Implementing Explainable Machine Learning Models for Practical Prediction of Early Neonatal Hypoglycemia
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Feature Selection and Model Building
2.3. Model Performance Measurement
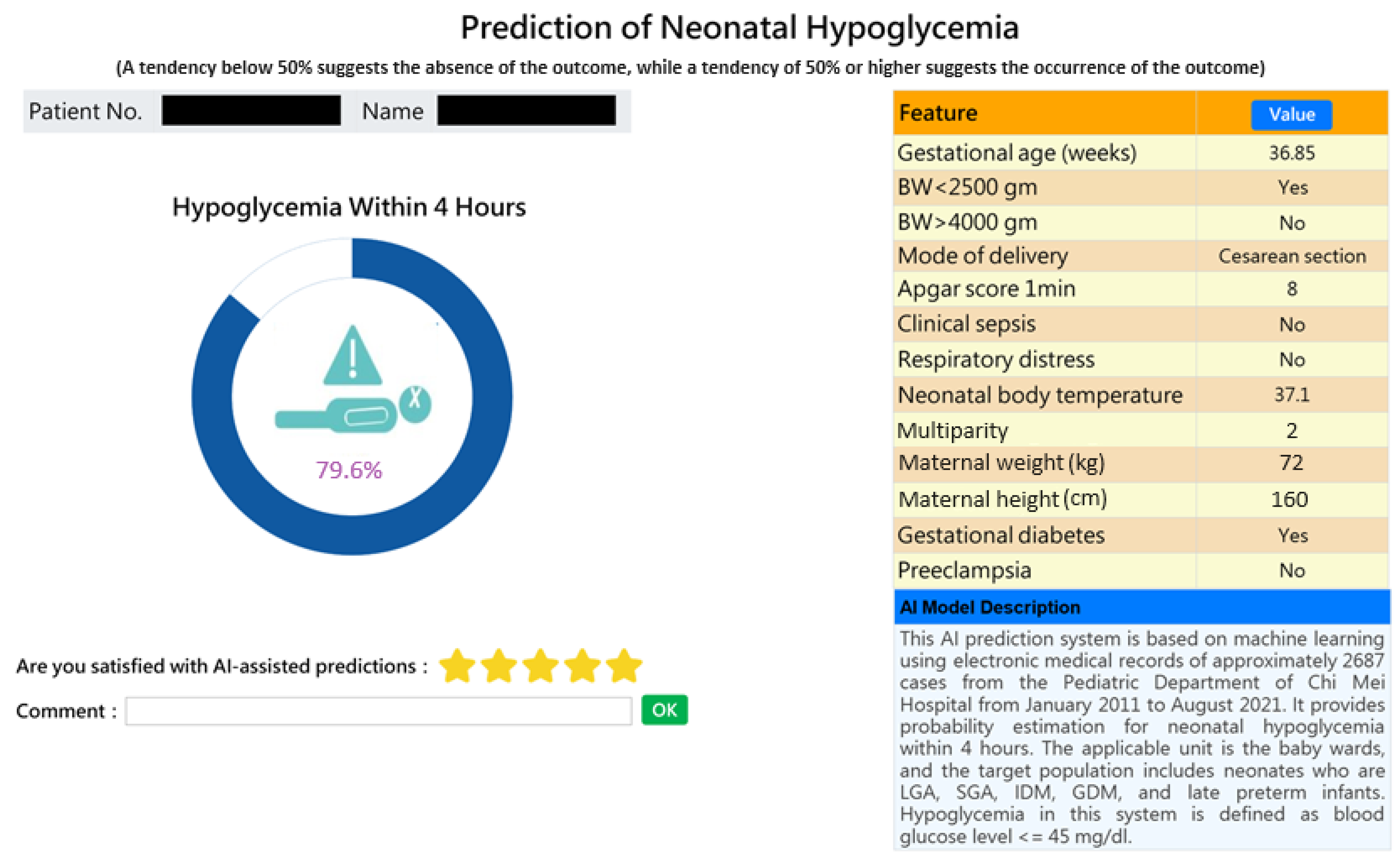
2.4. Model Deployment with HIS
2.5. Clinical Validation of AI Assistance
3. Results
3.1. Demographics of the Hypoglycemic and Non-Hypoglycemic Groups
3.2. Correlation between Feature Variables and Hypoglycemia
3.3. The Selected Features
3.4. The Predictive Models Using the 12 Feature Variables
3.5. Explainability of the Best Prediction Model
3.6. Computer-Assisted Prediction Application Development
3.7. Result of Clinical Validation of AI Assistance
4. Discussion
4.1. Main Findings and Contribution
4.2. Feature Importance Results and Clinical Implication
4.3. Comparison with Related Studies
4.4. Integrating the Model into the Existing Hospital Information System (HIS)
4.5. Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Algorithm | Hyper-Parameters |
|---|---|
| Logistic Regression (LR) | LR__C: [0.001, 0.01, 0.1, 1, 10, 100] LR__max_iter: [100, 200, 300, 400, 500, 1000] LR__penalty: [l1, l2] LR__class_weight: [‘balanced’, None] LR__solver: [‘lbfgs’, ‘saga’, ‘liblinear’] |
| Random Forest (RF) | n_estimators: [100, 200, 300, 400, 500] max_depth: [None, 4, 5, 6, 8, 10, 20, 30] class_weight: [‘balanced’, {0: 1, 1: 3}] min_samples_split: [6, 8, 10, 12, 15, 20, 30] min_samples_leaf: [5, 6, 8, 10, 15] max_features: [‘auto’, ‘sqrt’, ‘log2’] |
| Support Vector Machine (SVM) | SVM__kernel: [rbf, linear] SVM__gamma: [1e-3, 1e-4] SVM__C: [1, 10, 100, 500] shrinking: True, False |
| LightGBM | learning_rate: [0.001, 0.01, 0.1, 1] n_estimators: [100, 200, 300, 400, 500] max_depth: [4, 5, 10, 12, 15, 20, 30] random_state: [8, 16, 42, 57, 66] class_weight: [‘balanced’] importance_type: [‘gain’] |
| MLP (Multi-layer Perceptron) | hidden_layer_sizes: [(50, 30)] learning_rate_init: [1e-3, 1e-2, 1e-1] max_iter: [200, 100, 50] early_stopping: [True, False] alpha: [1e-3, 1e-4, 1e-5] |
| XGBoost | n_estimators: [100, 200, 300, 400, 500] learning_rate: [0.001, 0.01, 0.05, 0.1] gamma: [0, 5, 10] scale_pos_weight: [1, 2, 3] |
| Voting | Voting: [hard, soft] |
| Stacking | final_estimator: [Logistic regression] |
| AdaBoost | base_estimator: [DecisionTreeClassifier] max_depth: [1, 2, 3, 5] n_estimators: [50, 100, 200, 400] learning_rate: [0.001, 0.01, 0.1, 1.0] |






References
- Johnson, T.S. Hypoglycemia and the Full-Term Newborn: How Well Does Birth Weight for Gestational Age Predict Risk? J. Obstet. Gynecol. Neonatal. Nurs. 2003, 32, 48–57. [Google Scholar] [CrossRef] [PubMed]
- Cole, M.D. New Factors Associated with the Incidence of Hypoglycemia: A Research Study. Neonatal. Netw. 1991, 10, 47–50. [Google Scholar]
- Cole, M.D.; Peevy, K. Hypoglycemia in Normal Neonates Appropriate for Gestational Age. J. Perinatol. 1994, 14, 118–120. [Google Scholar]
- Srinivasan, G.; Pildes, R.S.; Cattamanchi, G.; Voora, S.; Lilien, L.D. Plasma Glucose Values in Normal Neonates: A New Look. J. Pediatr. 1986, 109, 114–117. [Google Scholar] [CrossRef] [PubMed]
- Singhal, P.K.; Singh, M.; Paul, V.K.; Deorari, A.K.; Ghorpade, M.G.; Malhotra, A. Neonatal Hypoglycemia—Clinical Profile and Glucose Requirements. Indian Pediatr. 1992, 29, 167–171. [Google Scholar] [PubMed]
- Adamkin, D.H. Postnatal Glucose Homeostasis in Late-Preterm and Term Infants. Pediatrics 2011, 127, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Yunarto, Y.; Sarosa, G. Risk Factors of Neonatal Hypoglycemia. Paediatr. Indones. 2019, 59, 252–256. [Google Scholar] [CrossRef]
- Bromiker, R.; Perry, A.; Kasirer, Y.; Einav, S.; Klinger, G.; Levy-Khademi, F. Early Neonatal Hypoglycemia: Incidence and Risk Factors. A Cohort Study Using Universal Point of Care Screening. J. Matern. Fetal Neonatal Med. 2019, 32, 786–792. [Google Scholar] [CrossRef]
- Wang, L.Y.; Wang, L.Y.; Wang, Y.L.; Ho, C.H. Early Neonatal Hypoglycemia in Term and Late Preterm Small for Gestational Age Newborns. Pediatr. Neonatol. 2023, 64, 538–546. [Google Scholar] [CrossRef]
- Armentrout, D.; Caple, J. Newborn Hypoglycemia. J. Pediatr. Health Care 1999, 13, 2–6. [Google Scholar] [CrossRef]
- Aynsley-Green, A. Glucose: A Fuel for Thought. J. Pediatr. Child Health 1991, 27, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Cornblath, M.; Hawdon, J.M.; Williams, A.F.; Aynsley-Green, A.; Ward-Platt, M.P.; Schwartz, R. Controversies Regarding Definition of Neonatal Hypoglycemia: Suggested Operational Thresholds. Pediatrics 2000, 105, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Lucas, A.; Morley, R.; Cole, T.J. Adverse Neurodevelopmental Outcome of Moderate Hypoglycemia. BMJ 1988, 297, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Pildes, R.S.; Cornblath, M.; Warren, I.; Page-El, E.; Di Menza, S.; Merritt, D.M.; Peeva, A. A Prospective Controlled Study of Neonatal Hypoglycemia. Pediatrics 1974, 54, 5–14. [Google Scholar] [PubMed]
- WHO. Guidelines on Drawing Blood: Best Practices in Phlebotomy; WHO Document Production Services: Geneva, Switzerland, 2010. [Google Scholar]
- Wiens, J.; Shenoy, E.S. Machine Learning for Healthcare: On the Verge of a Major Shift in Healthcare Epidemiology. Clin. Infect. Dis. 2018, 66, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.A.; Battegay, M.; Juchler, F.; Vogt, J.E.; Widmer, A.F. Introduction to Machine Learning in Digital Healthcare Epidemiology. Infect. Control Hosp. Epidemiol. 2018, 38, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
- Zihni, E.; Madai, V.I.; Livne, M.; Galinovic, I.; Khalil, A.A.; Fiebach, J.B.; Frey, D. Opening the black box of artificial intelligence for clinical decision support: A study predicting stroke outcome. PLoS ONE 2020, 15, e0231166. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.I.; Hsu, C.C.; Kao, Y.; Chen, C.-J.; Kuo, Y.-W.; Hsu, S.-L.; Liu, T.-L.; Lin, H.-J.; Wang, J.-J.; Liu, C.-F.; et al. Real-time AI prediction for major adverse cardiac events in emergency department patients with chest pain. Scand. J. Trauma Resusc. Emerg. Med. 2020, 28, 93. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.-H.; Hsu, C.-C.; Chen, C.-J.; Hsu, S.-L.; Liu, T.-L.; Lin, H.-J.; Wang, J.J.; Liu, C.-F.; Huang, C.-C. Predicting outcomes in older ED patients with influenza in real time using a big data-driven and machine learning approach to the hospital information system. BMC Geriatr. 2021, 21, 280. [Google Scholar] [CrossRef]
- Chen, Y.M.; Kao, Y.; Hsu, C.C.; Chen, C.J.; Ma, Y.S.; Shen, Y.T.; Liu, T.L.; Hsu, S.L.; Lin, H.J.; Wang, J.J. Real-time interactive artificial intelligence of things–based prediction for adverse outcomes in adult patients with pneumonia in the emergency department. Acad. Emerg. Med. 2021, 28, 1277–1285. [Google Scholar] [CrossRef]
- Moser, E.; Narayan, G. Improving breast cancer care coordination and symptom management by using AI driven predictive toolkits. Breast 2020, 50, 25–29. [Google Scholar] [CrossRef]
- Awan, S.E.; Sohel, F.; Sanfilippo, F.M.; Bennamoun, M.; Dwivedi, G. Machine learning in heart failure: Ready for prime time. Curr. Opin. Cardiol. 2018, 33, 190–198. [Google Scholar] [CrossRef]
- Golas, S.B.; Shibahara, T.; Agboola, S.; Otaki, H.; Sato, J.; Nakae, T.; Hisamitsu, T.; Kojima, G.; Felsted, J.; Kakarmath, S.; et al. A machine learning model to predict the risk of 30-day readmissions in patients with heart failure: A retrospective analysis of electronic medical records data. BMC Med. Inform. Decis. Mak. 2018, 18, 44. [Google Scholar] [CrossRef] [PubMed]
- Liao, K.M.; Liu, C.F.; Chen, C.J.; Shen, Y.T. Machine learning approaches for predicting acute respiratory failure, ventilator dependence, and mortality in chronic obstructive pulmonary disease. Diagnostics 2021, 20, 2396. [Google Scholar] [CrossRef] [PubMed]
- Fasihfar, Z.; Rokhsati, H.; Sadeghsalehi, H.; Ghaderzadeh, M.; Gheisari, M. AI-driven malaria diagnosis: Developing a robust model for accurate detection and classification of malaria parasites. Iran. J. Blood Cancer 2023, 15, 112–124. [Google Scholar] [CrossRef]
- Ghaderzadeh, M.; Asadi, F.; Ghorbani, N.R.; Almasi, S.; Taami, T. Toward artificial intelligence (AI) applications in the determination of COVID-19 infection severity: Considering AI as a disease control strategy in future. Iran. J. Blood Cancer 2023, 15, 93–111. [Google Scholar] [CrossRef]
- Gheisari, M.; Ghaderzadeh, M.; Li, H.; Taami, T.; Fernández-Campusano, C.; Sadeghsalehi, H.; Afzaal Abbasi, A. Mobile Apps for COVID-19 Detection and Diagnosis for Future Pandemic Control: Multidimensional Systematic Review. JMIR Mhealth Uhealth 2024, 12, e58810. [Google Scholar] [CrossRef]
- Teji, J.S.; Jain, S.; Gupta, S.K.; Suri, J.S. Machine Learning-Based Paradigm for Prediction of Neonatal and Infant Risk of Death. Comput. Biol. Med. 2022, 147, 105639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Xiao, Q.; Luo, J. Infant Death Prediction Using Machine Learning: A Population-Based Retrospective Study. Comput. Biol. Med. 2023, 165, 107423. [Google Scholar] [CrossRef]
- Malak, J.S.; Zeraati, H.; Nayeri, F.S.; Safdari, R.; Shahraki, A.D. Neonatal Intensive Care Decision Support Systems Using Artificial Intelligence Techniques: A Systematic Review. Artif. Intell. Rev. 2019, 52, 2685–2794. [Google Scholar] [CrossRef]
- Mani, S.; Ozdas, A.; Aliferis, C.; Varol, H.A.; Chen, Q.; Carnevale, R.; Chen, Y.; Romano-Keeler, J. Medical Decision Support Using Machine Learning for Early Detection of Late-Onset Neonatal Sepsis. J. Am. Med. Inform. Assoc. 2014, 21, 326–336. [Google Scholar] [CrossRef]
- Hamilton, E.F.; Dyachenko, A.; Ciampi, A.; Maurel, K.; Warrick, P.A.; Garite, T.J. Estimating Risk of Severe Neonatal Morbidity in Preterm Births Under 32 Weeks of Gestation. J. Matern. Fetal. Neonatal. Med. 2020, 33, 73–80. [Google Scholar] [CrossRef]
- Mago, N.; Srivastava, S.; Shirwaikar, R.D.; Acharya, U.D.; Lewis, L.E.S.; Shivakumar, M. Prediction of Apnea of Prematurity in Neonates Using Support Vector Machines and Random Forests. In Proceedings of the 2016 International Conference on Contemporary Computing and Informatics (IC3I), Greater Noida, India, 14–17 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 693–697. [Google Scholar]
- Warman, P.I.; Seas, A.; Satyadev, N.; Adil, S.M.; Kolls, B.J.; Haglund, M.M.; Dunn, T.W.; Fuller, A.T. Machine Learning for Predicting In-Hospital Mortality After Traumatic Brain Injury in Both High-Income and Low- and Middle-Income Countries. Neurosurgery 2020, 90, 605–612. [Google Scholar] [CrossRef]
- Liu, C.L.; Xu, Y.Q.; Wu, H.; Chen, S.S.; Guo, J.J. Correlation and Interaction Visualization of Altmetric Indicators Extracted from Scholarly Social Network Activities: Dimensions and Structure. J. Med. Internet Res. 2013, 15, e259–e277. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-Sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Hur, C.; Wi, J.A.; Kim, Y.B. Facilitating the Development of Deep Learning Models with Visual Analytics for Electronic Health Records. Int. J. Environ. Res. Public Health 2020, 17, 8303. [Google Scholar] [CrossRef]
- Mienye, I.D.; Sun, Y. A Survey of Ensemble Learning: Concepts, Algorithms, Applications, and Prospects. IEEE Access 2022, 10, 99129–99149. [Google Scholar] [CrossRef]
- Zweig, M.H.; Campbell, G. Receiver-Operating Characteristic (ROC) Plots: A Fundamental Evaluation Tool in Clinical Medicine. Clin. Chem. 1993, 39, 561–567. [Google Scholar] [CrossRef]
- Bradley, A.P. The Use of the Area Under the ROC Curve in the Evaluation of Machine Learning Algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver Operating Characteristic (ROC) Curve Analysis for Medical Diagnostic Test Evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. [Google Scholar]
- Ogami, C.; Tsuji, Y.; Seki, H.; Kawano, H.; To, H.; Matsumoto, Y.; Hosono, H. An Artificial Neural Network-Pharmacokinetic Model and its Interpretation Using Shapley Additive Explanations. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 760–768. [Google Scholar] [CrossRef]
- Tsai, W.C.; Liu, C.F.; Lin, H.J.; Hsu, C.C.; Ma, Y.S.; Chen, C.J.; Huang, C.C.; Chen, C.C. Design and Implementation of a Comprehensive AI Dashboard for Real-Time Prediction of Adverse Prognosis of ED Patients. Healthcare 2022, 10, 1498. [Google Scholar] [CrossRef]
- Amarasingham, R.; Patzer, R.E.; Huesch, M.; Nguyen, N.Q.; Xie, B. Implementing Electronic Health Care Predictive Analytics: Considerations and Challenges. Health Aff. 2014, 33, 1148–1154. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- Betts, K.S.; Kisely, S.; Alati, R. Predicting Neonatal Respiratory Distress Syndrome and Hypoglycaemia Prior to Discharge: Leveraging Health Administrative Data and Machine Learning. J. Biomed. Inform. 2021, 114, 103651. [Google Scholar] [CrossRef]
- Shukla, V.; Battarbee, A.N.; Melvin, R.; Godwin, R. Prediction of neonatal hypoglycemia risk from maternal continuous glucose monitoring data using artificial intelligence. Am. J. Obstet. Gynecol. 2023, 228, S759–S760. [Google Scholar] [CrossRef]
- Gerard, J.; Stuebe, A.; Patterson, E.S. Using machine learning to develop a predictive model of infant hypoglycemia based on maternal and infant variables in an electronic health record. In Proceedings of the International Symposium on Human Factors and Ergonomics in Health Care, Orlando, FL, USA, 26–29 March 2023; Volume 12. [Google Scholar]
- Alemu, B.T.; Olayinka, O.; Baydoun, H.A.; Hoch, M.; Akpinar-Elci, M. Neonatal Hypoglycemia in Diabetic Mothers: A Systematic Review. Curr. Pediatr. Res. 2017, 21, 42–53. [Google Scholar]




| Variables | Euglycemia (n = 1374, 73.1%) | Hypoglycemia (n = 506, 26.9%) | p-Value |
|---|---|---|---|
| Sex, n (%) | 0.372 | ||
| male | 718 (52.26) | 252 (49.80) | |
| female | 656 (47.74) | 254 (50.20) | |
| Gestational age, mean (SD) | 38.64 (1.33) | 37.82 (1.56) | <0.001 |
| BW < 2500 gm, n (%) | 326 (23.73) | 191 (37.75) | <0.001 |
| BW > 4000 gm, n (%) | 66 (4.80) | 30 (5.93) | 0.387 |
| Mode of delivery, n (%) | <0.001 | ||
| Vaginal delivery | 825 (60.04) | 193 (38.14) | |
| Cesarean section | 549 (39.96) | 313 (61.86) | |
| Head circumference, mean (SD) | 33.30 (1.77) | 33.27 (1.84) | 0.826 |
| Chest circumference, mean (SD) | 31.45 (2.27) | 31.36 (2.73) | 0.514 |
| Birth length, mean (SD) | 49.19 (2.91) | 48.91 (3.32) | 0.102 |
| Apgar score 1 min, mean (SD) | 7.95 (0.29) | 7.89 (0.43) | 0.005 |
| Apgar score 5 min, mean (SD) | 8.99 (0.13) | 8.97 (0.18) | 0.065 |
| Clinical sepsis, n (%) | 49 (3.57) | 33 (6.52) | 0.008 |
| Respiratory distress, n (%) | 219 (15.94) | 157 (31.03) | <0.001 |
| Polycythemia, n (%) | 13 (0.95) | 17 (3.36) | <0.001 |
| Body temperature, mean (SD) | 36.55 (0.58) | 36.40 (0.64) | <0.001 |
| Maternal age, mean (SD) | 32.12 (4.78) | 32.45 (4.74) | 0.181 |
| Maternal weight, mean (SD) | 67.48 (11.55) | 69.88 (11.92) | <0.001 |
| Maternal height, mean (SD) | 159.30 (5.44) | 159.96 (5.69) | 0.025 |
| Multiparity, mean (SD) | 1.54 (0.79) | 1.60 (0.76) | 0.090 |
| Prior delivery of SGA, n (%) | 93 (6.77) | 29 (5.73) | 0.481 |
| Prior delivery of LGA, n (%) | 51 (3.71) | 24 (4.74) | 0.379 |
| Gestational diabetes, n (%) | 186 (13.54) | 87 (17.19) | 0.055 |
| Preeclampsia, n (%) | 49 (3.57) | 29 (5.73) | 0.050 |
| HBsAg (+), n (%) | 85 (6.19) | 34 (6.72) | 0.753 |
| PROM > 24 h, n (%) | 24 (1.75) | 15 (2.96) | 0.144 |
| Variables | Correlation Coefficients |
|---|---|
| Gestational age | −0.2449 |
| Mode of delivery | 0.2133 |
| Respiratory distress | 0.1711 |
| BW-2500 | 0.1403 |
| Maternal weight | 0.1178 |
| Body temperature | −0.1144 |
| Apgar score 1 min | −0.1070 |
| Polycythemia | 0.0833 |
| Apgar score 5 min | −0.0707 |
| Gestational diabetes | 0.0678 |
| Maternal height | 0.0677 |
| Multiparity | 0.0650 |
| BW-4000 | 0.0573 |
| Maternal age | 0.0539 |
| Preeclampsia | 0.0506 |
| Sex | −0.0489 |
| Prior delivery of LGA | 0.0456 |
| Clinical sepsis | 0.0325 |
| Prior delivery of SGA | −0.0321 |
| Birth length | −0.0309 |
| PROM 24 h | 0.0295 |
| Chest circumference | −0.0292 |
| Head circumference | 0.0185 |
| HBSAg (+) | 0.0065 |
| Predictive Models | Accuracy | Sensitivity | Specificity | F1 Score | Precision | AUC |
|---|---|---|---|---|---|---|
| Stacking | 0.689 | 0.682 | 0.692 | 0.541 | 0.448 | 0.739 |
| Random Forest | 0.658 | 0.682 | 0.649 | 0.517 | 0.417 | 0.732 |
| Voting | 0.675 | 0.682 | 0.673 | 0.530 | 0.434 | 0.732 |
| AdaBoost | 0.646 | 0.682 | 0.632 | 0.509 | 0.405 | 0.723 |
| XGBoost | 0.647 | 0.691 | 0.631 | 0.513 | 0.408 | 0.722 |
| Logistic Regression | 0.675 | 0.687 | 0.671 | 0.532 | 0.434 | 0.721 |
| MLP | 0.675 | 0.682 | 0.673 | 0.530 | 0.434 | 0.721 |
| LightGBM | 0.646 | 0.682 | 0.632 | 0.509 | 0.405 | 0.717 |
| SVM | 0.656 | 0.650 | 0.658 | 0.504 | 0.411 | 0.713 |
| Study | This Study | Betts et al. [47] | Shukla et al. [48] | Gerard. et al. [49] |
|---|---|---|---|---|
| Study design and setting | Retrospective study Routine administrative data on neonates born ≥35 weeks of gestational age. | Retrospective study Routine administrative data on neonates born <39 weeks of gestational age. | Retrospective study Maternal continuous glucose monitoring (CGM) data for neonates born to mothers with diabetes. | Retrospective study Electronic health record (EHR) neonates |
| Sample size | 2687 | 154,755 | 90 | 13,476 |
| machine learning algorithms | Logistic Regression, Random Forest, Light GBM, XG Boost, MLP | Gradient boosted trees, Logistic regression | Multiple Representations Sequence Miner (MrSQM) framework | Logistic regression |
| Feature variables | The 13 variables consist of maternal and neonatal clinical data routinely collected and recorded immediately after birth. | The 528 variables include all available maternal clinical, demographic, and lifestyle data, as well as neonatal clinical data routinely collected and recorded immediately after birth | 1 variable maternal continuous glucose monitoring (CGM) data | Maternal data (All acute and chronic diagnoses for maternal patients and diagnosed issues in newborn patients) and neonatal data (all conditions billed for during care) |
| outcome | early neonatal hypoglycemia | neonatal hypoglycemia | neonatal hypoglycemia, | neonatal hypoglycemia |
| Testing results | AUC of 0.735 | AUC = 0.832 | AUC of 0.74 | p < 0.001 |
| Best predicting model | Random Forest | Gradient boosted trees | Multiple Representations Sequence Miner (MrSQM) framework | binary logistic regression model |
| Real world implementation | Yes | None | None | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.-Y.; Wang, L.-Y.; Sung, M.-I.; Lin, I.-C.; Liu, C.-F.; Chen, C.-J. Implementing Explainable Machine Learning Models for Practical Prediction of Early Neonatal Hypoglycemia. Diagnostics 2024, 14, 1571. https://doi.org/10.3390/diagnostics14141571
Wang L-Y, Wang L-Y, Sung M-I, Lin I-C, Liu C-F, Chen C-J. Implementing Explainable Machine Learning Models for Practical Prediction of Early Neonatal Hypoglycemia. Diagnostics. 2024; 14(14):1571. https://doi.org/10.3390/diagnostics14141571
Chicago/Turabian StyleWang, Lin-Yu, Lin-Yen Wang, Mei-I Sung, I-Chun Lin, Chung-Feng Liu, and Chia-Jung Chen. 2024. "Implementing Explainable Machine Learning Models for Practical Prediction of Early Neonatal Hypoglycemia" Diagnostics 14, no. 14: 1571. https://doi.org/10.3390/diagnostics14141571
APA StyleWang, L.-Y., Wang, L.-Y., Sung, M.-I., Lin, I.-C., Liu, C.-F., & Chen, C.-J. (2024). Implementing Explainable Machine Learning Models for Practical Prediction of Early Neonatal Hypoglycemia. Diagnostics, 14(14), 1571. https://doi.org/10.3390/diagnostics14141571








