kinase inhibitors. Biochem. Pharmacol. 2020, 183, 114348. [Google Scholar] [CrossRef] [PubMed]
- Daver, N.; Schlenk, R.F.; Russell, N.H.; Levis, M.J. Targeting FLT3 mutations in AML: Review of current knowledge and evidence. Leukemia 2019, 33, 299–312. [Google Scholar] [CrossRef]
- Levis, M.; Perl, A.E. Gilteritinib: Potent targeting of FLT3 mutations in AML. Blood Adv. 2020, 4, 1178–1191. [Google Scholar] [CrossRef] [PubMed]
- Saadi, M.I.; Zarei, T.; Ramzi, M.; Arandi, N. Mutation of the DNMT3A and IDH1/2 genes in Iranian acute myeloid leukemia patients with normal karyotype (CN-AML): Association with other gene mutation and clinical and laboratory characteristics. J. Hematop. 2018, 11, 29–36. [Google Scholar] [CrossRef]
- Goh, W.C.; Idris, B.; Kandasamy, R.; Shamsuddin, S.; Jaafar, H.; Ahmad, F. Pcr-rflp method enhance dna sequencing of idh1 somatic mutations detection in gliomas. Gulhane Med. J. 2019, 61, 167. [Google Scholar] [CrossRef]
- Sheffield, B.S.; Eaton, K.; Emond, B.; Lafeuille, M.-H.; Hilts, A.; Lefebvre, P.; Morrison, L.; Stevens, A.L.; Ewara, E.M.; Cheema, P. Cost Savings of Expedited Care with Upfront Next-Generation Sequencing Testing versus Single-Gene Testing among Patients with Metastatic Non-Small Cell Lung Cancer Based on Current Canadian Practices. Curr. Oncol. 2023, 30, 2348–2365. [Google Scholar] [CrossRef]
- Bestvina, C.M.; Waters, D.; Morrison, L.; Emond, B.; Lafeuille, M.-H.; Hilts, A.; Lefebvre, P.; He, A.; Vanderpoel, J. Cost of genetic testing, delayed care, and suboptimal treatment associated with polymerase chain reaction versus next-generation sequencing biomarker testing for genomic alterations in metastatic non-small cell lung cancer. J. Med. Econ. 2024, 27, 292–303. [Google Scholar] [CrossRef]
- Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. Blood 2016, 128, 462–463. [CrossRef] [PubMed]
- Sargas, C.; Ayala, R.; Chillón, M.C.; Larráyoz, M.J.; Carrillo-Cruz, E.; Bilbao, C.; Yébenes-Ramírez, M.; Llop, M.; Rapado, I.; García-Sanz, R.; et al. Networking for advanced molecular diagnosis in acute myeloid leukemia patients is possible: The PETHEMA NGS-AML project. Haematologica 2020, 106, 3079–3089. [Google Scholar] [CrossRef]
- Grimwade, D.; Hills, R.K.; Moorman, A.V.; Walker, H.; Chatters, S.; Goldstone, A.H.; Wheatley, K.; Harrison, C.J.; Burnett, A.K. Refinement of cytogenetic classification in acute myeloid leukemia: Determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood 2010, 116, 354–365. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Abdel-Wahab, O.; Guglielmelli, P.; Patel, J.; Caramazza, D.; Pieri, L.; Finke, C.M.; Kilpivaara, O.; Wadleigh, M.; et al. IDH1 and IDH2 mutation studies in 1473 patients with chronic-, fibrotic- or blast-phase essential thrombocythemia, polycythemia vera or myelofibrosis. Leukemia 2010, 24, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, Y.; Kiyoi, H.; Nakano, Y.; Suzuki, R.; Kodera, Y.; Miyawaki, S.; Asou, N.; Kuriyama, K.; Yagasaki, F.; Shimazaki, C.; et al. Activating mutation of D835 within the activation loop of FLT3 in human hematologic malignancies. Blood 2001, 97, 2434–2439. [Google Scholar] [CrossRef]
- Gibson, N.J. The use of real-time PCR methods in DNA sequence variation analysis. Clin. Chim. Acta 2005, 363, 32–47. [Google Scholar] [CrossRef]
- Furtado, L.V.; Weigelin, H.C.; Elenitoba-Johnson, K.S.J.; Betz, B.L. Detection of MPL Mutations by a Novel Allele-Specific PCR-Based Strategy. J. Mol. Diagn. 2013, 15, 810–818. [Google Scholar] [CrossRef]
- Warshawsky, I.; Mularo, F. Locked nucleic acid probes for enhanced detection of FLT3 D835/I836, JAK2 V617F and NPM1 mutations. J. Clin. Pathol. 2011, 64, 905–910. [Google Scholar] [CrossRef]
- Patel, K.P.; Barkoh, B.A.; Chen, Z.; Ma, D.; Reddy, N.; Medeiros, L.J.; Luthra, R. Diagnostic testing for IDH1 and IDH2 variants in acute myeloid leukemia. J. Mol. Diagn. 2011, 13, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, A.; Moradabadi, A.; Noroozi-Aghideh, A. Comparison of high-resolution melting analysis with direct sequencing for detection of FLT3-TKD, FLT3-ITD and WT1 mutations in acute myeloid leukemia. Cancer Treat. Res. Commun. 2021, 28, 100432. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Pandzic, T.; Ladenvall, C.; Engvall, M.; Mattsson, M.; Hermanson, M.; Cavelier, L.; Ljungström, V.; Baliakas, P. Five Percent Variant Allele Frequency Is a Reliable Reporting Threshold for TP53 Variants Detected by Next Generation Sequencing in Chronic Lymphocytic Leukemia in the Clinical Setting. HemaSphere 2022, 6, e761. [Google Scholar] [CrossRef]

| Gene | Exon | Nº of Patients | Nucleotide Change | Mutation Detected * | VAF (%) a |
|---|---|---|---|---|---|
| IDH1 | 4 | 2 | CGT>TGT | p.Arg132Cys | 2.40% |
| CGT>TGT | p.Arg132Cys | 1.80% | |||
| IDH2 | 4 | 3 | CGG>CAG | p.Arg140Gln | 3.20% |
| CGG>CAG | p.Arg140Gln | 3.10% | |||
| AGG>AAG | p.Arg172Lys | 4.00% | |||
| FLT3 | 20 | 7 | GAT>TAT | p.Asp835Tyr | 2.50% |
| GAT>TAT | p.Asp835Tyr | 3.00% | |||
| GAT>TAT | p.Asp835Tyr | 1.40% | |||
| GAT>TAT | p.Asp835Tyr | 4.70% | |||
| GAT>CAT | p.Asp835His | 3.90% | |||
| delGATATC | p.Asp835_Ile836del | 4.20% | |||
| AAC>AAA | p.Asn841Lys | 2.20% |
| Method | Gene | PPA, %[TP/(TP+FN)] | 95% CI, % | NPA, %[TN/(TN+FP)] |
|---|---|---|---|---|
| HRM vs. NGS | IDH1 | 98.32[117/117+2] | 97.64–99 | 100[1244/1244+0] |
| IDH2 | 98.25[168/168+3] | 97.41–98.82 | 100[1244/1244+0] | |
| FLT3 | 91.76[78/78+7] | 90.33–93.19 | 100[1341/1341+0] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gil, J.V.; de las Heras, S.; Miralles, A.; Sargas, C.; Llop, M.; Rodríguez-Veiga, R.; Torres-Miñana, L.; Boluda, B.; Cano-Ferri, I.; Acuña-Cruz, E.; et al. Feasible and Rapid Screening of IDH1/2 and FLT3-TKD2 Mutations by High-Resolution Melting for Patients with Acute Myeloid Leukemia. Diagnostics 2025, 15, 1230. https://doi.org/10.3390/diagnostics15101230
Gil JV, de las Heras S, Miralles A, Sargas C, Llop M, Rodríguez-Veiga R, Torres-Miñana L, Boluda B, Cano-Ferri I, Acuña-Cruz E, et al. Feasible and Rapid Screening of IDH1/2 and FLT3-TKD2 Mutations by High-Resolution Melting for Patients with Acute Myeloid Leukemia. Diagnostics. 2025; 15(10):1230. https://doi.org/10.3390/diagnostics15101230
Chicago/Turabian StyleGil, José Vicente, Sandra de las Heras, Alberto Miralles, Claudia Sargas, Marta Llop, Rebeca Rodríguez-Veiga, Laura Torres-Miñana, Blanca Boluda, Isabel Cano-Ferri, Evelyn Acuña-Cruz, and et al. 2025. "Feasible and Rapid Screening of IDH1/2 and FLT3-TKD2 Mutations by High-Resolution Melting for Patients with Acute Myeloid Leukemia" Diagnostics 15, no. 10: 1230. https://doi.org/10.3390/diagnostics15101230
APA StyleGil, J. V., de las Heras, S., Miralles, A., Sargas, C., Llop, M., Rodríguez-Veiga, R., Torres-Miñana, L., Boluda, B., Cano-Ferri, I., Acuña-Cruz, E., Navarro, I., Lloret-Madrid, P., Montesinos, P., & Barragán, E. (2025). Feasible and Rapid Screening of IDH1/2 and FLT3-TKD2 Mutations by High-Resolution Melting for Patients with Acute Myeloid Leukemia. Diagnostics, 15(10), 1230. https://doi.org/10.3390/diagnostics15101230






