Menstrual Blood as a Diagnostic Specimen for Human Papillomavirus Genotyping and Genital Tract Infection Using Next-Generation Sequencing as a Novel Diagnostic Tool
Abstract
1. Introduction
2. Method and Materials
2.1. Patients
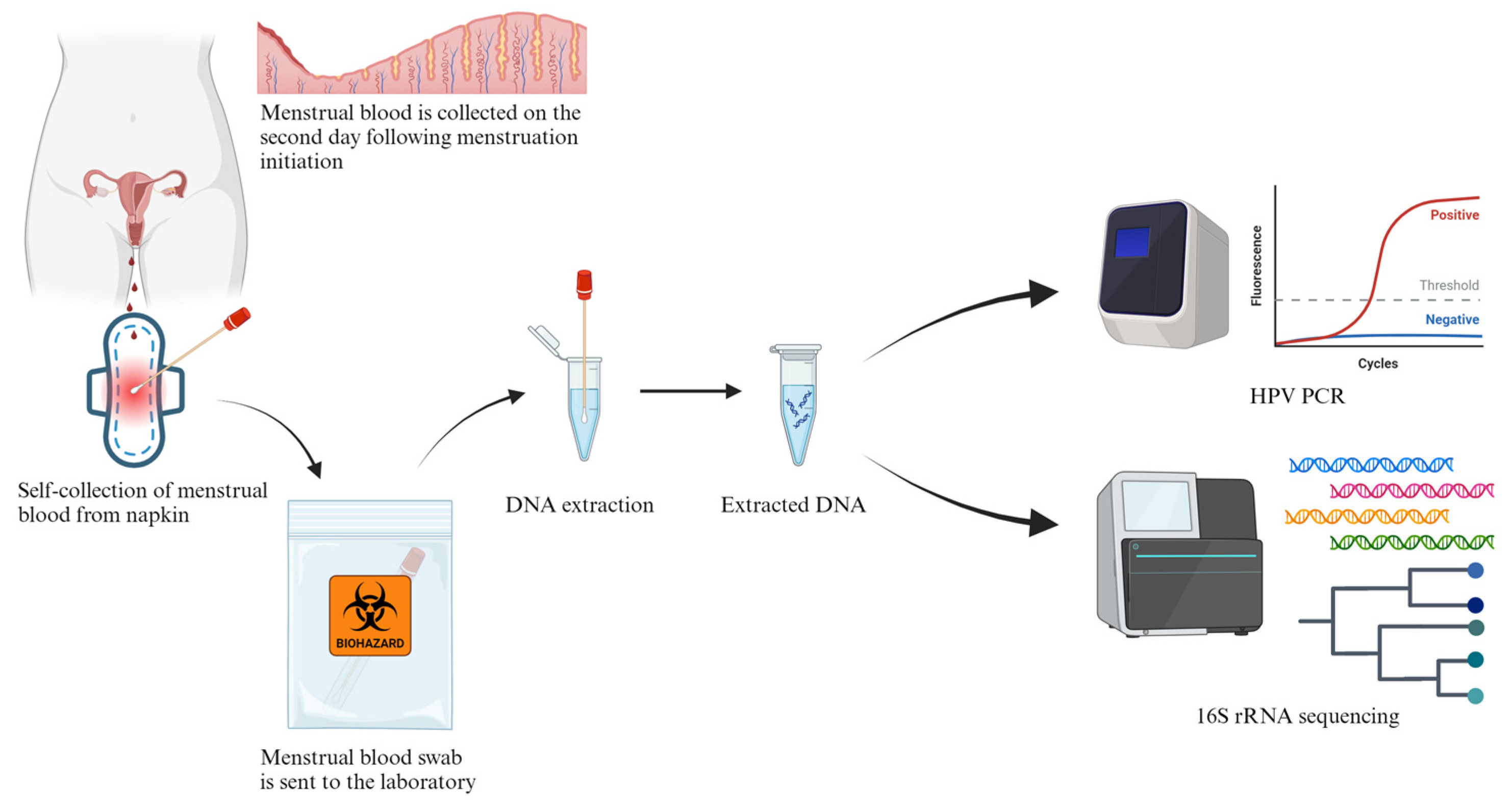
2.2. MB Collection and DNA Extraction
2.3. PacBio 16S rRNA HiFi Long-Read Sequencing
2.4. 16S rRNA Gene Analysis
2.5. Menstrual Blood HPV PCR
3. Results
3.1. Patients’ Characteristics
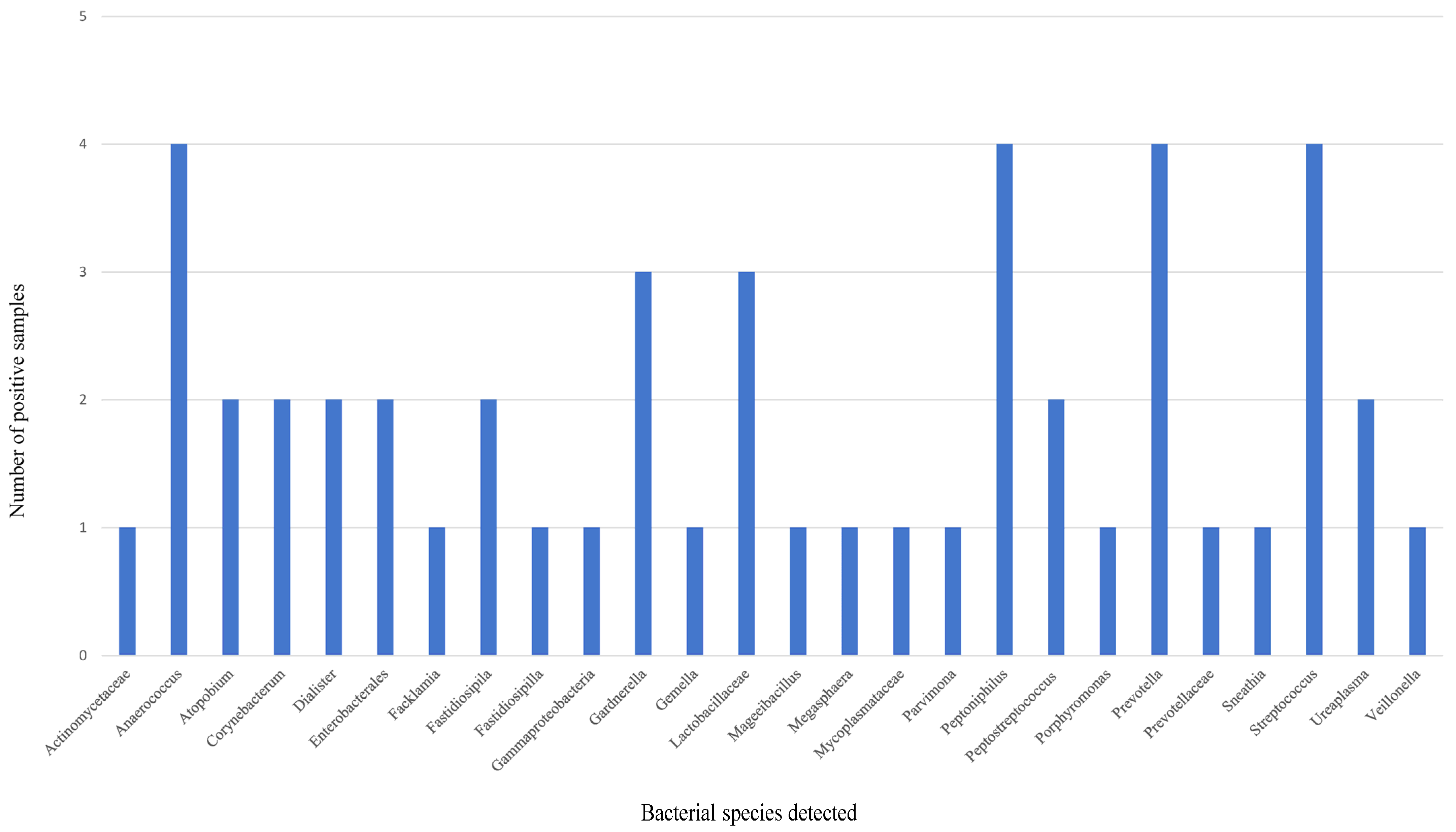
3.2. mNGS Analysis of MB
3.3. HPV DNA Detection of MB
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ngalame, A.N.; Mubiana-Mbewe, M.; Dionne, J.A. Genital Tract Infections in Women, Pregnancy and Neonates. Obstet. Gynecol. Clin. N. Am. 2022, 49, 751–769. [Google Scholar] [CrossRef]
- Volkova, L.V.; Pashov, A.I.; Omelchuk, N.N. Cervical Carcinoma: Oncobiology and Biomarkers. Int. J. Mol. Sci. 2021, 22, 12571. [Google Scholar] [CrossRef]
- Adolfsson, A.; Granevik, K.; Paulson, K. The reasons why women do not participate in the papsmear screening and testing program in Sweden. Adv. Sex. Med. 2012, 2, 31–37. [Google Scholar] [CrossRef][Green Version]
- Lv, H.; Hu, Y.; Cui, Z.; Jia, H. Human menstrual blood: A renewable and sustainable source of stem cells for regenerative medicine. Stem Cell Res. Ther. 2018, 9, 325. [Google Scholar] [CrossRef]
- Chakravarti, P.; Maheshwari, A.; Tahlan, S.; Kadam, P.; Bagal, S.; Gore, S.; Panse, N.; Deodhar, K.; Chaturvedi, P.; Dikshit, R.; et al. Diagnostic accuracy of menstrual blood for human papillomavirus detection in cervical cancer screening: A systematic review. Ecancermedicalscience 2022, 16, 1427. [Google Scholar] [CrossRef]
- Wong, S.C.C.; Au, T.C.C.; Chan, S.C.S.; Ng, L.P.W.; Tsang, H.F. Menstrual Blood Human Papillomavirus DNA and TAP1 Gene Polymorphisms as Potential Biomarkers for Screening and Monitoring of Cervical Squamous Intraepithelial Lesion. J. Infect. Dis. 2018, 218, 1739–1745. [Google Scholar] [CrossRef]
- Wong, S.C.C.; Au, T.C.C.; Chan, S.C.S.; Chan, C.M.L.; Lam, M.Y.Y.; Zee, B.C.Y.; Pong, W.M.; Chan, A.T.C. Human papillomavirus DNA detection in menstrual blood from patients with cervical intraepithelial neoplasia and condyloma acuminatum. J. Clin. Microbiol. 2010, 48, 709–713. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.M.; Yeung, M.H.Y.; Wong, A.N.N.; Tsang, H.F.; Yu, A.C.S.; Yim, A.K.Y.; Wong, S.C.C. Targeted Sequencing Approach and Its Clinical Applications for the Molecular Diagnosis of Human Diseases. Cells 2023, 12, 493. [Google Scholar] [CrossRef] [PubMed]
- Tsang, H.F.; Yu, A.C.S.; Jin, N.; Yim, A.K.Y.; Leung, W.M.S.; Lam, K.W.; Cho, W.C.S.; Chiou, J.; Wong, S.C.C. The clinical application of metagenomic next-generation sequencing for detecting pathogens in bronchoalveolar lavage fluid: Case reports and literature review. Expert Rev. Mol. Diagn. 2022, 22, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857, Erratum in Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014, 42, D633–D642. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4680–4687. [Google Scholar] [CrossRef]
- Burmeister, C.A.; Khan, S.F.; Schäfer, G.; Mbatani, N.; Adams, T.; Moodley, J.; Prince, S. Cervical cancer therapies: Current challenges and future perspectives. Tumour Virus Res. 2022, 13, 200238. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Molina-Pineda, A.; López-Cardona, M.G.; Limón-Toledo, L.P.; Cantón-Romero, J.C.; Martínez-Silva, M.G.; Ramos-Sánchez, H.V.; Flores-Miramontes, M.G.; de la Mata-González, P.; Jave-Suárez, L.F.; Aguilar-Lemarroy, A. High frequency of HPV genotypes 59, 66, 52, 51, 39 and 56 in women from Western Mexico. BMC Infect. Dis. 2020, 20, 889. [Google Scholar] [CrossRef]
- Peng, Q.; Wang, L.; Zuo, L.; Gao, S.; Jiang, X.; Han, Y.; Lin, J.; Peng, M.; Wu, N.; Tang, Y.; et al. HPV E6/E7: Insights into their regulatory role and mechanism in signaling pathways in HPV-associated tumor. Cancer Gene Ther. 2024, 31, 9–17. [Google Scholar] [CrossRef]
- Johnson, C.A.; James, D.; Marzan, A.; Armaos, M. Cervical Cancer: An Overview of Pathophysiology and Management. Semin. Oncol. Nurs. 2019, 35, 166–174. [Google Scholar] [CrossRef]
- Wood, J.; Stoltzfus, K.C.; Popalis, M.; Moss, J.L. Perspectives on Self-Sampling for Cancer Screening From Staff at Federally Qualified Health Centers in Rural and Segregated Counties: A Preliminary Qualitative Study. Cancer Control 2022, 29, 10732748221102819. [Google Scholar] [CrossRef]
- Chee, W.J.Y.; Chew, S.Y.; Than, L.T.L. Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb. Cell Fact. 2020, 19, 203. [Google Scholar] [CrossRef]
- Zheng, N.; Guo, R.; Wang, J.; Zhou, W.; Ling, Z. Contribution of Lactobacillus iners to Vaginal Health and Diseases: A Systematic Review. Front. Cell Infect. Microbiol. 2021, 11, 792787. [Google Scholar] [CrossRef]
- Boskey, E.R.; Cone, R.A.; Whaley, K.J.; Moench, T.R. Origins of vaginal acidity: High D/L lactate ratio is consistent with bacteria being the primary source. Hum. Reprod. 2001, 16, 1809–1813. [Google Scholar] [CrossRef]
- O’Hanlon, D.E.; Come, R.A.; Moench, T.R. Vaginal pH measured in vivo: Lactobacilli determine pH and lactic acid concentration. BMC Microbiol. 2019, 19, 13. [Google Scholar] [CrossRef]
- Mitra, A.; MacIntyre, D.A.; Marchesi, J.R.; Lee, Y.S.; Bennett, P.R.; Kyrgiou, M. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: What do we know and where are we going next? Microbiome 2016, 4, 58. [Google Scholar] [CrossRef]
- Shannon, B.; Gajer, P.; Yi, T.J.; Ma, B.; Humphrys, M.S.; Thomas-Pavanel, J.; Chieza, L.; Janakiram, P.; Saunders, M.; Tharao, W.; et al. Distinct Effects of the Cervicovaginal Microbiota and Herpes Simplex Type 2 Infection on Female Genital Tract Immunology. J. Infect. Dis. 2017, 215, 1366–1375. [Google Scholar] [CrossRef]
- Eastment, M.C.; McClelland, R.S. Vaginal microbiota and susceptibility to HIV. AIDS 2018, 32, 687–698. [Google Scholar] [CrossRef]
- Sharifian, K.; Shoja, Z.; Jalilvand, S. The interplay between human papillomavirus and vaginal microbiota in cervical cancer development. Virol. J. 2023, 20, 73. [Google Scholar] [CrossRef]
- Gajer, P.; Brotman, R.M.; Bai, G.; Sakamoto, J.; Schütte, U.M.; Zhong, X.; Koenig, S.S.; Fu, L.; Ma, Z.; Zhou, X.; et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012, 4, 132ra52. [Google Scholar] [CrossRef]
- Lee, J.E.; Lee, S.; Lee, H.; Song, Y.M.; Lee, K.; Han, M.J.; Sung, J.; Ko, G. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS ONE 2013, 8, e63514. [Google Scholar] [CrossRef]
- Yu, J.; Zhou, Y.; Luo, H.; Su, X.; Gan, T.; Wang, J.; Ye, Z.; Deng, Z.; He, J. Mycoplasma genitalium infection in the female reproductive system: Diseases and treatment. Front. Microbiol. 2023, 14, 1098276. [Google Scholar] [CrossRef]
- Naicker, M.; Dessai, F.; Singh, R.; Mitchev, N.; Tinarwo, P.; Abbai, N.S. Mycoplasma hominis does not share common risk factors with other genital pathogens: Findings from a South African pregnant cohort. S. Afr. J. Infect. Dis. 2021, 36, 207. [Google Scholar] [CrossRef]
- Yuan, X.Y.; Liu, H.Z.; Liu, J.F.; Sun, Y.; Song, Y. Pathogenic mechanism, detection methods and clinical significance of group B Streptococcus. Future Microbiol. 2021, 16, 671–685. [Google Scholar] [CrossRef]
- Rabe, L.K.; Winterscheid, K.K.; Hillier, S.L. Association of viridans group streptococci from pregnant women with bacterial vaginosis and upper genital tract infection. J. Clin. Microbiol. 1988, 26, 1156–1160. [Google Scholar] [CrossRef]
- Viet, N.T.; Van Du, V.; Thuan, N.D.; Van Tong, H.; Toan, N.L.; Van Mao, C.; Van Tuan, N.; Pallerla, S.R.; Nurjadi, D.; Velavan, T.P.; et al. Maternal Vaginal Colonization and Extended-Spectrum Beta-Lactamase-Producing Bacteria in Vietnamese Pregnant Women. Antibiotics 2021, 10, 572. [Google Scholar] [CrossRef]
- Flokas, M.E.; Karanika, S.; Alevizakos, M.; Mylonakis, E. Prevalence of ESBL-Producing Enterobacteriaceae in Pediatric Bloodstream Infections: A Systematic Review and Meta-Analysis. PLoS ONE 2017, 12, e0171216. [Google Scholar] [CrossRef]
- Dramowski, A.; Cotton, M.F.; Rabie, H.; Whitelaw, A. Trends in paediatric bloodstream infections at a South African referral hospital. BMC Pediatr. 2015, 15, 33. [Google Scholar] [CrossRef]
- Weinroth, M.D.; Belk, A.D.; Dean, C.; Noyes, N.; Dittoe, D.K.; Rothrock, M.J., Jr.; Ricke, S.C.; Myer, P.R.; Henniger, M.T.; Ramírez, G.A.; et al. Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. J. Anim. Sci. 2022, 100, skab346. [Google Scholar] [CrossRef]
- Zhao, N.; Cao, J.; Xu, J.; Liu, B.; Liu, B.; Chen, D.; Xia, B.; Chen, L.; Zhang, W.; Zhang, Y.; et al. Targeting RNA with Next- and Third-Generation Sequencing Improves Pathogen Identification in Clinical Samples. Adv. Sci. 2021, 8, e2102593. [Google Scholar] [CrossRef]
- Ma, Z.; Gharizadeh, B.; Cai, X.; Li, M.; Fellner, M.D.; Basiletti, J.A.; Correa, R.M.; Colucci, M.C.; Baldoni, G.; Vacchino, M.; et al. A comprehensive HPV-STI NGS assay for detection of 29 HPV types and 14 non-HPV sexually transmitted infections. Infect. Agent. Cancer 2022, 17, 9. [Google Scholar] [CrossRef]
- Basiletti, J.A.; Valls, J.; Poklépovich, T.; Fellner, M.D.; Rol, M.; Alonso, R.; Correa, R.M.; Colucci, M.C.; Rodriguez de la Pena, M.; Falabella, P.G.; et al. Human papillomavirus genotyping using next generation sequencing (NGS) in cervical lesions: Genotypes by histologic grade and their relative proportion in multiple infections. PLoS ONE 2022, 17, e0278117. [Google Scholar] [CrossRef]
- Arroyo Mühr, L.S.; Guerendiain, D.; Cuschieri, K.; Sundström, K. Human Papillomavirus Detection by Whole-Genome Next-Generation Sequencing: Importance of Validation and Quality Assurance Procedures. Viruses 2021, 13, 1323. [Google Scholar] [CrossRef] [PubMed]
- Budukh, A.; Palayekar, V.; Maheshwari, A.; Deodhar, K.; Purwar, P.; Bagal, S.; Vadigoppula, A.; Lokhande, M.; Panse, N.; Dikshit, R.; et al. Menstrual pad, a cervical cancer screening tool, a population-based study in rural India. Eur. J. Cancer Prev. 2018, 27, 546–552. [Google Scholar] [CrossRef]
- Torres-Rojas, F.I.; Mendoza-Catalán, M.A.; Alarcón-Romero, L.D.C.; Parra-Rojas, I.; Paredes-Solís, S.; Leyva-Vázquez, M.A.; Cortes-Arciniega, J.E.; Bracamontes-Benítez, C.J.; Illades-Aguiar, B. HPV molecular detection from urine versus cervical samples: An alternative for HPV screening in indigenous populations. PeerJ 2021, 9, e11564. [Google Scholar] [CrossRef]
- Téblick, L.; Van Keer, S.; De Smet, A.; Van Damme, P.; Laeremans, M.; Rios Cortes, A.; Beyers, K.; Vankerckhoven, V.; Matheeussen, V.; Mandersloot, R.; et al. Impact of Collection Volume and DNA Extraction Method on the Detection of Biomarkers and HPV DNA in First-Void Urine. Molecules 2021, 26, 1989. [Google Scholar] [CrossRef]
- Poljak, M.; Cuschieri, K.; Alemany, L.; Vorsters, A. Testing for Human Papillomaviruses in Urine, Blood, and Oral Specimens: An Update for the Laboratory. J. Clin. Microbiol. 2023, 61, e0140322. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Luo, Z.Y.; Lin, F.; Li, L.J.; Lu, M.; Xie, L.X.; Yang, L.Y. Comparison of Urine and Genital Samples for Detecting Human Papillomavirus (HPV) in Clinical Patients. Obstet. Gynecol. Int. 2023, 2023, 7483783. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | Number of Reads | Number of Bases (bp) | N50 Read Length (bp) | Median Read Quality (Phred) |
|---|---|---|---|---|
| ST04 | 551,743 | 823,059,442 | 1491 | 42.1 |
| ST06 | 78,529 | 117,584,092 | 1489 | 43 |
| ST08 | 370,588 | 557,280,881 | 1503 | 39.9 |
| ST10 | 357,940 | 541,838,371 | 1512 | 39.1 |
| ST12 | 295,312 | 446,999,092 | 1523 | 41.8 |
| ST13 | 47,963 | 72,772,473 | 1532 | 43.9 |
| Sample ID | Age | Cytological Diagnosis | Histological Diagnosis | HPV PCR Results in Pap Smear | HPV DNA Detected in MB | Bacterial DNA Detected in MB by mNGS |
|---|---|---|---|---|---|---|
| ST04 | 34 | ASC-US | CIN1 | High-risk HPV DNA detected | HPV 16 and 18 DNA detected | Sneathia (19%); Prevotella (18%); Gardnerella (7%); Fastidiosipila (6%); Megasphaera (6%); Parvimonas (5%); Atopobium (4%); Mageeibacillus (2%); Peptoniphilus (2%); Porphyromonas (2%); Dialister (2%); Gemella (2%); Peptostreptococcus (1%); Actinomycetaceae (0.7%) |
| ST06 | 45 | ASC-US | CIN1/2 HPV infection | High-risk HPV DNA detected | HPV DNA not detected | Gardnerella (46%); Anaerococcus (16%); Prevotella (7%); Peptoniphilus (4%); Dialister (3%); Peptostreptococcus (2%); Atopobium (2%); Streptococcus (1%); Mycoplasmataceae (0.7%) |
| ST08 | 43 | ASC-H | CIN2 | Not done | HPV DNA not detected | Gammaproteobacteria (36%); Enterobacterales (11%); Corynebacterum (6%); Fastidiosipilla (5%); Anaerococcus (0.8%); Facklamia (1%) |
| ST10 | 33 | LSIL | HPV infection | Not done | High-risk HPV DNA detected (not 16 & 18) | Anaerococcus (6%); Streptococcus (5%); Lactobacillaceae (3%); Prevotellaceae (3%); Corynebacterum (2%); Peptoniphilus (1%); Prevotella (1%) |
| ST12 | 39 | LSIL ASC-H | CIN3 HPV infection | High-risk HPV DNA not detected | HPV 16 DNA detected | Veillonella (33%); Peptoniphilus (6%); Streptococcus (5%); Lactobacillaceae (2%); Anaerococcus (2%); Prevotella (2%); Corynebacterum (2%); Ureaplasma (1%); Enterobacterales (1%) |
| ST13 | 45 | LSIL | CIN1 HPV infection | High-risk HPV DNA detected | High-risk HPV DNA detected (not 16 and 18) | Gardnerella (37%); Lactobacillaceae (11%); Streptococcus (2%); Ureaplasma (2%); |
| HPV DNA Detected in MB | HPV DNA Not Detected in MB | Sensitivity | PPV | ||
|---|---|---|---|---|---|
| HPV PCR using Pap smear | HPV DNA detected | 2 a | 1 b | 66.7% | 66.7% |
| HPV DNA not detected | 1 c | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsang, H.-F.; Cheung, Y.-S.; Yu, C.-S.A.; Chan, C.-S.S.; Wong, C.-B.T.; Yim, K.-Y.A.; Pei, X.; Wong, S.-C.C. Menstrual Blood as a Diagnostic Specimen for Human Papillomavirus Genotyping and Genital Tract Infection Using Next-Generation Sequencing as a Novel Diagnostic Tool. Diagnostics 2024, 14, 686. https://doi.org/10.3390/diagnostics14070686
Tsang H-F, Cheung Y-S, Yu C-SA, Chan C-SS, Wong C-BT, Yim K-YA, Pei X, Wong S-CC. Menstrual Blood as a Diagnostic Specimen for Human Papillomavirus Genotyping and Genital Tract Infection Using Next-Generation Sequencing as a Novel Diagnostic Tool. Diagnostics. 2024; 14(7):686. https://doi.org/10.3390/diagnostics14070686
Chicago/Turabian StyleTsang, Hin-Fung, Yui-Shing Cheung, Chi-Shing Allen Yu, Chung-Sum Sammy Chan, Chi-Bun Thomas Wong, Kay-Yuen Aldrin Yim, Xiaomeng Pei, and Sze-Chuen Cesar Wong. 2024. "Menstrual Blood as a Diagnostic Specimen for Human Papillomavirus Genotyping and Genital Tract Infection Using Next-Generation Sequencing as a Novel Diagnostic Tool" Diagnostics 14, no. 7: 686. https://doi.org/10.3390/diagnostics14070686
APA StyleTsang, H.-F., Cheung, Y.-S., Yu, C.-S. A., Chan, C.-S. S., Wong, C.-B. T., Yim, K.-Y. A., Pei, X., & Wong, S.-C. C. (2024). Menstrual Blood as a Diagnostic Specimen for Human Papillomavirus Genotyping and Genital Tract Infection Using Next-Generation Sequencing as a Novel Diagnostic Tool. Diagnostics, 14(7), 686. https://doi.org/10.3390/diagnostics14070686







