Enhancing Precision in HIV Treatment: Validation of a Robust Next-Generation Sequencing System for Drug Resistance Mutation Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Dideoxynucleoside Sanger Sequencing
2.3. NGS and Data Analysis
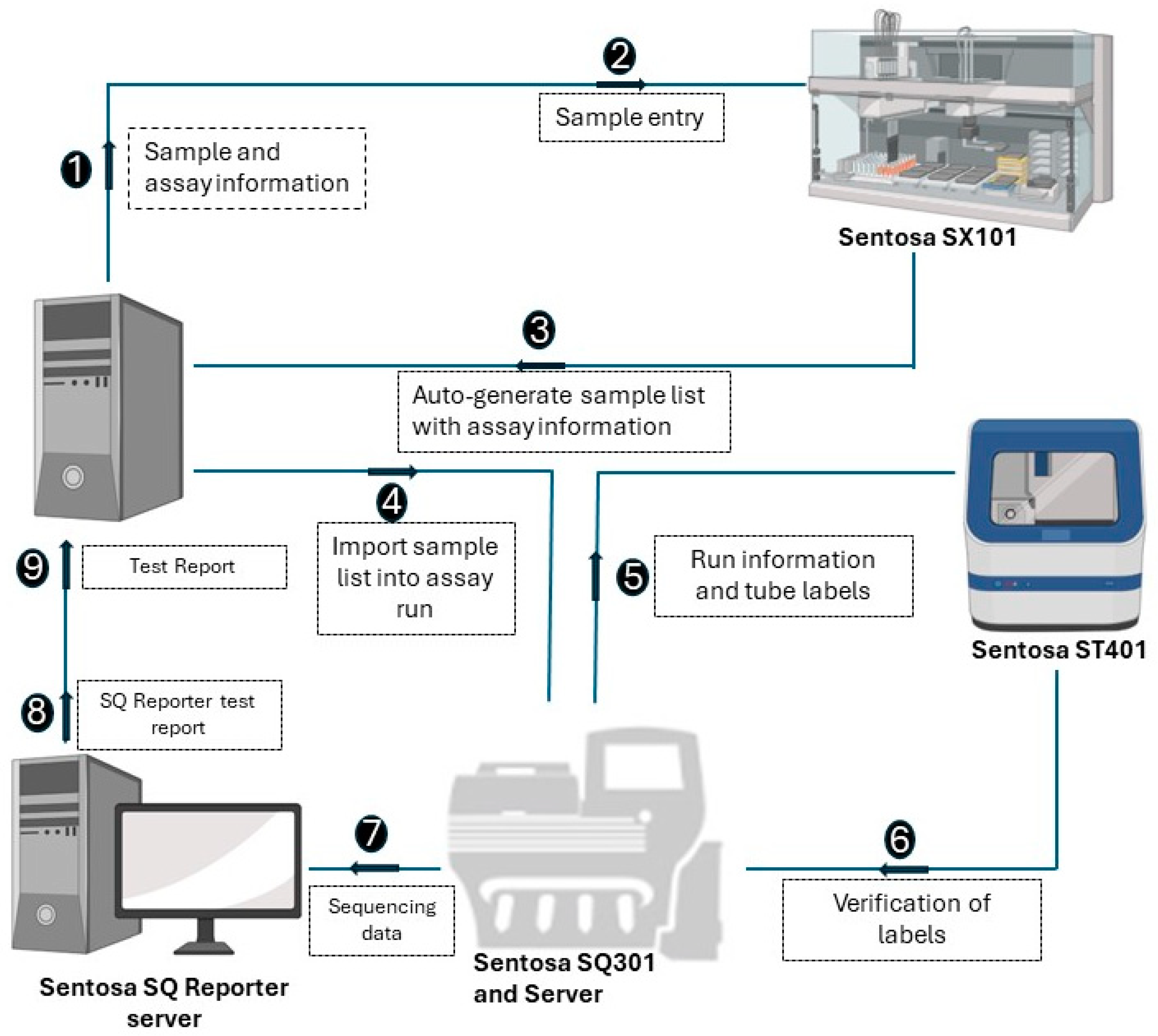
2.4. Post NGS Drug Susceptibility Reporting Workflow
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Performance Metrics of Vela NGS Sequencing
3.3. Comparison of Vela NGS System and Sanger Sequencing
3.4. Precision Analysis of Vela NGS System
3.4.1. Determination of Limit of Detection (LOD) and Intra-Assay Reproducibility
3.4.2. Inter-Assay Accuracy
4. Discussion
5. Limitations and Suggestions
5.1. Sample Size Considerations
5.2. Focus on Subtype B
5.3. Generalizability Issues
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Health Sector Strategies on, Respectively, HIV, Viral Hepatitis and Sexually Transmitted Infections for the Period 2022–2030. 2022. Available online: https://apps.who.int/iris/handle/10665/360348 (accessed on 1 March 2024).
- UNAIDS Fact Sheet. Available online: https://www.unaids.org/en/resources/fact-sheet (accessed on 10 January 2024).
- Gutiérrez-Sevilla, J.E.; Cárdenas-Bedoya, J.; Escoto-Delgadillo, M.; Zúñiga-González, G.M.; Pérez-Ríos, A.M.; Gómez-Meda, B.C.; González-Enríquez, G.C.; Figarola-Centurión, I.; Chavarría-Avila, E.; Torres-Mendoza, B.M. Genomic instability in people living with HIV. Mutat. Res. Genet. Toxicol. Environ. Mutagenes. 2021, 865, 503336. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Li, G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef] [PubMed]
- Galli, L.; Parisi, M.R.; Poli, A.; Menozzi, M.; Fiscon, M.; Garlassi, E.; Francisci, D.; Di Biagio, A.; Sterrantino, G.; Fornabaio, C.; et al. Burden of disease in PWH harboring a multidrug-resistant virus: Data from the PRESTIGIO registry. Open Forum Infect. Dis. 2020, 7, ofaa456. [Google Scholar] [CrossRef] [PubMed]
- HIV Drug Resistance Report 2021. 24 November 2021. Available online: https://www.who.int/publications/i/item/9789240038608 (accessed on 2 January 2024).
- Dessilly, G.; Goeminne, L.; Vandenbroucke, A.T.; Dufrasne, F.E.; Martin, A.; Kabamba-Mukadi, B. First evaluation of the Next-Generation Sequencing platform for the detection of HIV-1 drug resistance mutations in Belgium. PLoS ONE 2018, 13, e0209561. [Google Scholar] [CrossRef] [PubMed]
- Alidjinou, E.K.; Coulon, P.; Hallaert, C.; Robineau, O.; Meybeck, A.; Huleux, T.; Ajana, F.; Hober, D.; Bocket, L. Routine drug resistance testing in HIV-1 proviral DNA, using an automated next- generation sequencing assay. J. Clin. Virol. 2019, 121, 104207. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Sandstrom, P.; Paredes, R.; Harrigan, P.R.; Brumme, C.J.; Avila Rios, S.; Noguera-Julian, M.; Parkin, N.; Kantor, R. Are we ready for NGS HIV drug resistance testing? The second “winnipeg consensus” symposium. Viruses 2020, 12, 586. [Google Scholar] [CrossRef] [PubMed]
- Ávila-Ríos, S.; Parkin, N.; Swanstrom, R.; Paredes, R.; Shafer, R.; Ji, H.; Kantor, R. Next-generation sequencing for HIV drug resistance testing: Laboratory, clinical, and implementation considerations. Viruses 2020, 12, 617. [Google Scholar] [CrossRef] [PubMed]
- May, S.; Adamska, E.; Tang, J. Evaluation of Vela Diagnostics HIV-1 genotyping assay on an automated next generation sequencing platform. J. Clin. Virol. 2020, 127, 104376. [Google Scholar] [CrossRef] [PubMed]
- Gholami, M.; Rouzbahani, N.; Samiee, S.; Tayeri, K.; Ghorban, K.; Dehkharghani, A.D.; Gholami, A.A.; Moshiri, F.; Sattari, A.; Dadmanesh, M.; et al. HIV-1 drug resistance mutations detection and HIV-1 subtype G report by using next-generation sequencing platform. Microb. Pathog. 2020, 146, 104221. [Google Scholar] [CrossRef] [PubMed]
- Raymond, S.; Nicot, F.; Abravanel, F.; Minier, L.; Carcenac, R.; Lefebvre, C.; Harter, A.; Martin-Blondel, G.; Delobel, P.; Izopet, J. Performance evaluation of the Vela Dx Sentosa next-generation sequencing system for HIV-1 DNA genotypic resistance. J. Clin. Virol. 2020, 122, 104229. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.; Volkova, I.; Sahoo, M.K.; Tzou, P.L.; Shafer, R.W.; Pinsky, B.A. Prospective evaluation of the Vela Diagnostics next-generation sequencing platform for HIV-1 genotypic resistance testing. J. Mol. Diagn. 2019, 21, 961–970. [Google Scholar] [CrossRef] [PubMed]
- ViroSeq TM HIV-1 Integrase Genotyping Kit Product Inserts; Abbott Molecular Inc.: Des Plaines, IL, USA. Available online: www.abbottmolecular.com (accessed on 20 April 2018).
- ViroSeq TM HIV-1 Genotyping System v2.0 Product Insert; Abbott Molecular Inc.: Des Plaines, IL, USA. Available online: www.abbottmolecular.com (accessed on 20 April 2018).
- Delaugerre, C.; Mouroux, M.; Yvon-Groussin, A.; Simon, A.; Angleraud, F.; Huraux, J.M.; Agut, H.; Katlama, C.; Calvez, V. Prevalence and conditions of selection of E44D/A and V118I human immunodeficiency virus type 1 reverse transcriptase mutations in clinical practice. Antimicrob. Agents Chemother. 2001, 45, 946–948. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Neubert, J.; Michalsky, N.; Laws, H.J.; Borkhardt, A.; Jensen, B.; Lübke, N. HIV-1 Subtype diversity and prevalence of primary drug resistance in a single-center pediatric cohort in Germany. Intervirology 2017, 59, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, A.; Kodama, E.N.; Sarafianos, S.G.; Schuckmann, M.M.; Sakagami, Y.; Matsuoka, M.; Takiguchi, M.; Gatanaga, H.; Oka, S. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 2008, 82, 3261–3270. [Google Scholar] [CrossRef] [PubMed]
- Grossman, Z.; Istomin, V.; Averbuch, D.; Lorber, M.; Risenberg, K.; Levi, I.; Chowers, M.; Burke, M.; Bar Yaacov, N.; Schapiro, J.M.; et al. Genetic variation at NNRTI resistance-associated positions in patients infected with HIV-1 subtype C. AIDS 2004, 18, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Singhroy, D.N.; Wainberg, M.A.; Mesplède, T. Combination of the R263K and M184I/V resistance substitutions against dolutegravir and lamivudine decreases HIV replicative capacity. Antimicrob. Agents Chemother. 2015, 59, 2882–2885. [Google Scholar] [CrossRef] [PubMed]
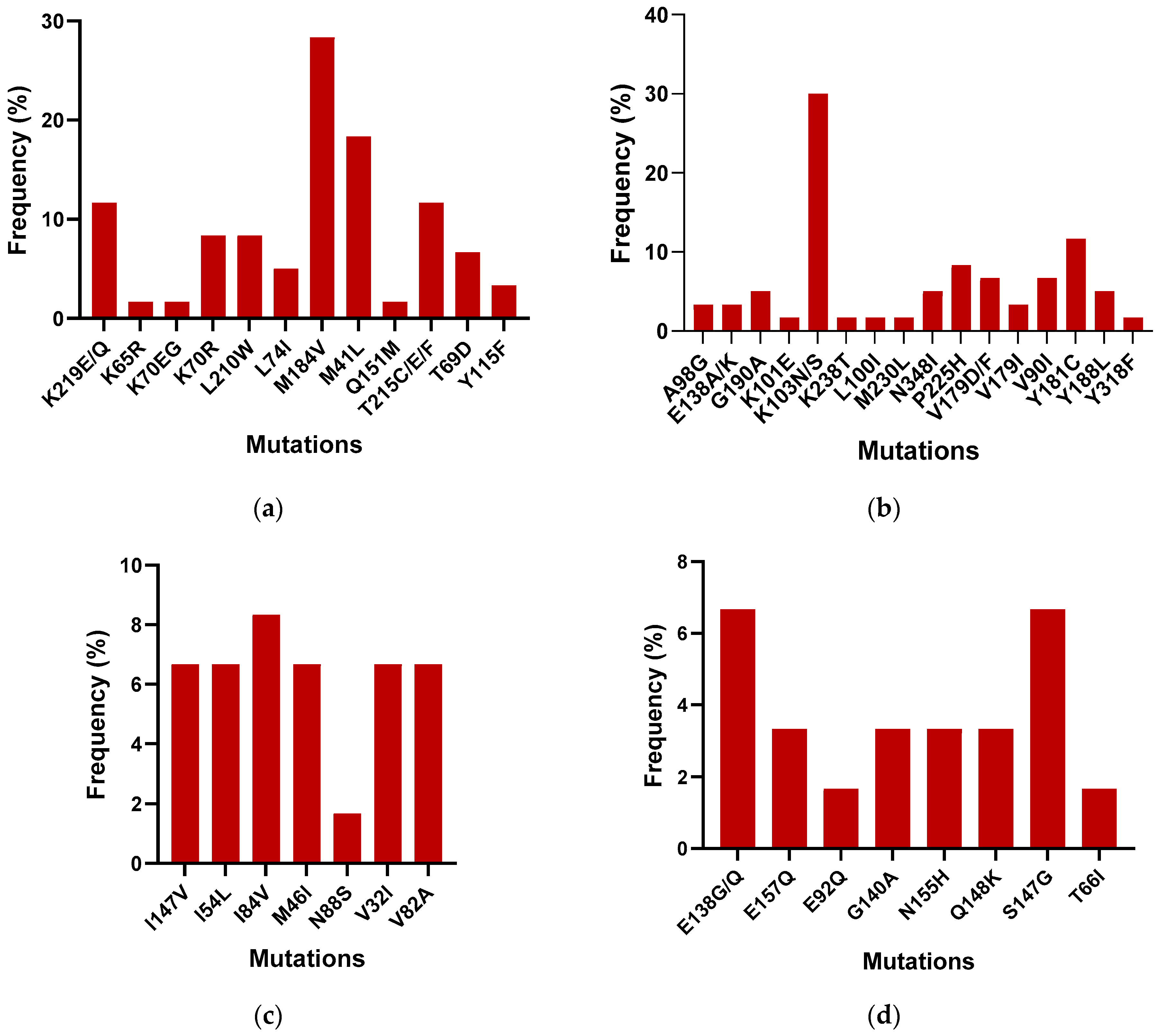

| Mutation | No. of Patient Samples Positive for the Mutation | Gene Regions | |
|---|---|---|---|
| Vela NGS | Abbott ViroSeq | ||
| E138G/Q | 4 | 4 | IN |
| E157Q | 2 | 2 | IN |
| E92Q | 1 | 0 | IN |
| G140A | 2 | 3 | IN |
| K156N | 1 | 1 | IN |
| L74M | 1 | 1 | IN |
| N155H | 2 | 2 | IN |
| Q148K | 2 | 3 | IN |
| S147G | 4 | 4 | IN |
| T66I | 1 | 1 | IN |
| T97A | 1 | 1 | IN |
| A98G | 2 | 2 | NNRTI |
| E138A/K | 2 | 2 | NNRTI |
| G190A | 3 | 5 | NNRTI |
| H221Y | 2 | 0 | NNRTI |
| K101E | 1 | 1 | NNRTI |
| K103N/S | 18 | 21 | NNRTI |
| K238T | 1 | 1 | NNRTI |
| L100I | 1 | 2 | NNRTI |
| M230L | 1 | 1 | NNRTI |
| N348I | 3 | 0 | NNRTI |
| P225H | 5 | 3 | NNRTI |
| V106I | 3 | 4 | NNRTI |
| V108I | 1 | 1 | NNRTI |
| V179D/F | 4 | 4 | NNRTI |
| V179I | 2 | 0 | NNRTI |
| V90I | 4 | 4 | NNRTI |
| Y181C | 7 | 5 | NNRTI |
| Y188L | 3 | 3 | NNRTI |
| Y318F | 1 | 1 | NNRTI |
| A62V | 1 | 1 | NRTI |
| D67N | 8 | 7 | NRTI |
| E44D | 2 | 0 | NRTI |
| F116Y | 1 | 1 | NRTI |
| F77L | 1 | 1 | NRTI |
| K219E/Q | 7 | 6 | NRTI |
| K65R | 1 | 1 | NRTI |
| K70EG | 1 | 0 | NRTI |
| K70R | 5 | 4 | NRTI |
| L210S | 1 | 0 | NRTI |
| L210W | 5 | 5 | NRTI |
| L74I | 3 | 4 | NRTI |
| M184I | 4 | 4 | NRTI |
| M184V | 17 | 18 | NRTI |
| M41L | 11 | 11 | NRTI |
| Q151M | 1 | 1 | NRTI |
| T215C/E/F | 7 | 7 | NRTI |
| T69D | 4 | 5 | NRTI |
| T69N | 3 | 3 | NRTI |
| T69S | 2 | 3 | NRTI |
| V75I | 1 | 1 | NRTI |
| Y115F | 2 | 2 | NRTI |
| I147V | 4 | 3 | PI |
| I54L | 4 | 4 | PI |
| I84V | 5 | 5 | PI |
| L10FI | 1 | 4 | PI |
| L90M | 1 | 1 | PI |
| M46I | 4 | 4 | PI |
| N88S | 1 | 1 | PI |
| V32I | 4 | 4 | PI |
| V82A | 4 | 4 | PI |
| Run ID | Sample ID | Viral Load (cp/mL) | NRTI | NNRTI | PI | INT |
|---|---|---|---|---|---|---|
| 1_ENEWL | 18-1628 | 2000 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Other: M36I, L63P | Other: K156N |
| 1_ENEWL | 18-1629 | 1000 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Other: M36I, L63P | K156N |
| 1_ENEWL | 18-1630 | 750 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Other: M36I, L63P | K156N |
| 1_ENEWL | 18-1631 | 500 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Other: M36I, L63P | K156N |
| 1_ENEWL | 18-1632 | 250 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Other: M36I, L63P | K156N |
| 1_ENEWL | 18-1633 | 100 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Other: M36I, L63P | K156N |
| Run ID | Sample ID | Viral Load | NRTI | NNRTI | PI | INT |
|---|---|---|---|---|---|---|
| 1_ENEWL | 18-1628 (17-5913) | 2000 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Others: M36I, L63P, I62V | Other: K156N |
| 2_LKED2 | 18-4707 (17-5913) | 2000 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Others: M36I, L63P, I62V | Other: K156N |
| 3_QPWOE | 18-4757 (17-5913) | 2000 | K65R, Y115F | K101E, E138Q, V179F, Y181C | Others: M36I, L63P | Other: K156N |
| 1_ENEWL | 18-1635 (17-6049) | 4000 | M184V | Other: V901I, V118I | Other: V77I | E92Q Other: K156N |
| 2_LKED2 | 18-4708 (17-6049) | 8277 | M184V | Other: V901I, V118I | Other: V77I | K156N |
| 3_QPWOE | 18-4758 (17-6049) | 8277 | M184V | Other: V901I, V118I | Other: V77I | K156N |
| 1_ENEWL | 18-1636 (17-6050) | 10,000 | - | V179D, G190A, Y318F Other: V90I, V106I | Other: M36I, L63P | Other: T206S |
| 2_LKED2 | 18-4709 (17-6050) | 10,000 | - | V179D, G190A, Y318F Other: V90I, V106I | Other: M36I, L63P | Other: G193E, T206S |
| 3_QPWOE | 18-4759 (17-6050) | 10,000 | - | V179D, G190A, Y318F Other: V90I, V106I | Other: M36I, L63P | Other: T206S |
| 1_ENEWL | 18-1639 (17-7072) | 10,000 | M41L, T215E | - | Others: M36I, L63P, I64LM, I93L | - |
| 2_LKED2 | 18-4710 (17-7072) | 10,000 | M41L, T215E | - | Others: M36I, L63P, I64LM, I93L | - |
| 3_QPWOE | 18-4760 (17-7072) | 10,000 | M41L, T215E | - | Others: M36I, L63P, I64LM, I93L | - |
| PPA = TP/(TP + FN) (Sensitivity) | NPA = TN/(TN + FP) (Specificity) | PPV = TP/(TP + FP) | NPV = TN/(TN + FN) | |
|---|---|---|---|---|
| NRTI DR Mutation | 100% | 100% | 100% | 100% |
| NNRTI DR Mutation | 100% | 100% | 100% | 100% |
| PI DR Mutation | 100% | 100% | 100% | 100% |
| Integrase | 100% | 100% | 100% | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vashisht, A.; Mondal, A.K.; Vashisht, V.; Ananth, S.; Alptekin, A.; Jones, K.; Farmaha, J.K.; Kolhe, R. Enhancing Precision in HIV Treatment: Validation of a Robust Next-Generation Sequencing System for Drug Resistance Mutation Analysis. Diagnostics 2024, 14, 1766. https://doi.org/10.3390/diagnostics14161766
Vashisht A, Mondal AK, Vashisht V, Ananth S, Alptekin A, Jones K, Farmaha JK, Kolhe R. Enhancing Precision in HIV Treatment: Validation of a Robust Next-Generation Sequencing System for Drug Resistance Mutation Analysis. Diagnostics. 2024; 14(16):1766. https://doi.org/10.3390/diagnostics14161766
Chicago/Turabian StyleVashisht, Ashutosh, Ashis K. Mondal, Vishakha Vashisht, Sudha Ananth, Ahmet Alptekin, Kimya Jones, Jaspreet K. Farmaha, and Ravindra Kolhe. 2024. "Enhancing Precision in HIV Treatment: Validation of a Robust Next-Generation Sequencing System for Drug Resistance Mutation Analysis" Diagnostics 14, no. 16: 1766. https://doi.org/10.3390/diagnostics14161766
APA StyleVashisht, A., Mondal, A. K., Vashisht, V., Ananth, S., Alptekin, A., Jones, K., Farmaha, J. K., & Kolhe, R. (2024). Enhancing Precision in HIV Treatment: Validation of a Robust Next-Generation Sequencing System for Drug Resistance Mutation Analysis. Diagnostics, 14(16), 1766. https://doi.org/10.3390/diagnostics14161766





