Cross-Modality Medical Image Segmentation via Enhanced Feature Alignment and Cross Pseudo Supervision Learning
Abstract
1. Introduction
- We propose a novel, unsupervised domain adaptation framework for medical image segmentation, comprising a feature alignment sub-network and a pseudo-supervised, dual-stream segmentation sub-network. The feature alignment sub-network facilitates the alignment of features between the source and target domains, while the pseudo-supervised, dual-stream segmentation sub-network achieves segmentation of cross-modal images and promotes the learning process of the feature sub-network;
- For the first time, cross-pseudo-supervision is introduced to mitigate the impact of low-quality pseudo-labels, thereby insulating the segmentation network from fluctuations in the feature alignment sub-network;
- Given that medical images present clear structures and shapes, incorporating a self-attention module into the feature alignment sub-network significantly improves the learning process;
- Our method was evaluated on two challenging tasks, and the results indicate that it is increasingly approaching the performance of fully supervised models.
2. Related Works
2.1. Cross-Modality Medical Image Segmentation
2.2. Single-Source Domain Adaptation
2.3. Feature Alignment
2.4. Cross-Pseudo-Supervision
3. Method
3.1. Datasets
3.2. Overall Framework Architecture
3.3. Cross-Modality Feature Alignment Sub-Network
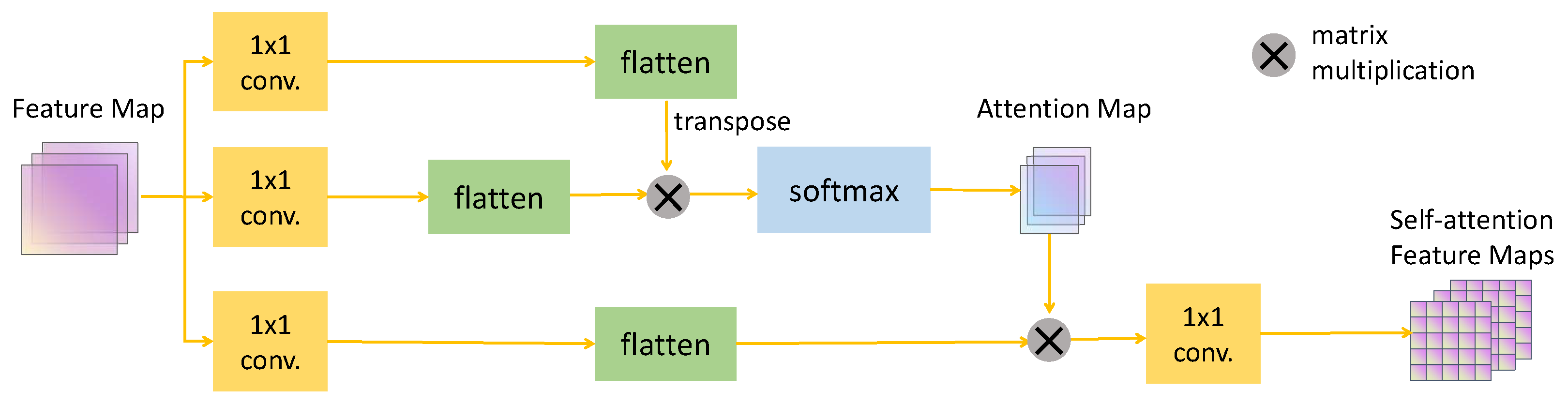
Self-Attention Module
3.4. Cross-Pseudo-Supervised Dual-Stream Segmentation Sub-Network
3.5. Total Loss
4. Experimental Results and Discussion
4.1. Experimental Setup
4.2. Comparison Results
4.2.1. Results on Abdominal Organ Segmentation
4.2.2. Results on Brain-Tumor Segmentation
4.3. Ablation Study
4.4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rubak, S.; Sandbæk, A.; Lauritzen, T.; Christensen, B. Motivational interviewing: A systematic review and meta-analysis. Br. J. Gen. Pract. 2005, 55, 305–312. [Google Scholar] [PubMed]
- Umirzakova, S.; Whangbo, T.K. Detailed feature extraction network-based fine-grained face segmentation. Knowl.-Based Syst. 2022, 250, 109036. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Watanabe, K.; Ushiku, Y.; Harada, T. Maximum classifier discrepancy for unsupervised domain adaptation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3723–3732. [Google Scholar]
- Xie, Q.; Li, Y.; He, N.; Ning, M.; Ma, K.; Wang, G.; Lian, Y.; Zheng, Y. Unsupervised Domain Adaptation for Medical Image Segmentation by Disentanglement Learning and Self-Training. IEEE Trans. Med. Imaging 2022, 43, 4–14. [Google Scholar] [CrossRef]
- Bousmalis, K.; Silberman, N.; Dohan, D.; Erhan, D.; Krishnan, D. Unsupervised pixel-level domain adaptation with generative adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 3722–3731. [Google Scholar]
- Zhao, H.; Li, H.; Maurer-Stroh, S.; Guo, Y.; Deng, Q.; Cheng, L. Supervised segmentation of un-annotated retinal fundus images by synthesis. IEEE Trans. Med. Imaging 2018, 38, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation. IEEE Trans. Med. Imaging 2020, 39, 2494–2505. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Saenko, K. Deep coral: Correlation alignment for deep domain adaptation. In Proceedings of the Computer Vision–ECCV 2016 Workshops, Amsterdam, The Netherlands, 8–10 15–16 October 2016; pp. 443–450. [Google Scholar]
- Mancini, M.; Porzi, L.; Bulo, S.R.; Caputo, B.; Ricci, E. Boosting domain adaptation by discovering latent domains. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 3771–3780. [Google Scholar]
- Tsai, Y.H.; Hung, W.C.; Schulter, S.; Sohn, K.; Yang, M.H.; Chandraker, M. Learning to adapt structured output space for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7472–7481. [Google Scholar]
- Huo, Y.; Xu, Z.; Moon, H.; Bao, S.; Assad, A.; Moyo, T.K.; Savona, M.R.; Abramson, R.G.; Landman, B.A. Synseg-net: Synthetic segmentation without target modality ground truth. IEEE Trans. Med. Imaging 2018, 38, 1016–1025. [Google Scholar] [CrossRef]
- Han, X.; Qi, L.; Yu, Q.; Zhou, Z.; Zheng, Y.; Shi, Y.; Gao, Y. Deep symmetric adaptation network for cross-modality medical image segmentation. IEEE Trans. Med. Imaging 2021, 41, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhuang, X. CF Distance: A New Domain Discrepancy Metric and Application to Explicit Domain Adaptation for Cross-Modality Cardiac Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 4274–4285. [Google Scholar] [CrossRef]
- Zhang, H.; Goodfellow, I.; Metaxas, D.; Odena, A. Self-attention generative adversarial networks. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 7354–7363. [Google Scholar]
- Chen, X.; Yuan, Y.; Zeng, G.; Wang, J. Semi-supervised semantic segmentation with cross pseudo supervision. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 2613–2622. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment anything. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Paris, France, 1–6 October 2023; pp. 4015–4026. [Google Scholar]
- Chen, X.; Lian, C.; Wang, L.; Deng, H.; Kuang, T.; Fung, S.; Gateno, J.; Yap, P.T.; Xia, J.J.; Shen, D. Anatomy-regularized representation learning for cross-modality medical image segmentation. IEEE Trans. Med. Imaging 2020, 40, 274–285. [Google Scholar] [CrossRef]
- Zhang, D.; Huang, G.; Zhang, Q.; Han, J.; Han, J.; Yu, Y. Cross-modality deep feature learning for brain tumor segmentation. Pattern Recognit. 2021, 110, 107562. [Google Scholar] [CrossRef]
- Li, K.; Yu, L.; Wang, S.; Heng, P.A. Towards cross-modality medical image segmentation with online mutual knowledge distillation. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 775–783. [Google Scholar]
- Wang, M.; Deng, W. Deep visual domain adaptation: A survey. Neurocomputing 2018, 312, 135–153. [Google Scholar] [CrossRef]
- Pei, C.; Wu, F.; Huang, L.; Zhuang, X. Disentangle domain features for cross-modality cardiac image segmentation. Med. Image Anal. 2021, 71, 102078. [Google Scholar] [PubMed]
- Pei, C.; Wu, F.; Yang, M.; Pan, L.; Ding, W.; Dong, J.; Huang, L.; Zhuang, X. Multi-Source Domain Adaptation for Medical Image Segmentation. IEEE Trans. Med. Imaging 2023, 43, 1640–1651. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Chikontwe, P.; Won, D.; Luna, M.; Park, S.H. Structure-preserving image translation for multi-source medical image domain adaptation. Pattern Recognit. 2023, 144, 109840. [Google Scholar] [CrossRef]
- Hu, S.; Liao, Z.; Zhang, J.; Xia, Y. Domain and content adaptive convolution based multi-source domain generalization for medical image segmentation. IEEE Trans. Med. Imaging 2022, 42, 233–244. [Google Scholar] [CrossRef]
- Zhao, Z.; Xu, K.; Li, S.; Zeng, Z.; Guan, C. Mt-uda: Towards unsupervised cross-modality medical image segmentation with limited source labels. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; pp. 293–303. [Google Scholar]
- Xia, Y.; Yang, D.; Yu, Z.; Liu, F.; Cai, J.; Yu, L.; Zhu, Z.; Xu, D.; Yuille, A.; Roth, H. Uncertainty-aware multi-view co-training for semi-supervised medical image segmentation and domain adaptation. Med. Image Anal. 2020, 65, 101766. [Google Scholar] [CrossRef]
- Liu, X.; Xing, F.; Shusharina, N.; Lim, R.; Jay Kuo, C.C.; El Fakhri, G.; Woo, J. Act: Semi-supervised domain-adaptive medical image segmentation with asymmetric co-training. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Vancouver, BC, Canada, 8–12 October 2022; pp. 66–76. [Google Scholar]
- Bateson, M.; Kervadec, H.; Dolz, J.; Lombaert, H.; Ayed, I.B. Source-free domain adaptation for image segmentation. Med. Image Anal. 2022, 82, 102617. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Zhang, X.; Ye, C. Source-free domain adaptation for medical image segmentation via selectively updated mean teacher. In Proceedings of the International Conference on Information Processing in Medical Imaging, San Carlos de Bariloche, Argentina, 18–23 June 2023; pp. 225–236. [Google Scholar]
- Yang, C.; Guo, X.; Chen, Z.; Yuan, Y. Source free domain adaptation for medical image segmentation with fourier style mining. Med. Image Anal. 2022, 79, 102457. [Google Scholar] [CrossRef]
- Long, M.; Cao, Z.; Wang, J.; Jordan, M.I. Conditional adversarial domain adaptation. Adv. Neural Inf. Process. Syst. 2018, 31. [Google Scholar]
- Csurka, G. Domain adaptation for visual applications: A comprehensive survey. arXiv 2017, arXiv:1702.05374. [Google Scholar]
- Ouyang, C.; Kamnitsas, K.; Biffi, C.; Duan, J.; Rueckert, D. Data efficient unsupervised domain adaptation for cross-modality image segmentation. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; pp. 669–677. [Google Scholar]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative adversarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Ganin, Y.; Ustinova, E.; Ajakan, H.; Germain, P.; Larochelle, H.; Laviolette, F.; Marchand, M.; Lempitsky, V. Domain-adversarial training of neural networks. J. Mach. Learn. Res. 2016, 17, 2030–2096. [Google Scholar]
- Liu, D.; Zhang, D.; Song, Y.; Zhang, F.; O’Donnell, L.; Huang, H.; Chen, M.; Cai, W. Pdam: A panoptic-level feature alignment framework for unsupervised domain adaptive instance segmentation in microscopy images. IEEE Trans. Med. Imaging 2020, 40, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Ouali, Y.; Hudelot, C.; Tami, M. Semi-supervised semantic segmentation with cross-consistency training. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12674–12684. [Google Scholar]
- Ibrahim, M.S.; Vahdat, A.; Ranjbar, M.; Macready, W.G. Semi-supervised semantic image segmentation with self-correcting networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 12715–12725. [Google Scholar]
- Feng, Z.; Zhou, Q.; Cheng, G.; Tan, X.; Shi, J.; Ma, L. Semi-supervised semantic segmentation via dynamic self-training and classbalanced curriculum. arXiv 2020, arXiv:2004.08514. [Google Scholar]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. Multi-Atlas Labeling Beyond the Cranial Vault—Workshop and Challenge. 2017. Available online: https://www.synapse.org/Synapse:syn3193805/wiki/217789 (accessed on 1 July 2024).
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Yao, H.; Hu, X.; Li, X. Enhancing pseudo label quality for semi-supervised domain-generalized medical image segmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtual Event, 22 February–1 March 2022; Volume 36, pp. 3099–3107. [Google Scholar]
- Sutter, T.; Daunhawer, I.; Vogt, J. Multimodal generative learning utilizing jensen-shannon-divergence. Adv. Neural Inf. Process. Syst. 2020, 33, 6100–6110. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
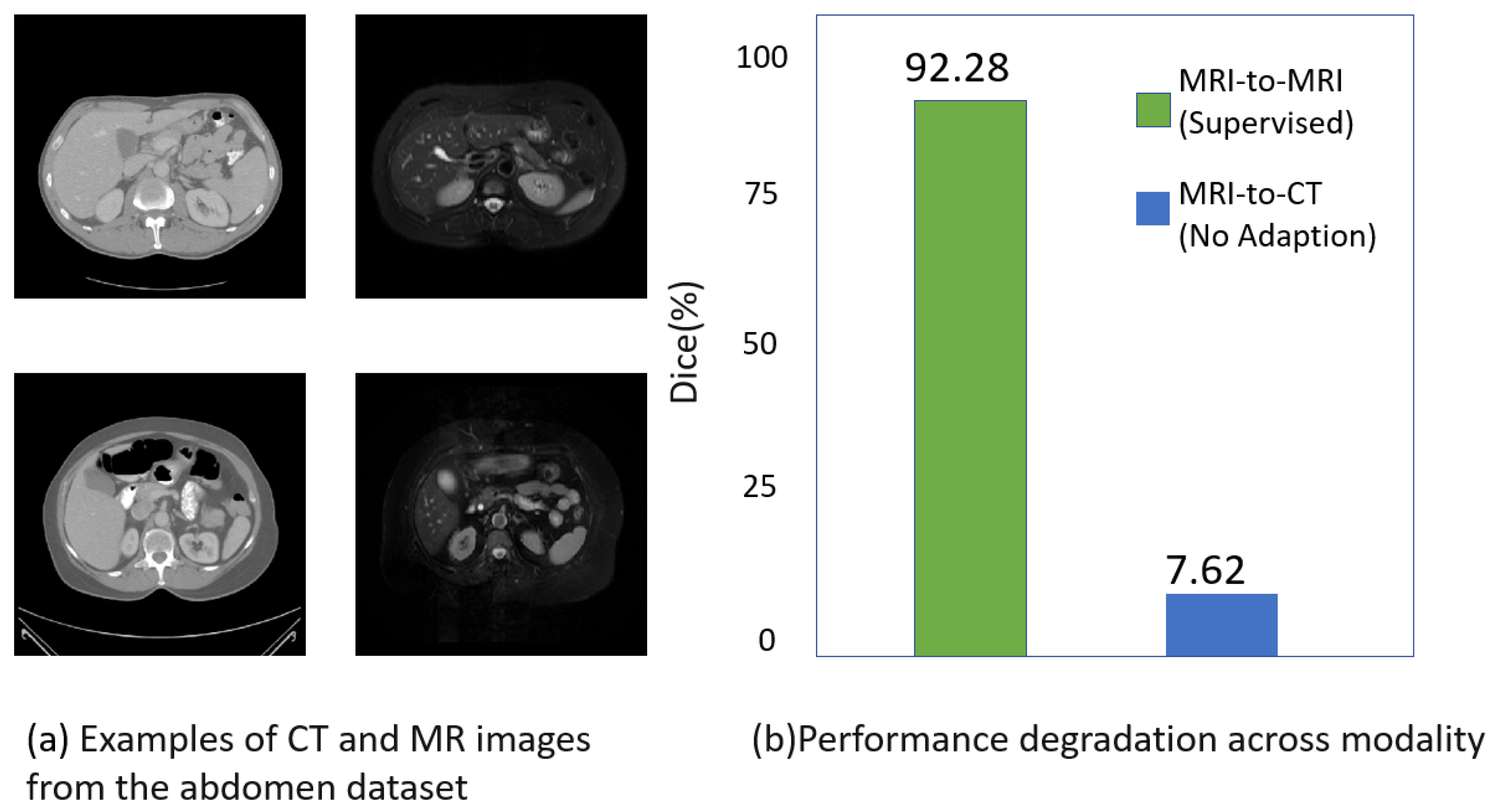

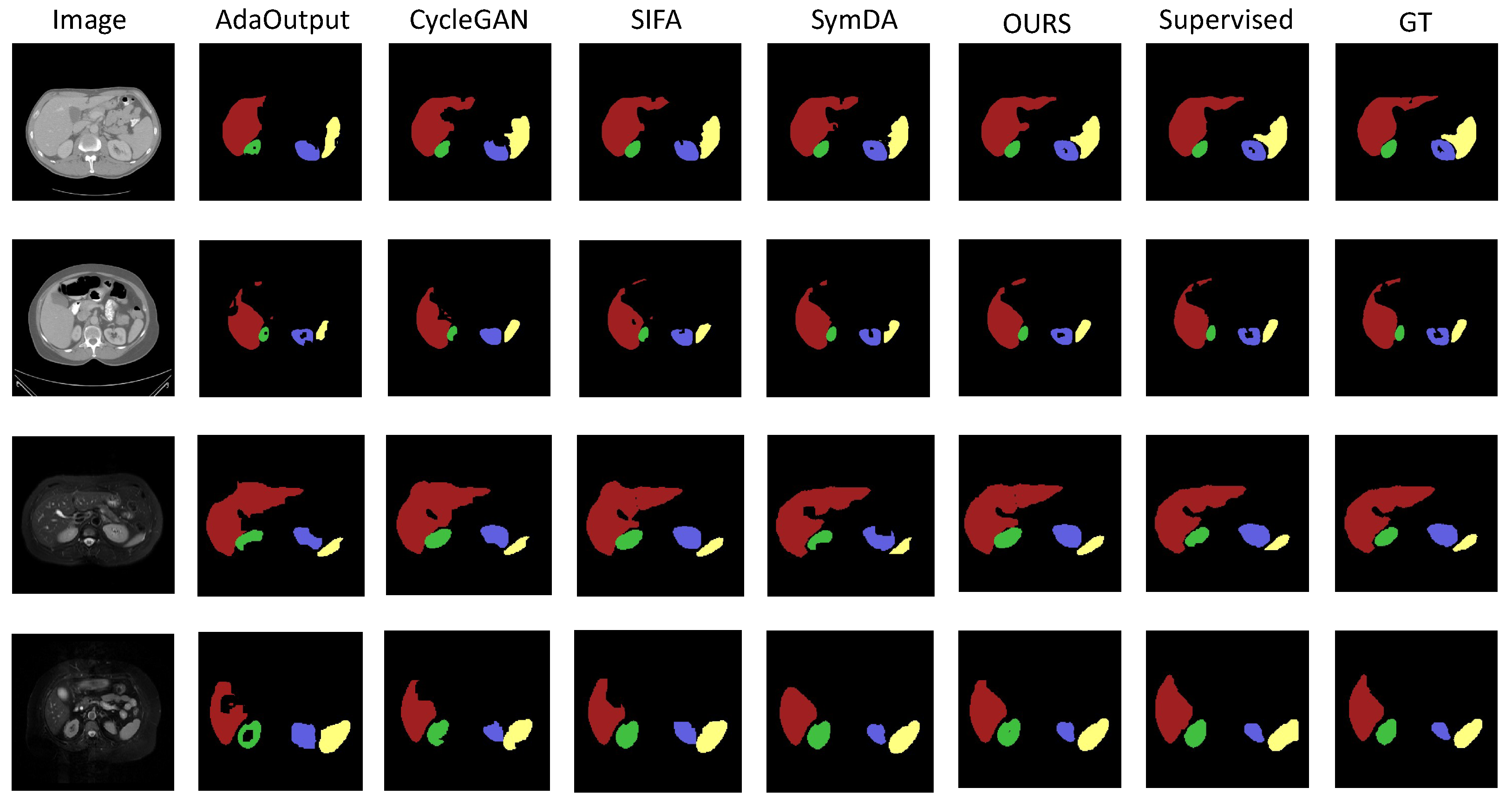
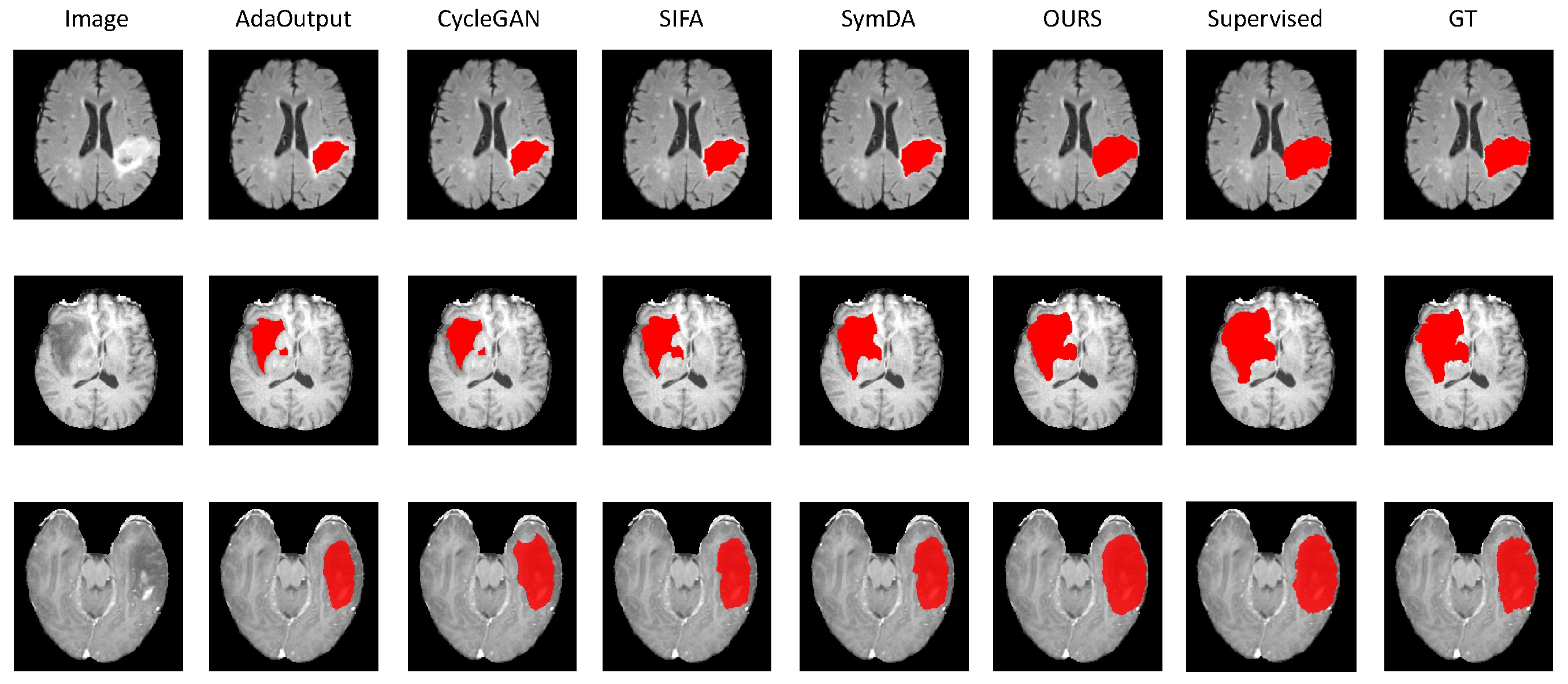
| Abdominal MRI→Abdominal CT | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | Dice (%) | ASD (voxel) | ||||||||
| Liver | R. kidney | L. kidney | Spleen | Average | Liver | R. kidney | L. kidney | Spleen | Average | |
| ST | 93.68 ± 3.57 | 91.74 ± 0.38 | 91.74 ± 0.18 | 92.60 ± 0.19 | 91.83 ± 0.57 | 1.11 ± 0.12 | 1.74 ± 0.23 | 0.92 ± 0.23 | 1.26 ± 0.04 | 1.22 ± 0.09 |
| NA | 23.43 ± 2.54 | 5.33 ± 2.20 | 1.42 ± 0.95 | 3.09 ± 1.25 | 7.98 ± 0.27 | 24.07 ± 2.05 | 55.64 ± 3.52 | 60.26 ± 4.19 | 71.48 ± 2.01 | 52.61 ± 3.26 |
| AdaOutput [11] | 83.56 ± 1.34 | 80.21 ± 0.77 | 79.84 ± 1.14 | 80.71 ± 0.49 | 80.83 ± 0.59 | 1.63 ± 0.06 | 1.21 ± 0.05 | 1.64 ± 0.16 | 1.63 ± 0.07 | 1.53 ± 0.02 |
| CycleGAN [43] | 83.19 ± 1.42 | 78.54 ± 1.62 | 78.24 ± 1.68 | 81.19 ± 1.35 | 80.63 ± 0.58 | 1.68 ± 0.09 | 1.22 ± 0.12 | 1.21 ± 0.05 | 1.86 ± 0.07 | 1.50 ± 0.06 |
| SIFA [8] | 83.93 ± 0.78 | 83.15 ± 1.73 | 84.46 ± 1.45 | 82.71 ± 0.59 | 83.14 ± 0.64 | 1.14 ± 0.06 | 1.17 ± 0.11 | 1.47 ± 0.06 | 1.59 ± 0.06 | 1.34 ± 0.01 |
| SymDA [13] | 87.21 ± 1.06 | 85.07 ± 0.56 | 84.52 ± 0.51 | 86.00 ± 0.02 | 85.95 ± 0.36 | 1.37 ± 0.21 | 1.13 ± 0.13 | 1.35 ± 0.08 | 1.66 ± 0.15 | 1.38 ± 0.08 |
| OURS | 89.41 ± 0.18 | 86.26 ± 0.28 | 87.48 ± 0.50 | 85.77 ± 0.47 | 87.07 ± 0.50 | 1.74 ± 0.12 | 0.89 ± 0.09 | 1.71 ± 0.20 | 2.66 ± 0.22 | 1.75 ± 0.08 |
| Abdominal CT→Abdominal MRI | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | Dice (%) | ASD (voxel) | ||||||||
| Liver | R. kidney | L. kidney | Spleen | Average | Liver | R. kidney | L. kidney | Spleen | Average | |
| ST | 92.15 ± 0.31 | 91.50 ± 0.32 | 91.26 ± 0.19 | 93.57 ± 0.33 | 92.04 ± 0.35 | 1.22 ± 0.06 | 2.18 ± 0.16 | 0.91 ± 0.06 | 0.57 ± 0.05 | 1.22 ± 0.08 |
| NA | 8.45 ± 0.09 | 0.79 ± 0.33 | 0.69 ± 0.42 | 6.07 ± 0.91 | 3.83 ± 0.43 | 37.23 ± 0.86 | 65.68 ± 2.00 | 56.43 ± 1.13 | 44.97 ± 2.24 | 53.78 ± 10.66 |
| AdaOutput [11] | 86.25 ± 1.08 | 82.40 ± 0.53 | 83.15 ± 0.47 | 83.87 ± 0.12 | 83.60 ± 1.87 | 1.95 ± 0.03 | 1.62 ± 0.27 | 3.47 ± 0.38 | 2.07 ± 0.23 | 2.35 ± 0.73 |
| CycleGAN [43] | 86.24 ± 0.17 | 83.47 ± 1.25 | 81.52 ± 1.04 | 81.47 ± 1.00 | 84.48 ± 1.45 | 2.12 ± 0.11 | 3.38 ± 0.18 | 2.11 ± 0.19 | 2.78 ± 0.15 | 2.54 ± 0.61 |
| SIFA [8] | 86.68 ± 0.11 | 87.45 ± 0.31 | 86.39 ± 0.19 | 85.99 ± 0.11 | 86.83 ± 0.43 | 1.64 ± 0.12 | 0.78 ± 0.19 | 1.54 ± 0.04 | 2.62 ± 0.25 | 1.32 ± 0.38 |
| SymDA [13] | 89.63 ± 0.31 | 86.68 ± 0.07 | 85.56 ± 0.29 | 88.74 ± 0.13 | 87.62 ± 2.06 | 1.58 ± 0.16 | 1.87 ± 0.08 | 3.07 ± 0.16 | 1.94 ± 0.03 | 2.17 ± 0.67 |
| OURS | 90.92 ± 0.04 | 88.35 ± 0.04 | 87.56 ± 0.03 | 91.21 ± 0.15 | 88.28 ± 1.42 | 1.05 ± 0.03 | 0.90 ± 0.03 | 1.00 ± 0.04 | 0.75 ± 0.18 | 0.98 ± 0.06 |
| Method | Dice (%) | Hausdorff Distance (mm) | ||||||
|---|---|---|---|---|---|---|---|---|
| T1 | FLAIR | T1CE | Average | T1 | FLAIR | T1CE | Average | |
| ST | 75.47 ± 0.44 | 85.21 ± 1.01 | 72.52 ± 0.47 | 77.07 ± 0.57 | 9.04 ± 0.41 | 5.52 ± 0.42 | 9.90 ± 0.35 | 8.82 ± 0.38 |
| NA | 4.82 ± 1.64 | 23.42 ± 5.20 | 13.09 ± 3.04 | 13.11 ± 3.16 | 56.05 ± 8.45 | 29.47 ± 3.27 | 49.53 ± 7.44 | 45.35 ± 7.18 |
| AdaOutput [11] | 44.39 ± 0.93 | 61.09 ± 1.06 | 33.38 ± 2.02 | 46.62 ± 1.27 | 24.67 ± 2.00 | 20.23 ± 1.27 | 33.13 ± 3.24 | 26.01 ± 2.16 |
| CycleGAN [43] | 36.76 ± 1.44 | 66.15 ± 1.10 | 43.26 ± 0.25 | 48.06 ± 1.11 | 27.22 ± 2.09 | 20.31 ± 1.55 | 23.04 ± 0.84 | 23.52 ± 1.67 |
| SIFA [8] | 52.70 ± 1.13 | 68.54 ± 2.39 | 58.53 ± 1.90 | 59.26 ± 1.87 | 19.47 ± 1.04 | 17.15 ± 1.48 | 15.27 ± 1.71 | 17.30 ± 1.37 |
| SymDA [13] | 57.09 ± 0.76 | 81.33 ± 0.53 | 62.08 ± 0.62 | 66.50 ± 0.63 | 14.27 ± 0.56 | 8.63 ± 0.46 | 13.71 ± 1.34 | 12.87 ± 0.98 |
| OURS | 60.07 ± 0.23 | 82.28 ± 0.55 | 66.20 ± 0.60 | 69.18 ± 0.46 | 12.95 ± 0.26 | 8.53 ± 0.38 | 10.93 ± 0.95 | 10.47 ± 0.53 |
| Method | Self_Att | ||||||
|---|---|---|---|---|---|---|---|
| SegOnly | ✓ | ||||||
| FA | ✓ | ✓ | ✓ | ✓ | |||
| FA+CPS | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| FA+DACPS | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| FACPS | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Method | Liver | L.Kid | R.Kid | Spleen | Average |
|---|---|---|---|---|---|
| SegOnly | 8.35 | 0.96 | 0.25 | 5.34 | 3.75 |
| FA | 78.59 | 68.69 | 71.22 | 63.26 | 70.44 |
| FA+CPS | 83.12 | 79.26 | 80.08 | 82.71 | 81.29 |
| FA+DACPS | 90.24 | 87.07 | 85.31 | 90.79 | 88.35 |
| FACPS | 92.89 | 87.52 | 88.31 | 91.03 | 89.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Wu, Z.; Zheng, H.; Huang, L.; Ding, W.; Pan, L.; Yin, L. Cross-Modality Medical Image Segmentation via Enhanced Feature Alignment and Cross Pseudo Supervision Learning. Diagnostics 2024, 14, 1751. https://doi.org/10.3390/diagnostics14161751
Yang M, Wu Z, Zheng H, Huang L, Ding W, Pan L, Yin L. Cross-Modality Medical Image Segmentation via Enhanced Feature Alignment and Cross Pseudo Supervision Learning. Diagnostics. 2024; 14(16):1751. https://doi.org/10.3390/diagnostics14161751
Chicago/Turabian StyleYang, Mingjing, Zhicheng Wu, Hanyu Zheng, Liqin Huang, Wangbin Ding, Lin Pan, and Lei Yin. 2024. "Cross-Modality Medical Image Segmentation via Enhanced Feature Alignment and Cross Pseudo Supervision Learning" Diagnostics 14, no. 16: 1751. https://doi.org/10.3390/diagnostics14161751
APA StyleYang, M., Wu, Z., Zheng, H., Huang, L., Ding, W., Pan, L., & Yin, L. (2024). Cross-Modality Medical Image Segmentation via Enhanced Feature Alignment and Cross Pseudo Supervision Learning. Diagnostics, 14(16), 1751. https://doi.org/10.3390/diagnostics14161751





