A Novel Hybrid Machine Learning-Based System Using Deep Learning Techniques and Meta-Heuristic Algorithms for Various Medical Datatypes Classification
Abstract
1. Introduction
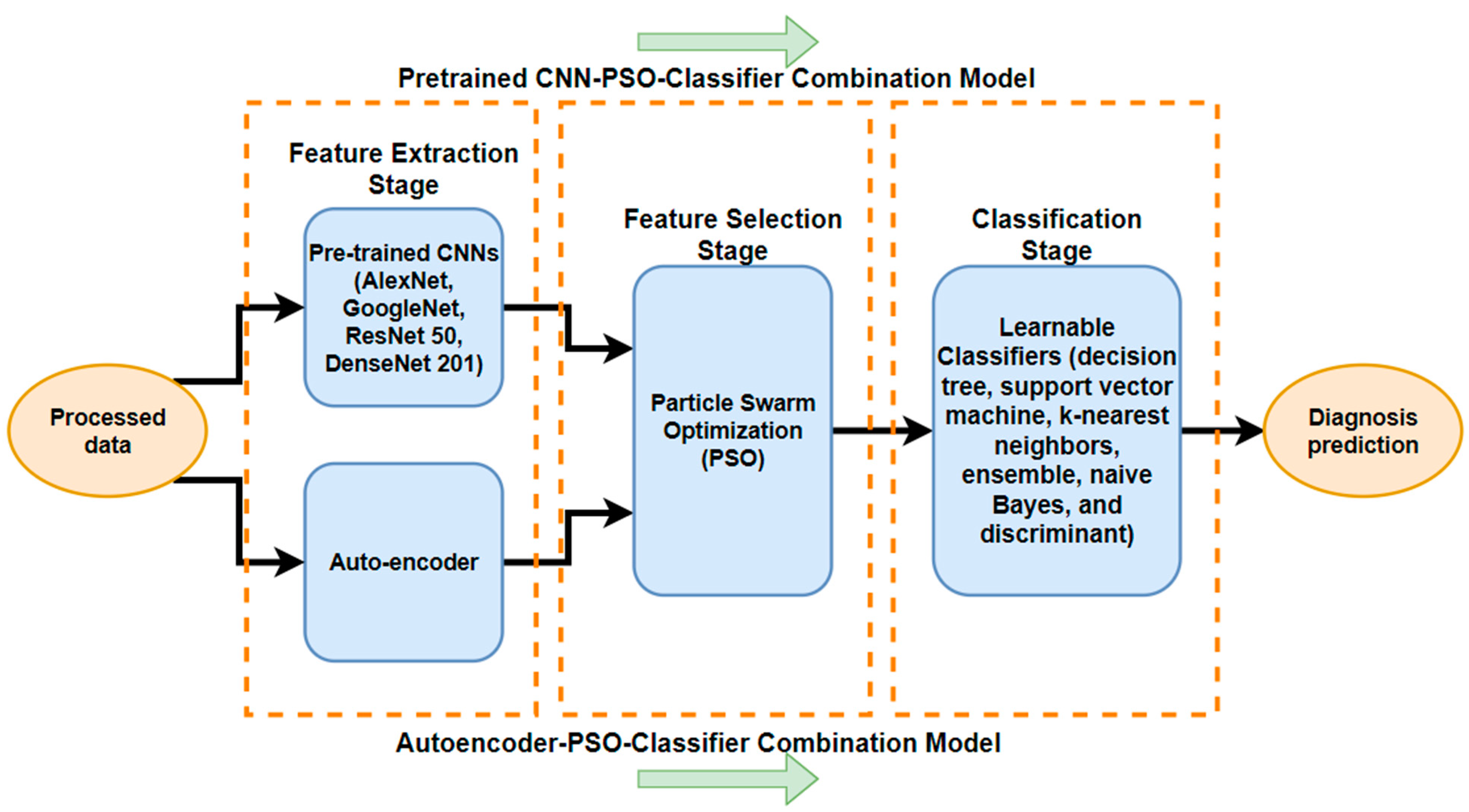
- Development of interpretable models: We focus on developing models that not only provide accurate predictions but also offer insights into features by extracting features from different medical image datasets using several deep learning techniques; autoencoders; and pre-trained CNNs (namely, AlexNet, GoogLeNet, ResNet50, and DenseNet201). These are coupled with the next step of the PSO meta-heuristic algorithm, and the last step is that the results are predicted by a different classifier (SVM, KNN, DT, etc.). Ultimately, we utilize the particle swarm optimization (PSO) method to enhance detection precision by selecting the most significant and effective features while eliminating redundant ones from different datasets.
- The results of this study were proven through the use of two different medical datasets, the first being for a brain tumor imaged with MRI, and the second being a completely different dataset for the lungs of COVID-19 patients that were imaged with CXR.
- We demonstrate the novelty and superiority of this proposed feature selection combination over existing diagnostic baseline models.
- We validate the effectiveness of the PSO algorithm in feature selection compared to genetic algorithms (GAs) and ant colony optimization (ACO) for various medical datasets, including MRI and X-ray images. This algorithm showed superiority over other heuristic algorithms in different datasets.
- This proposed method will aid in improving understanding and interpretation by medical professionals.
2. Related Works
3. Material and Methods
3.1. Datasets
3.1.1. COVID-19 Dataset
3.1.2. Brain Tumor Dataset
3.1.3. Dataset Preprocessing
3.2. Methodology
3.2.1. Feature Extraction with CNN
- Alexnet: Input size is 227 × 227 × 3, and the number of features is 4096.
- Googlenet: Input size is 224 × 224 × 3, and the number of features is 1000.
- Resnet50: Input size is 224 × 224 × 3, and the number of features is 2048.
- Densenet201: Input size is 224 × 224 × 3, and the number of features is 1920.
3.2.2. Feature Extraction with Autoencoders
Sparse Autoencoders
Sparsity Regularization
L2 Regularization
Deep Sparse Autoencoders
3.2.3. Particle Swarm Optimization Algorithm for Feature Selection
PSO Explanation and Literature
PSO for Feature Selection
4. Results and Discussion
4.1. Autoencoder with PSO for COVID-19 Dataset
4.2. Autoencoder with PSO for Brain Tumor Dataset
4.3. Pre-Trained CNN with PSO for COVID-19 Dataset
4.4. Pre-Trained CNN with PSO for Brain Tumor Dataset
4.5. Comparison Tables
5. Conclusions
6. Limitations
- Its reliance on supervised learning with labeled data limits automation and potential inability to fully exploit deep learning’s potential.
- The constraint of a small dataset in the medical field restricts the generalizability and robustness of an approach.
- The need for human participation in the diagnostic prediction process hinders scalability and real-time application in clinical settings.
- The deep learning models employed are computationally intensive, requiring substantial hardware resources for training and inference, which may limit their applicability in resource-constrained settings.
- The complexity of segmenting brain tumors and COVID-19 lesions, which may require specialized expertise and resources.
7. Future Work
- We should aim to include a larger and more diverse dataset, covering various demographics and imaging devices, to improve the robustness and generalizability of the models.
- We should address data labeling challenges, as the process can be time-consuming and error-prone, affecting model quality.
- Involving close collaboration with healthcare professionals to ensure the method’s relevance, feasibility, and usability in clinical practice can lead to locating the exact segmentation of the tumor.
- After the classification of these tumors, the segmentation of tumors is needed in order to perform exact detection: knowing the shape and size will aid the doctors in identifying the level at which the tumor is.
- Focus on optimizing model architectures and hyperparameters to enhance performance while reducing computational requirements, making the models more feasible for real-time clinical use.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Parameter | Abbreviation |
| Accuracy | ACC |
| Sensitivity, Recall, hit rate, True Positive Rate | TPR |
| Specificity, Selectivity, True Negative Rate | TNR |
| False positive rate | FPR |
| False negative rate | FNR |
| Precision, Positive Predictive Value | PPV |
| Negative Predictive Value | NPV |
| F1 Score | F1 Score |
| Misclassification rate | MR |
References
- Devi, A. Brain tumor detection. IJITR 2015, 3, 1950–1952. [Google Scholar]
- Bhattacharyya, D.; Kim, T. Brain tumor detection using MRI image analysis. In Proceedings of the International Conference on Ubiquitous Computing and Multimedia Applications, Daejeon, Republic of Korea, 13–15 April 2011; Springer: Berlin/Heidelberg, Germany, 2011; pp. 307–314. [Google Scholar]
- Zhang, Y.-D.; Wu, L. An MR brain images classifier via principal component analysis and kernel support vector machine. Prog. Electromagn. Res. 2012, 130, 369–388. [Google Scholar] [CrossRef]
- Lippi, G.; Plebani, M.; Henry, B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: A meta-analysis. Clin. Chim. Acta 2020, 506, 145–148. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, X.; Chen, Z.; Duan, J.; Hashimoto, K.; Yang, L.; Liu, C.; Yang, C. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain. Behav. Immun. 2020, 87, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Yang, Y.; Du, Y.; Peng, F.; Hu, P.; Wang, R.; Yin, M.; Li, T.; Tu, L.; Sun, J. Clinical pathway for early diagnosis of COVID-19: Updates from experience to evidence-based practice. Clin. Rev. Allergy Immunol. 2020, 59, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Lalmuanawma, S.; Hussain, J.; Chhakchhuak, L. Applications of machine learning and artificial intelligence for COVID-19 (SARS-CoV-2) pandemic: A review. Chaos Solitons Fractals 2020, 139, 110059. [Google Scholar] [CrossRef] [PubMed]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef] [PubMed]
- Al-Tawfiq, J.A.; Memish, Z.A. Diagnosis of SARS-CoV-2 infection based on CT scan vs. RT-PCR: Reflecting on experience from MERS-CoV. J. Hosp. Infect. 2020, 105, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.-S.; Cui, Z.; Xiao, R.; Gandomi, A.H.; Karamanoglu, M. Swarm Intelligence and Bio-Inspired Computation: Theory and Applications; Newnes: London, UK, 2013; ISBN 0124051774. [Google Scholar]
- Barolli, A.; Sakamoto, S.; Barolli, L.; Takizawa, M. Performance analysis of simulation system based on particle swarm optimization and distributed genetic algorithm for WMNs considering different distributions of mesh clients. In Proceedings of the International Conference on Innovative Mobile and Internet Services in Ubiquitous Computing, Matsue, Japan, 4–6 July 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 32–45. [Google Scholar]
- Dorigo, M.; Birattari, M.; Stutzle, T. Ant colony optimization. IEEE Comput. Intell. Mag. 2006, 1, 28–39. [Google Scholar] [CrossRef]
- Seetha, J.; Raja, S.S. Brain tumor classification using convolutional neural networks. Biomed. Pharmacol. J. 2018, 11, 1457. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. Mri-based brain tumor classification using ensemble of deep features and machine learning classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef] [PubMed]
- Gumaei, A.; Hassan, M.M.; Hassan, M.R.; Alelaiwi, A.; Fortino, G. A hybrid feature extraction method with regularized extreme learning machine for brain tumor classification. IEEE Access 2019, 7, 36266–36273. [Google Scholar] [CrossRef]
- Yang, G.; Zhang, Y.; Yang, J.; Ji, G.; Dong, Z.; Wang, S.; Feng, C.; Wang, Q. Automated classification of brain images using wavelet-energy and biogeography-based optimization. Multimed. Tools Appl. 2016, 75, 15601–15617. [Google Scholar] [CrossRef]
- Shankar, K.; Mohanty, S.N.; Yadav, K.; Gopalakrishnan, T.; Elmisery, A.M. Automated COVID-19 diagnosis and classification using convolutional neural network with fusion based feature extraction model. Cogn. Neurodyn. 2023, 17, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Júnior, D.A.D.; da Cruz, L.B.; Diniz, J.O.B.; da Silva, G.L.F.; Junior, G.B.; Silva, A.C.; de Paiva, A.C.; Nunes, R.A.; Gattass, M. Automatic method for classifying COVID-19 patients based on chest X-ray images, using deep features and PSO-optimized XGBoost. Expert Syst. Appl. 2021, 183, 115452. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Ashok, A.; Ansari, M.A. Brain tumor classification using hybrid model of PSO and SVM classifier. In Proceedings of the 2018 International Conference on Advances in Computing, Communication Control and Networking (ICACCCN), Greater Noida, India, 12–13 October 2018; pp. 1022–1026. [Google Scholar]
- Yu, J.-B. Evolutionary manifold regularized stacked denoising autoencoders for gearbox fault diagnosis. Knowl.-Based Syst. 2019, 178, 111–122. [Google Scholar] [CrossRef]
- El-Kenawy, E.-S.M.; Ibrahim, A.; Mirjalili, S.; Eid, M.M.; Hussein, S.E. Novel feature selection and voting classifier algorithms for COVID-19 classification in CT images. IEEE Access 2020, 8, 179317–179335. [Google Scholar] [CrossRef] [PubMed]
- Navaneeth, B.; Suchetha, M. PSO optimized 1-D CNN-SVM architecture for real-time detection and classification applications. Comput. Biol. Med. 2019, 108, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, K.; Kaya, Y.; Kuncan, M.; Ertunç, H.M. Brain tumor classification using modified local binary patterns (LBP) feature extraction methods. Med. Hypotheses 2020, 139, 109696. [Google Scholar] [CrossRef] [PubMed]
- Kutlu, H.; Avcı, E. A novel method for classifying liver and brain tumors using convolutional neural networks, discrete wavelet transform and long short-term memory networks. Sensors 2019, 19, 1992. [Google Scholar] [CrossRef] [PubMed]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef] [PubMed]
- Hashemzehi, R.; Mahdavi, S.J.S.; Kheirabadi, M.; Kamel, S.R. Detection of brain tumors from MRI images base on deep learning using hybrid model CNN and NADE. Biocybern. Biomed. Eng. 2020, 40, 1225–1232. [Google Scholar] [CrossRef]
- Khan, M.A.; Ashraf, I.; Alhaisoni, M.; Damaševičius, R.; Scherer, R.; Rehman, A.; Bukhari, S.A.C. Multimodal brain tumor classification using deep learning and robust feature selection: A machine learning application for radiologists. Diagnostics 2020, 10, 565. [Google Scholar] [CrossRef] [PubMed]
- Barstugan, M.; Ozkaya, U.; Ozturk, S. Coronavirus (COVID-19) classification using ct images by machine learning methods. arXiv 2020, arXiv:2003.09424. [Google Scholar]
- Pathak, Y.; Shukla, P.K.; Tiwari, A.; Stalin, S.; Singh, S. Deep transfer learning based classification model for COVID-19 disease. IRBM 2020, 43, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Huang, W.; Cao, S.; Yang, R.; Yang, W.; Yun, Z.; Wang, Z.; Feng, Q. Enhanced performance of brain tumor classification via tumor region augmentation and partition. PLoS ONE 2015, 10, e0140381. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-R.; Lee, L.-H. Contrastive approach towards text source classification based on top-bag-of-word similarity. In Proceedings of the 22nd Pacific Asia Conference on Language, Information and Computation, Cebu City, Philippines; 2008; pp. 404–410. [Google Scholar]
- Rui, W.; Xing, K.; Jia, Y. BOWL: Bag of word clusters text representation using word embeddings. In Proceedings of the International Conference on Knowledge Science, Engineering and Management, Passau, Germany, 5–7 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 3–14. [Google Scholar]
- Thejaswini, P.; Bhat, M.B.; Prakash, M.K. Detection and classification of tumour in brain MRI. Int. J. Eng. Manufact. (IJEM) 2019, 9, 11–20. [Google Scholar]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of coronavirus disease (COVID-19) based on deep features and Support Vector Machine. Int. J. Math. Eng. Manag. Sci. 2020, 5, 643–651. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef] [PubMed]
- Ismael, M.R.; Abdel-Qader, I. Brain tumor classification via statistical features and back-propagation neural network. In Proceedings of the 2018 IEEE international conference on electro/information technology (EIT), Rochester, MI, USA, 3–5 May 2018; pp. 252–257. [Google Scholar]
- Ahmed, S.; Frikha, M.; Hussein, T.D.H.; Rahebi, J. Face Recognition System using Histograms of Oriented Gradients and Convolutional Neural Network based on with Particle Swarm Optimization. In Proceedings of the 2021 International Conference on Electrical, Communication and Computer Engineering (ICECCE), Kuala Lumpur, Malaysia, 12–13 June 2021; pp. 1–5. [Google Scholar]
- Allagwail, S.; Gedik, O.S.; Rahebi, J. Face recognition with symmetrical face training samples based on local binary patterns and the Gabor filter. Symmetry 2019, 11, 157. [Google Scholar] [CrossRef]
- Ghassemi, N.; Shoeibi, A.; Rouhani, M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control 2020, 57, 101678. [Google Scholar] [CrossRef]
- Alashik, K.M.; Yildirim, R. Human Identity Verification From Biometric Dorsal Hand Vein Images Using the DL-GAN Method. IEEE Access 2021, 9, 74194–74208. [Google Scholar] [CrossRef]
- Hussin, S.H.S.; Yildirim, R. StyleGAN-LSRO Method for Person Re-Identification. IEEE Access 2021, 9, 13857–13869. [Google Scholar] [CrossRef]
- Özkaya, U.; Öztürk, Ş.; Barstugan, M. Coronavirus (COVID-19) classification using deep features fusion and ranking technique. In Big Data Analytics and Artificial Intelligence Against COVID-19: Innovation Vision and Approach; Springer: Berlin/Heidelberg, Germany, 2020; pp. 281–295. [Google Scholar]
- Mollalo, A.; Rivera, K.M.; Vahedi, B. Artificial neural network modeling of novel coronavirus (COVID-19) incidence rates across the continental United States. Int. J. Environ. Res. Public Health 2020, 17, 4204. [Google Scholar] [CrossRef] [PubMed]
- Gozes, O.; Frid-Adar, M.; Sagie, N.; Zhang, H.; Ji, W.; Greenspan, H. Coronavirus detection and analysis on chest ct with deep learning. arXiv 2020, arXiv:2004.02640. [Google Scholar]
- Yeşilkanat, C.M. Spatio-temporal estimation of the daily cases of COVID-19 in worldwide using random forest machine learning algorithm. Chaos Solitons Fractals 2020, 140, 110210. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, D. Covid-xpert: An ai powered population screening of COVID-19 cases using chest radiography images. arXiv 2020, arXiv:2004.03042. [Google Scholar]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef] [PubMed]
- Asif, S.; Zhao, M.; Li, Y.; Tang, F.; Zhu, Y. CGO-Ensemble: Chaos Game Optimization Algorithm-Based Fusion of Deep Neural Networks for Accurate Mpox Detection. Neural Netw. 2024, 173, 106183. [Google Scholar] [CrossRef]
- Asif, S.; Zhao, M.; Tang, F.; Zhu, Y.; Zhao, B. Metaheuristics optimization-based ensemble of deep neural networks for Mpox disease detection. Neural Netw. 2023, 167, 342–359. [Google Scholar] [CrossRef] [PubMed]
- Asif, S.; Zhao, M.; Chen, X.; Zhu, Y. BMRI-NET: A deep stacked ensemble model for multi-class brain tumor classification from MRI images. Interdiscip. Sci. Comput. Life Sci. 2023, 15, 499–514. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pei, Y.; Wang, S.-H.; manuel Gorrz, J.; Zhang, Y.-D. PSTCNN: Explainable COVID-19 diagnosis using PSO-guided self-tuning CNN. Biocell 2023, 47, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Punitha, S.; Stephan, T.; Kannan, R.; Mahmud, M.; Kaiser, M.S.; Belhaouari, S.B. Detecting COVID-19 from lung computed tomography images: A swarm optimized artificial neural network approach. IEEE Access 2023, 11, 12378–12393. [Google Scholar] [CrossRef]
- Rajeev, S.K.; Rajasekaran, M.P.; Vishnuvarthanan, G.; Arunprasath, T. A biologically-inspired hybrid deep learning approach for brain tumor classification from magnetic resonance imaging using improved gabor wavelet transform and Elmann-BiLSTM network. Biomed. Signal Process. Control 2022, 78, 103949. [Google Scholar] [CrossRef]
- Rajakumar, S.; Agalya, V.; Rajeswari, R.; Pachlor, R. Political exponential deer hunting optimization-based deep learning for brain tumor classification using MRI. Signal Image Video Process. 2023, 17, 3451–3459. [Google Scholar] [CrossRef]
- Geetha, M.; Srinadh, V.; Janet, J.; Sumathi, S. Hybrid archimedes sine cosine optimization enabled deep learning for multilevel brain tumor classification using mri images. Biomed. Signal Process. Control 2024, 87, 105419. [Google Scholar] [CrossRef]
- Zhu, H.; Wei, L.; Niu, P. The novel coronavirus outbreak in Wuhan, China. Glob. Health Res. Policy 2020, 5, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, A.C.; Lee, D.T.; Misquitta, K.K.; Uno, K.; Wald, S. COVID-19 detection from chest radiographs using machine learning and convolutional neural networks. medRxiv 2020. [Google Scholar] [CrossRef]
- Cheng, J.; Yang, W.; Huang, M.; Huang, W.; Jiang, J.; Zhou, Y.; Yang, R.; Zhao, J.; Feng, Y.; Feng, Q. Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation. PLoS ONE 2016, 11, e0157112. [Google Scholar] [CrossRef]
- Møller, M.F. A scaled conjugate gradient algorithm for fast supervised learning. Neural Netw. 1993, 6, 525–533. [Google Scholar] [CrossRef]
- Olshausen, B.A.; Field, D.J. Sparse coding with an overcomplete basis set: A strategy employed by V1? Vis. Res. 1997, 37, 3311–3325. [Google Scholar] [CrossRef]
- Fernandez-Viagas, V.; Ruiz, R.; Framinan, J.M. A new vision of approximate methods for the permutation flowshop to minimise makespan: State-of-the-art and computational evaluation. Eur. J. Oper. Res. 2017, 257, 707–721. [Google Scholar] [CrossRef]
- Beni, G. Swarm intelligence. Complex Soc. Behav. Syst. Game Theory Agent-Based Models 2020, 791–818. [Google Scholar] [CrossRef]
- Nguyen, B.H.; Xue, B.; Zhang, M. A survey on swarm intelligence approaches to feature selection in data mining. Swarm Evol. Comput. 2020, 54, 100663. [Google Scholar] [CrossRef]
- Niu, W.; Feng, Z.; Feng, B.; Xu, Y.; Min, Y. Parallel computing and swarm intelligence based artificial intelligence model for multi-step-ahead hydrological time series prediction. Sustain. Cities Soc. 2021, 66, 102686. [Google Scholar] [CrossRef]
- Cho, W.K.T. An evolutionary algorithm for subset selection in causal inference models. J. Oper. Res. Soc. 2017, 69, 630–644. [Google Scholar]
- Hassan, R.; Cohanim, B.; De Weck, O.; Venter, G. A comparison of particle swarm optimization and the genetic algorithm. In Proceedings of the Collection of Technical Papers—AIAA/ASME/ASCE/AHS/ASC Structures, Structural Dynamics and Materials Conference, Austin, TX, USA, 18–21 April 2005; Volume 2, pp. 1138–1150. [Google Scholar]
- Widhiarso, W.; Yohannes, Y.; Prakarsah, C. Brain tumor classification using gray level co-occurrence matrix and convolutional neural network. IJEIS (Indones. J. Electron. Instrum. Syst.) 2018, 8, 179–190. [Google Scholar] [CrossRef]
- Belaid, O.N.; Loudini, M. Classification of Brain Tumor by Combination of Pre-Trained VGG16 CNN. J. Inf. Technol. Manag. 2020, 12, 13–25. [Google Scholar]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N. Brain tumor type classification via capsule networks. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 3129–3133. [Google Scholar]
- Li, D.; Fu, Z.; Xu, J. Stacked-autoencoder-based model for COVID-19 diagnosis on CT images. Appl. Intell. 2021, 51, 2805–2817. [Google Scholar] [CrossRef]
- Khozeimeh, F.; Sharifrazi, D.; Izadi, N.H.; Joloudari, J.H.; Shoeibi, A.; Alizadehsani, R.; Gorriz, J.M.; Hussain, S.; Sani, Z.A.; Moosaei, H. CNN AE: Convolution Neural Network combined with Autoencoder approach to detect survival chance of COVID-19 patients. arXiv 2021, arXiv:2104.08954. [Google Scholar]
- Mohanaiah, P.; Sathyanarayana, P.; GuruKumar, L. Image texture feature extraction using GLCM approach. Int. J. Sci. Res. Publ. 2013, 3, 1–5. [Google Scholar]
- Latha, Y.L.M.; Prasad, M.V.N.K. GLCM based texture features for palmprint identification system. In Computational Intelligence in Data Mining-Volume 1; Springer: Berlin/Heidelberg, Germany, 2015; pp. 155–163. [Google Scholar]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Mohanty, A.K.; Beberta, S.; Lenka, S.K. Classifying benign and malignant mass using GLCM and GLRLM based texture features from mammogram. Int. J. Eng. Res. Appl. 2011, 1, 687–693. [Google Scholar]
- Ammar, M.; Mahmoudi, S.; Stylianos, D. A Set of Texture-Based Methods for Breast Cancer Response Prediction in Neoadjuvant Chemotherapy Treatment. In Soft Computing Based Medical Image Analysis; Elsevier: Amsterdam, The Netherlands, 2018; pp. 137–147. [Google Scholar]
- Sachdeva, J.; Kumar, V.; Gupta, I.; Khandelwal, N.; Ahuja, C.K. Segmentation, feature extraction, and multiclass brain tumor classification. J. Digit. Imaging 2013, 26, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Gárate-Escamila, A.K.; El Hassani, A.H.; Andrès, E. Classification models for heart disease prediction using feature selection and PCA. Inform. Med. Unlocked 2020, 19, 100330. [Google Scholar] [CrossRef]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. Covid-caps: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef]
- Wu, X.; Liu, S.; Cao, Y.; Li, X.; Yu, J.; Dai, D.; Ma, X.; Hu, S.; Wu, Z.; Liu, X. Speech emotion recognition using capsule networks. In Proceedings of the ICASSP 2019-2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 6695–6699. [Google Scholar]
- Lee, K.Y.; Park, J.-B. Application of particle swarm optimization to economic dispatch problem: Advantages and disadvantages. In Proceedings of the 2006 IEEE PES Power Systems Conference and Exposition, Atlanta, GA, USA, 29 October–1 November 2006; pp. 188–192. [Google Scholar]
- Selvi, V.; Umarani, R. Comparative analysis of ant colony and particle swarm optimization techniques. Int. J. Comput. Appl. 2010, 5, 1–6. [Google Scholar] [CrossRef]
- Kadhim, Y.A.; Khan, M.U.; Mishra, A. Deep learning-based computer-aided diagnosis (cad): Applications for medical image datasets. Sensors 2022, 22, 8999. [Google Scholar] [CrossRef] [PubMed]

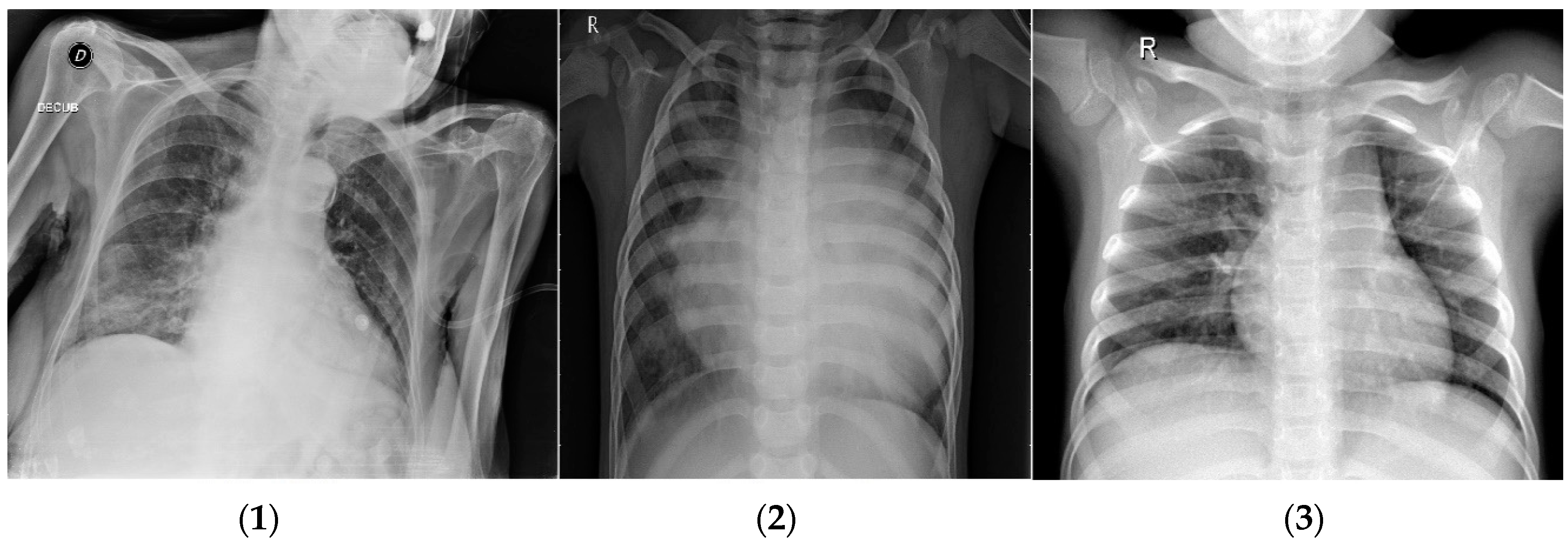







| Method | ACC | TPR | TNR | FPR | FNR | PPV | NPV | F1-Score | MR |
|---|---|---|---|---|---|---|---|---|---|
| Decision Tree | 98.05 | 93.33 | 98.47 | 1.53 | 6.67 | 84.48 | 99.40 | 88.68 | 1.95 |
| SVM | 98.83 | 97.19 | 98.98 | 1.02 | 2.81 | 89.65 | 99.74 | 93.27 | 1.17 |
| KNN | 97.90 | 92.38 | 98.39 | 1.61 | 7.62 | 83.62 | 99.31 | 87.78 | 2.10 |
| Ensemble | 98.29 | 94.33 | 98.64 | 1.36 | 5.67 | 86.2 | 99.48 | 90.08 | 1.71 |
| Naïve Bayes | 90.37 | 87.23 | 99.34 | 0.66 | 12.77 | 93.96 | 90.01 | 90.47 | 9.63 |
| Discriminant | 98.60 | 97.11 | 98.73 | 1.27 | 2.89 | 87.06 | 99.74 | 91.81 | 1.40 |
| Method | ACC | TPR | TNR | FPR | FNR | PPV | NPV | F1-Score | MR |
|---|---|---|---|---|---|---|---|---|---|
| Decision Tree | 94.12 | 89.47 | 96.21 | 3.79 | 10.53 | 91.39 | 95.31 | 90.42 | 5.88 |
| SVM | 95.75 | 89.6 | 98.78 | 1.22 | 10.4 | 97.31 | 95.08 | 93.30 | 4.25 |
| KNN | 99.51 | 98.41 | 99.94 | 0.06 | 1.59 | 99.98 | 99.29 | 99.19 | 0.49 |
| Ensemble | 94.61 | 87.68 | 98.04 | 1.96 | 12.32 | 95.69 | 94.14 | 91.51 | 5.39 |
| Naïve Bayes | 79.44 | 76.93 | 98.39 | 1.61 | 23.07 | 97.31 | 71.66 | 85.93 | 20.56 |
| Discriminant | 97.22 | 92.46 | 99.51 | 0.49 | 7.54 | 98.92 | 96.48 | 95.58 | 2.78 |
| Pre-Trained CNN with PSO (COVID-19 Dataset) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Classifiers | ACC | TPR | TNR | FPR | FNR | PPV | NPV | F1-SCORE | MR |
| Pre-trained CNN (AlexNet) + PSO | |||||||||
| Decision Tree | 96.42 | 89.74 | 97.63 | 2.37 | 10.26 | 90.05 | 98.46 | 89.89 | 3.58 |
| SVM | 99.45 | 99.92 | 99.40 | 0.60 | 0.08 | 96.95 | 99.11 | 98.41 | 0.55 |
| KNN | 98.52 | 99.10 | 98.40 | 1.60 | 0.90 | 93.33 | 98.4 | 96.13 | 1.48 |
| Ensemble | 98.91 | 98.74 | 98.81 | 1.19 | 1.26 | 97.42 | 98.52 | 98.08 | 1.09 |
| Naïve Bayes | 95.72 | 96.36 | 99.46 | 0.54 | 3.64 | 94.82 | 95.81 | 95.58 | 4.28 |
| Discriminant | 99.68 | 99.87 | 99.65 | 0.35 | 0.13 | 97.89 | 99.14 | 98.87 | 0.32 |
| Pre-trained CNN (GoogleNet) + PSO | |||||||||
| Decision Tree | 96.97 | 90.13 | 98.20 | 1.80 | 9.87 | 92.16 | 98.89 | 91.13 | 3.03 |
| SVM | 98.75 | 99.01 | 98.90 | 1.10 | 0.99 | 96.02 | 99.91 | 97.49 | 1.25 |
| KNN | 98.83 | 99.08 | 98.73 | 1.27 | 0.92 | 92.98 | 99.91 | 95.93 | 1.17 |
| Ensemble | 98.75 | 99.01 | 98.73 | 1.27 | 0.99 | 96.60 | 99.91 | 97.79 | 1.25 |
| Naïve Bayes | 98.52 | 96.40 | 99.23 | 0.77 | 3.60 | 93.05 | 99.14 | 94.70 | 1.48 |
| Discriminant | 98.83 | 99.02 | 98.81 | 1.19 | 0.98 | 97.19 | 99.91 | 98.10 | 1.17 |
| Pre-trained CNN (ResNet 50) + PSO | |||||||||
| Decision Tree | 97.51 | 92.85 | 97.89 | 2.11 | 7.15 | 90.29 | 99.40 | 91.55 | 2.49 |
| SVM | 99.76 | 99.89 | 99.74 | 0.26 | 0.11 | 97.41 | 99.18 | 98.63 | 0.24 |
| KNN | 98.99 | 98.13 | 99.06 | 0.94 | 1.87 | 94.38 | 99.82 | 96.22 | 1.01 |
| Ensemble | 99.22 | 99.81 | 99.15 | 0.85 | 0.19 | 96.49 | 99.60 | 98.12 | 0.78 |
| Naïve Bayes | 99.06 | 98.82 | 99.91 | 0.09 | 1.18 | 99.13 | 99.06 | 98.97 | 0.94 |
| Discriminant | 99.66 | 99.40 | 99.64 | 0.36 | 0.60 | 97.31 | 99.63 | 98.34 | 0.34 |
| Pre-trained CNN (DenseNet 201) + PSO | |||||||||
| Decision Tree | 97.82 | 94.84 | 98.14 | 1.86 | 5.16 | 92.63 | 99.57 | 93.72 | 2.18 |
| SVM | 99.37 | 99.09 | 99.40 | 0.60 | 0.91 | 96.25 | 99.91 | 97.65 | 0.63 |
| KNN | 98.75 | 99.01 | 98.73 | 1.27 | 0.99 | 94.61 | 99.91 | 96.76 | 1.25 |
| Ensemble | 98.91 | 98.46 | 98.81 | 1.19 | 1.54 | 96.84 | 98.91 | 97.64 | 1.09 |
| Naïve Bayes | 99.06 | 97.71 | 99.57 | 0.43 | 2.29 | 95.68 | 99.40 | 96.68 | 0.94 |
| Discriminant | 98.75 | 99.01 | 98.73 | 1.27 | 0.99 | 97.19 | 99.91 | 98.09 | 1.25 |
| Pre-Trained CNN with PSO (Brain Tumor Dataset) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Classifiers | ACC | TPR | TNR | FPR | FNR | PPV | NPV | F1-SCORE | MR |
| Pre-trained CNN (AlexNet) + PSO | |||||||||
| Decision Tree | 89.39 | 82.70 | 92.28 | 7.72 | 17.30 | 82.25 | 92.50 | 82.47 | 10.61 |
| SVM | 97.87 | 96.25 | 98.59 | 1.41 | 3.75 | 96.77 | 98.36 | 96.51 | 2.13 |
| KNN | 98.69 | 97.34 | 99.29 | 0.71 | 2.66 | 98.38 | 98.82 | 97.86 | 1.31 |
| Ensemble | 95.75 | 93.47 | 96.73 | 3.27 | 6.53 | 92.47 | 97.18 | 92.97 | 4.25 |
| Naïve Bayes | 88.09 | 86.93 | 96.33 | 3.67 | 13.07 | 92.47 | 92.98 | 89.61 | 11.91 |
| Discriminant | 97.55 | 95.72 | 98.35 | 1.65 | 4.28 | 96.23 | 98.12 | 95.97 | 2.45 |
| Pre-trained CNN (GoogleNet) + PSO | |||||||||
| Decision Tree | 87.76 | 81.00 | 90.55 | 9.45 | 19.00 | 77.95 | 92.03 | 79.45 | 12.24 |
| SVM | 96.41 | 93.15 | 97.87 | 2.13 | 6.85 | 95.16 | 96.95 | 94.14 | 3.59 |
| KNN | 93.80 | 87.23 | 97.32 | 2.68 | 12.77 | 94.08 | 93.67 | 90.53 | 6.20 |
| Ensemble | 92.98 | 85.92 | 96.37 | 3.63 | 14.08 | 91.93 | 93.44 | 88.82 | 7.02 |
| Naïve Bayes | 90.86 | 86.50 | 96.72 | 3.28 | 13.50 | 93.01 | 89.92 | 89.64 | 9.14 |
| Discriminant | 96.41 | 92.67 | 99.03 | 0.97 | 7.33 | 97.84 | 95.78 | 95.18 | 3.59 |
| Pre-trained CNN (ResNet 50) + PSO | |||||||||
| Decision Tree | 90.21 | 85.39 | 92.18 | 7.82 | 14.61 | 81.75 | 93.91 | 83.53 | 9.79 |
| SVM | 97.87 | 96.75 | 98.36 | 1.64 | 3.25 | 96.23 | 98.59 | 96.49 | 2.13 |
| KNN | 98.85 | 98.90 | 98.83 | 1.17 | 1.10 | 97.31 | 99.53 | 98.10 | 1.15 |
| Ensemble | 96.73 | 95.60 | 97.21 | 2.79 | 4.40 | 93.54 | 98.12 | 94.56 | 3.27 |
| Naïve Bayes | 90.51 | 88.72 | 97.95 | 2.05 | 11.28 | 95.69 | 91.08 | 92.07 | 9.49 |
| Discriminant | 97.22 | 94.96 | 99.27 | 0.73 | 5.04 | 98.38 | 97.42 | 96.64 | 2.78 |
| Pre-trained CNN (DenseNet 201) + PSO | |||||||||
| Decision Tree | 93.47 | 88.42 | 95.74 | 4.26 | 11.58 | 90.32 | 94.84 | 89.36 | 6.53 |
| SVM | 96.90 | 96.13 | 97.22 | 2.78 | 3.87 | 93.54 | 98.36 | 94.82 | 3.10 |
| KNN | 97.71 | 95.74 | 98.58 | 1.42 | 4.26 | 96.77 | 98.12 | 96.25 | 2.29 |
| Ensemble | 96.73 | 94.62 | 97.65 | 2.35 | 5.38 | 94.62 | 97.65 | 94.62 | 3.27 |
| Naïve Bayes | 93.31 | 95.49 | 96.84 | 3.16 | 4.51 | 93.01 | 96.64 | 94.23 | 6.69 |
| Discriminant | 96.90 | 94.96 | 98.11 | 1.89 | 5.04 | 95.69 | 97.42 | 95.32 | 3.10 |
| Ref. | Journal | Year | Methods | Datasets | Classes | Results |
|---|---|---|---|---|---|---|
| [19] | IEEE International Conference | 2018 | PSO + SVM | Brain Tumor | Benign, malignant | 95.23% |
| [17] | Springer Link | 2021 | CNN + (GLCM), (GLRM), (LBP) + PSO | COVID-19 | COVID-19, Pneumonia, normal | 98.06% |
| [8] | Journal Pre-proof | 2020 | CNN | COVID-19 | COVID-19, non-COVID-19 | 86.27% |
| [68] | Indonesian Journal of Electronics and Instrumentation Systems (IJEIS) | 2018 | GLCM + CNN | Brain Tumor | Meningioma, glioma, pituitary | 82.00% |
| [69] | The Information Technology Management (ICCMIT’20) | 2020 | GLCM + VGG16 + Softmax | Brain Tumor | Meningioma, glioma, pituitary | 96.50% |
| [70] | Scientific Reports—Computer Science | 2018 | Capsule networks (CapsNets) + Softmax | Brain Tumor | Meningioma, glioma, pituitary | 86.56% |
| [71] | Springer Link | 2020 | Stacked autoencoder + Softmax | COVID-19 | Positive, negative | 94.70% |
| [72] | Scientific Reports—Computer Science | 2021 | CNN + Autoencoder + SVM | COVID-19 (Private) | COVID-19, normal | 96.05% |
| Proposed Method | 2021 | CNN + PSO + SVM | COVID-19 | Meningioma, glioma, pituitary | 99.76% | |
| Proposed Method | 2021 | Autoencoder + PSO + kNN | Brain Tumor | COVID-19, pneumonia, normal | 99.51% |
| Dataset | Combined Methods | PSO | ACO | GA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Classifiers | Acc. | Time (h) | Classifiers | Acc. | Time (h) | Classifiers | Acc. | Time (h) | ||
| COVID-19 | Autoencoder | SVM | 98.83% | 1:15:00 | SVM | 98.68% | 0:27:00 | KNN | 97.98% | 1:00:00 |
| CNN (AlexNet) | Discriminant | 99.68% | 4:12:00 | Discriminant | 99.53% | 1:17:00 | KNN | 98.60% | 2:07:00 | |
| CNN (GoogleNet) | Discriminant | 98.83% | 1:00:00 | SVM | 98.91% | 0:21:00 | Naïve Bayes | 98.13% | 0:40:00 | |
| CNN (ResNet 50) | SVM | 99.76% | 2:05:00 | SVM | 99.61% | 0:43:00 | KNN | 98.60% | 1:05:00 | |
| CNN (DenseNet 201) | SVM | 99.37% | 2:00:00 | Naïve Bayes | 99.14% | 0:39:00 | KNN | 98.75% | 1:04:00 | |
| Brain Tumor | Autoencoder | KNN | 99.51% | 0:23:00 | KNN | 99.18% | 0:11:00 | Ensemble | 96.24% | 0:16:00 |
| CNN (AlexNet) | KNN | 98.69% | 0:58:00 | Discriminant | 98.69% | 0:12:00 | Ensemble | 94.61% | 0:30:00 | |
| CNN (GoogleNet) | Discriminant | 96.41% | 0:15:00 | Discriminant | 96.73% | 0:05:00 | KNN | 93.96% | 0:09:00 | |
| CNN (ResNet 50) | KNN | 98.85% | 0:30:00 | KNN | 97.87% | 0:11:00 | Ensemble | 97.06% | 0:16:00 | |
| CNN (DenseNet 201) | KNN | 97.71% | 0:27:00 | SVM | 98.20% | 0:09:00 | Ensemble | 96.57% | 0:18:00 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kadhim, Y.A.; Guzel, M.S.; Mishra, A. A Novel Hybrid Machine Learning-Based System Using Deep Learning Techniques and Meta-Heuristic Algorithms for Various Medical Datatypes Classification. Diagnostics 2024, 14, 1469. https://doi.org/10.3390/diagnostics14141469
Kadhim YA, Guzel MS, Mishra A. A Novel Hybrid Machine Learning-Based System Using Deep Learning Techniques and Meta-Heuristic Algorithms for Various Medical Datatypes Classification. Diagnostics. 2024; 14(14):1469. https://doi.org/10.3390/diagnostics14141469
Chicago/Turabian StyleKadhim, Yezi Ali, Mehmet Serdar Guzel, and Alok Mishra. 2024. "A Novel Hybrid Machine Learning-Based System Using Deep Learning Techniques and Meta-Heuristic Algorithms for Various Medical Datatypes Classification" Diagnostics 14, no. 14: 1469. https://doi.org/10.3390/diagnostics14141469
APA StyleKadhim, Y. A., Guzel, M. S., & Mishra, A. (2024). A Novel Hybrid Machine Learning-Based System Using Deep Learning Techniques and Meta-Heuristic Algorithms for Various Medical Datatypes Classification. Diagnostics, 14(14), 1469. https://doi.org/10.3390/diagnostics14141469









