An Intelligent Mechanism to Detect Multi-Factor Skin Cancer
Abstract
1. Introduction
2. Related Work
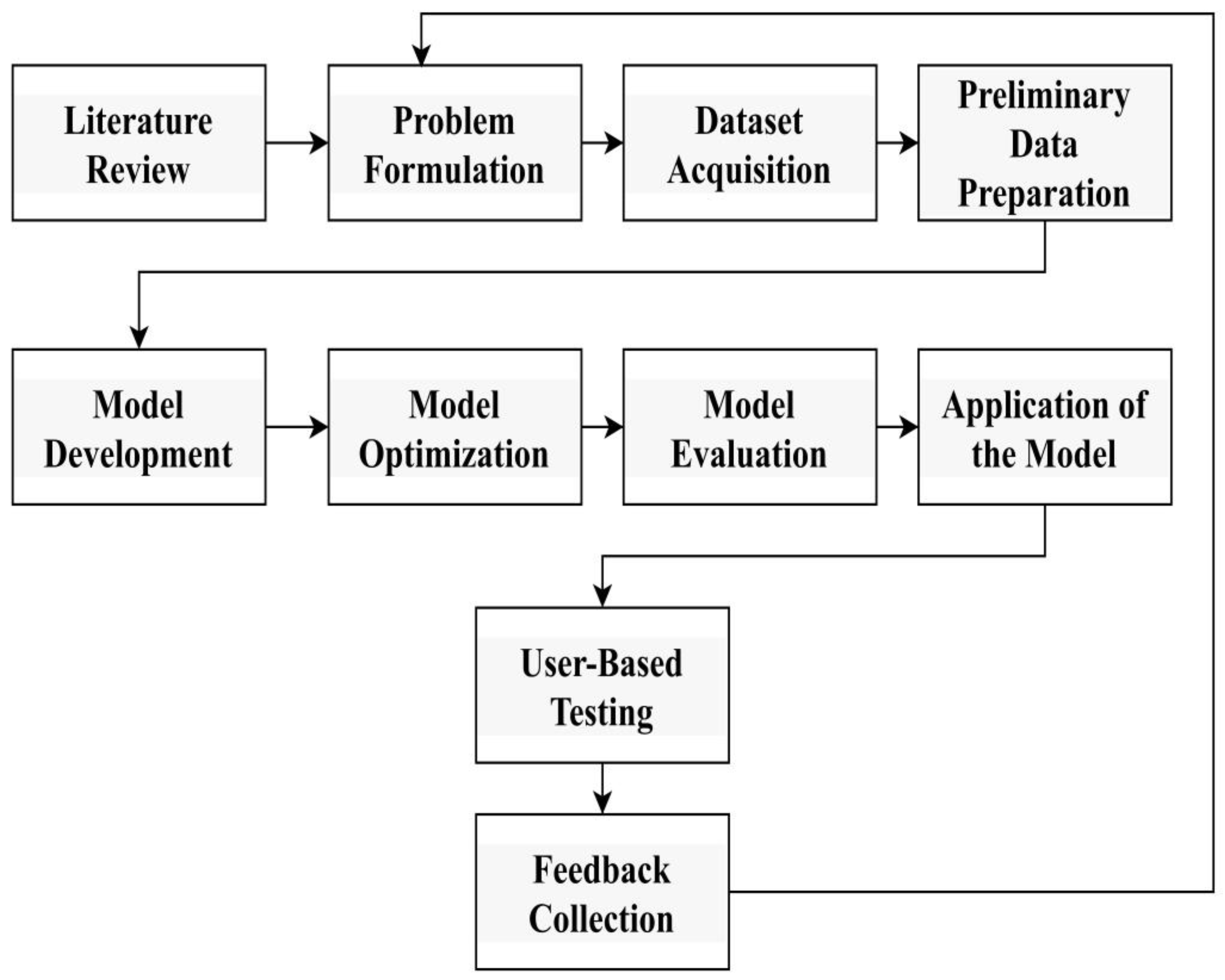
3. Research Methodology
- (a)
- Literature Review: A comprehensive review of the existing literature to understand research history, identify gaps, and gather insight into methods and technologies used in cancer diagnosis.
- (b)
- Problem Formulation: Establish clear research goals and describe the research, including the type of skin cancer to be tested and the expected results.
- (c)
- Dataset acquisition
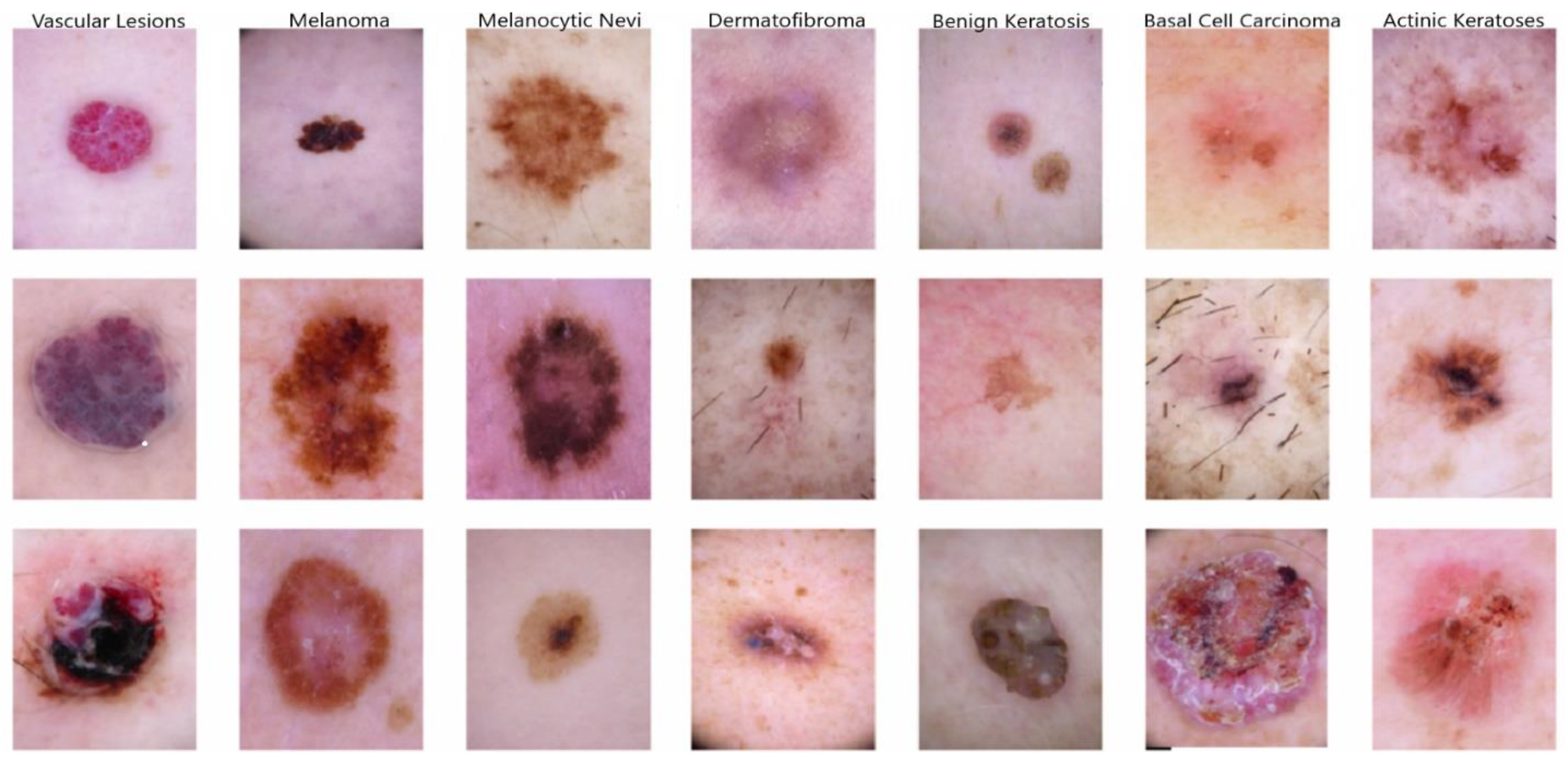
- Melanocytic nevi (NV);
- Benign keratosis-like lesions (BKLs);
- Dermatofibroma (DF);
- Vascular lesions (VASCs);
- Actinic keratoses and intraepithelial carcinoma (AKIEC);
- Basal cell carcinoma (BCC);
- Melanoma (MEL).
- Training data: involving 75% of the dataset;
- Validation data: making up 15% of the dataset;
- Testing data: accounting for 10% of the dataset.
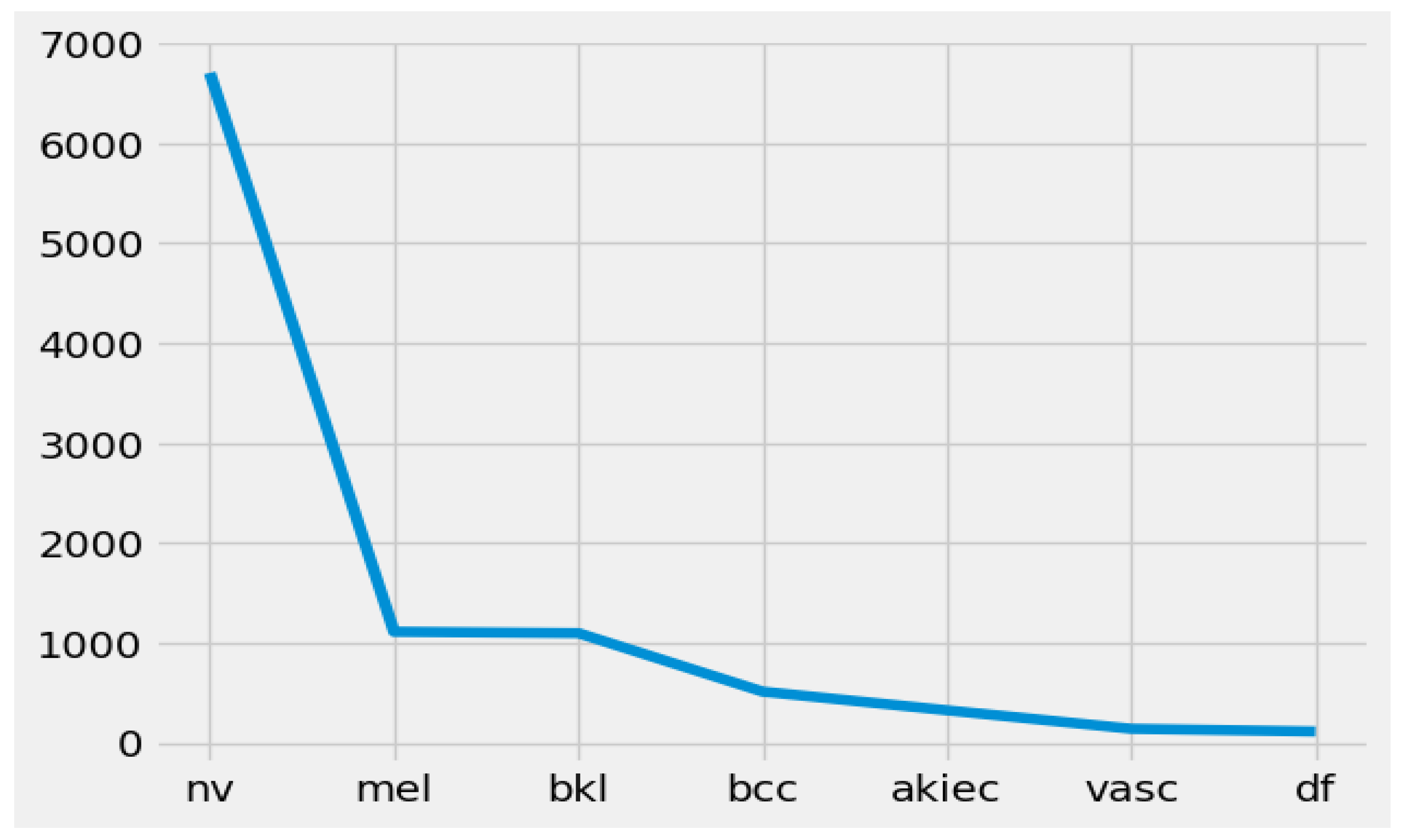
- Class-wise distribution of the HAM10000 dataset
- NV (melanocytic nevi): 6705 instances (images) have a place in this class. Melanocytic nevi are commonly benign moles or skin lesions.
- MEL (melanoma): This class comprises 1113 instances. Melanoma is a profoundly malignant type of skin cancer.
- BKLs (benign keratosis-like lesions): There are 1099 instances in this class. Benign keratosis-like lesions are skin conditions that look like keratosis but are benign (non-cancerous).
- BCC (Basal cell carcinoma): This class contains 514 instances. Basal cell carcinoma is a typical type of skin cancer that is generally non-lethal yet requires treatment.
- AKIEC (actinic keratoses and intraepithelial carcinoma): There are 327 instances in this class. Actinic keratoses and intraepithelial carcinoma address precancerous and early cancerous skin conditions.
- VASCs (vascular lesions): This class contains 142 instances. Vascular lesions involve abnormalities in veins inside the skin.
- DF (dermatofibroma): There are 115 instances in this class. Dermatofibroma is a benign skin condition portrayed by little, hard developments on the skin.
- (d)
- Preliminary data preparation
- Data resampling: The RandomOverSampler was used to balance the dataset by oversampling the minority class. This forestalls class imbalance issues [33].
- Data preparation for convolutional neural networks (CNNs): We reshaped the input data (XData) to have the shape (n_samples, 28, 28, 3) to match the expected input shape for a CNN that processes 3-channel (RGB) images [34].
- The pixel values were normalized by dividing by 255, which scales the pixel values between 0 and 1.
- Train, test, split: The dataset was split into training validation and testing sets using corresponding 75%, 15%, and 10% ratios. The chosen split best fits the requirements of the proposed model and ensures an appropriate balance among the training, validation, and testing phases. The shapes of the resulting training were printed into the test sets.
- (e)
- Targeting Deep Neural Model for Skin Cancer Detection
- (f)
- Model optimization
- (g)
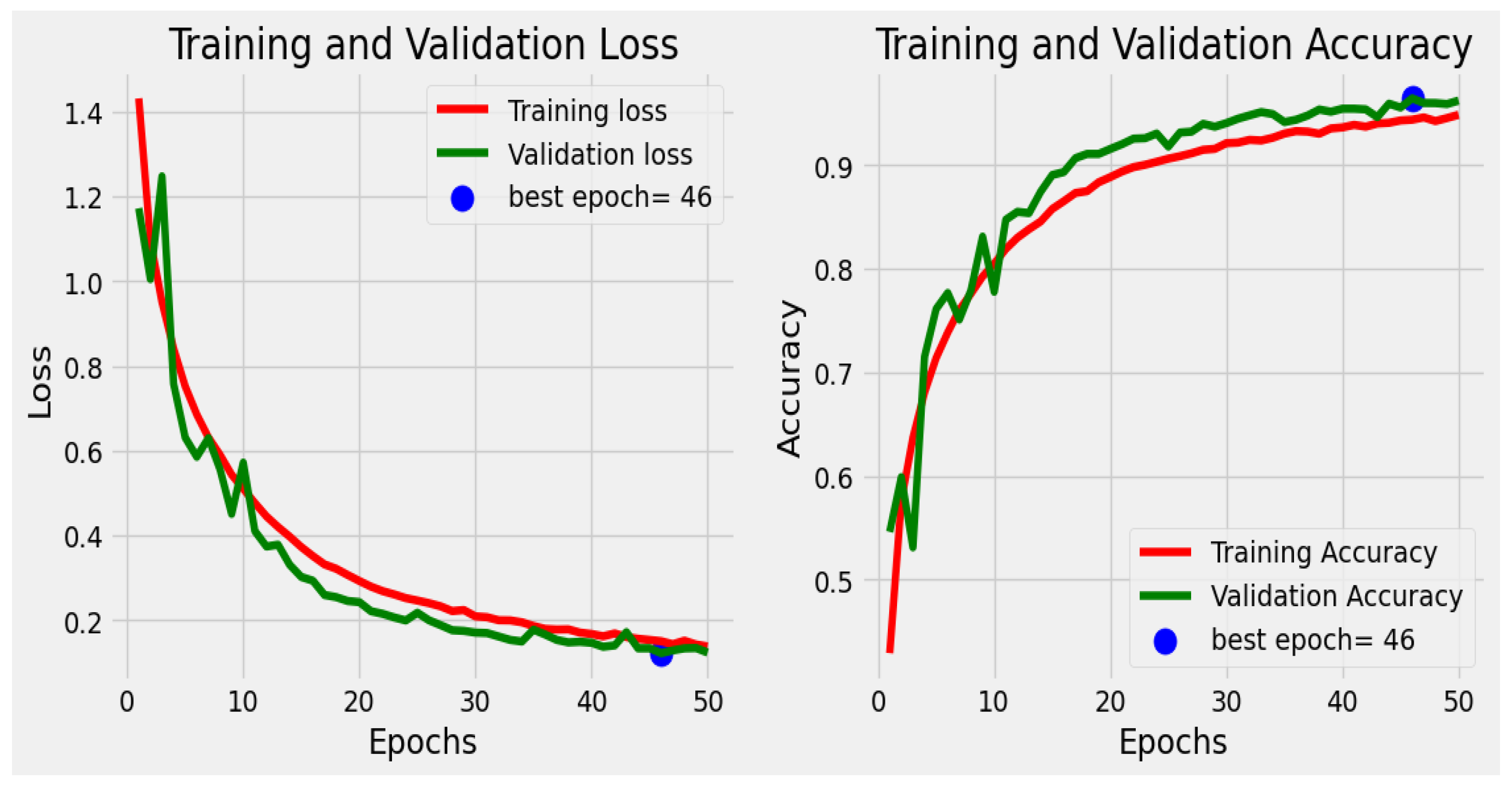
- Model Evaluation
- (h)
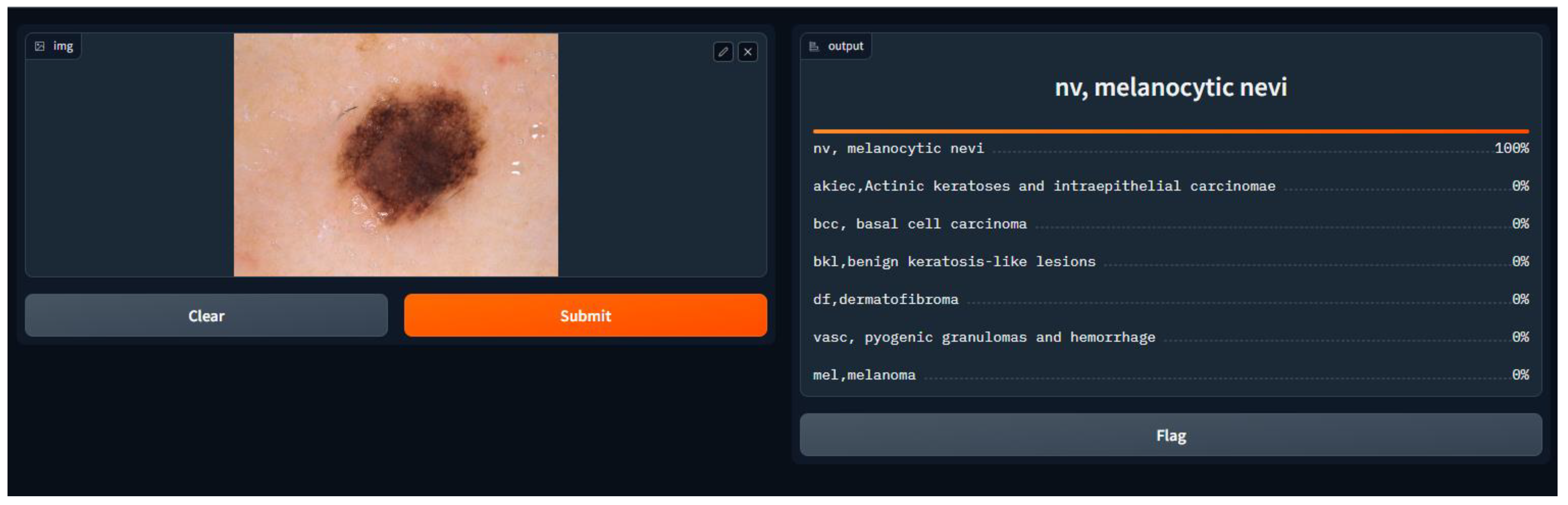
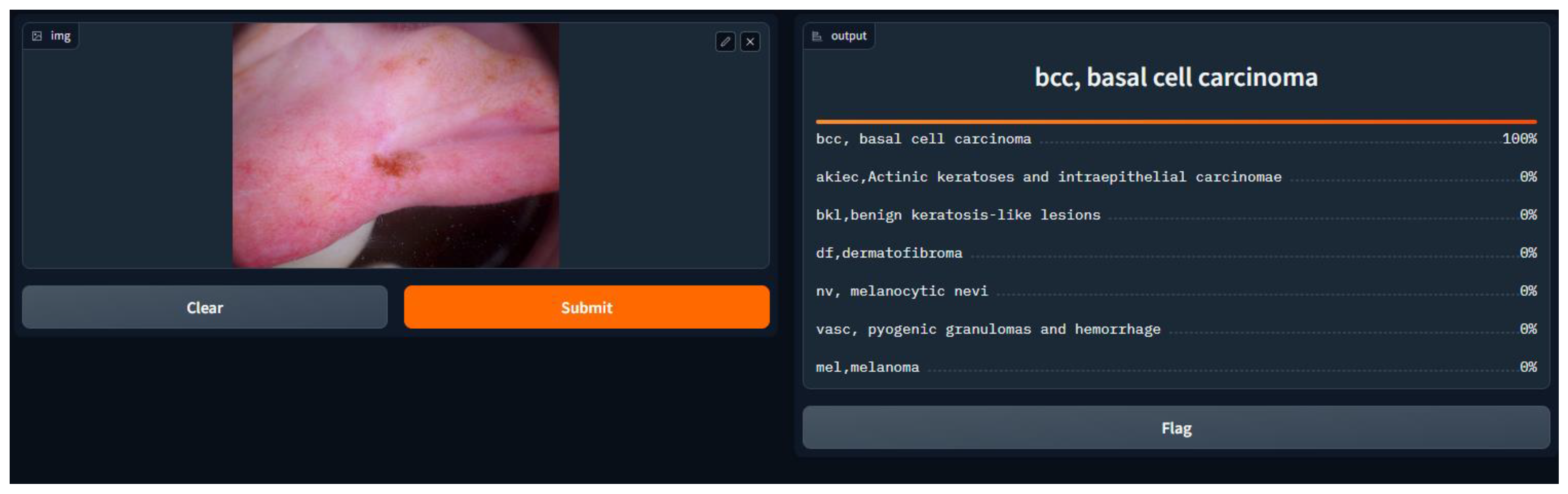
- Application of the Model
- import Gradio as gr, and here, we imported the Gradio library, which gives tools to make basic and intuitive UIs for machine learning models.
- gr.Interface(fn = predict, inputs = image, outputs = label, capture_session = True).launch(debug = True, share = True).
- fn = predict: This specifies the function that will be used for prediction, which is the prediction function we defined earlier.
- inputs = image: This specifies that the input component of the interface is the image input we defined earlier.
- outputs = label: This specifies that the output component of the interface is the label output we defined earlier.
- capture_session = True: This captures the TensorFlow session to optimize performance.
- Launch (debug = True, share = True): This launches the Gradio interface in debug mode, allowing us to test it locally, and empowers sharing to share the interface with others.
- (i)
- User-based testing via a web interface: User testing is performed through a web interface or interactive interface to evaluate the usability, performance, and user experience of cancer treatment development.
- (j)
- Send feedback through questions for further improvement: Address feedback from users and stakeholders through sample questions or surveys. Conduct research to identify areas for improvement, gather insight, and guide future iterations of the system.
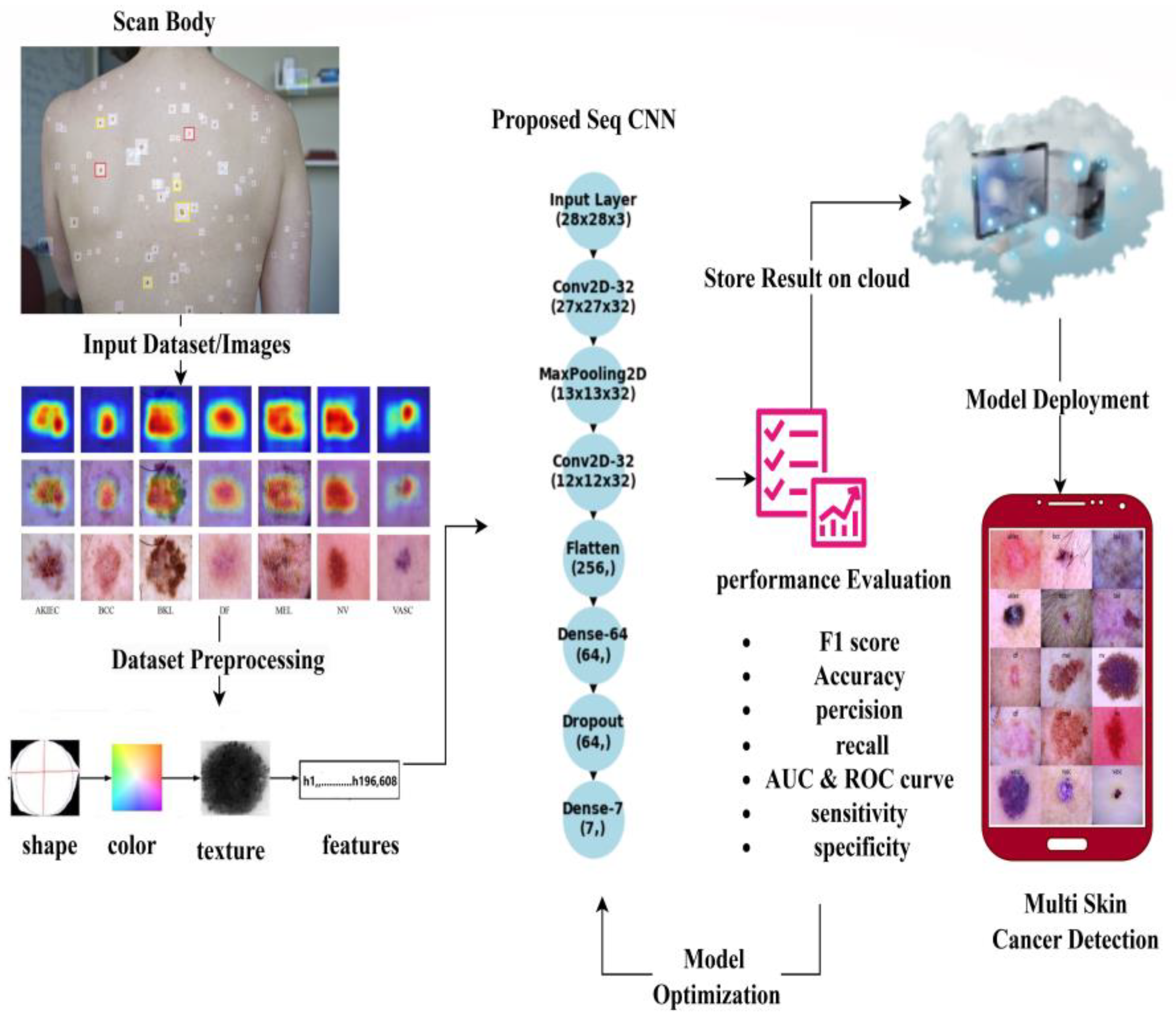
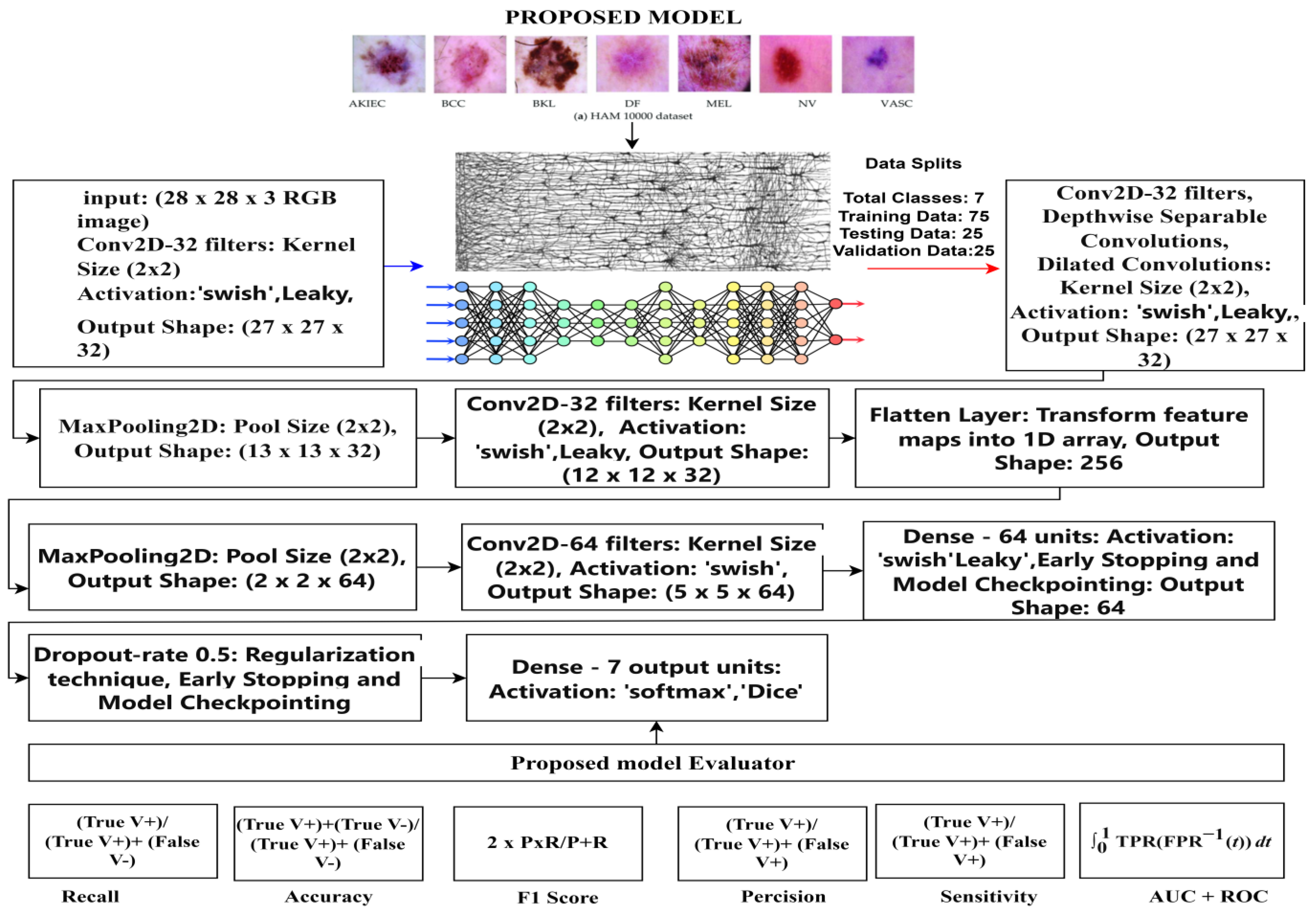
4. Proposed Model
- (a)
- Input Layer
- (b)
- Convolutional Layers
- Yi denotes the output feature map of the i-th convolutional layer. Wi represents the learned weights for the i-th convolutional filter. Additionally, bi is a biased term. f is the activation function, Leaky, and swishReLU (Rectified Linear Unit). After every convolution operation, a non-linear activation function was applied elementwise to the result.
- Pooling Layers
- b.
- Fully Connected Layers
- c.
- Layer of Output
- The output is the vector of anticipated probabilities, and w_output is the weight matrix related to the output layer. X is the input to the output layer. X_boutput is the inclination vector for the output layer, where output addresses the anticipated probabilities for each class.
5. Model Evaluation
6. Deployment
Discussion on the Findings of User Testing Based on Questionnaire Responses
- Ease of use: An overwhelming 94% of users assessed the tool’s ease of use as “Very Satisfied” (5 on a 5-point scale), suggesting an intuitive and user-friendly design.
- Image upload procedure: 78% of customers were very satisfied (rating 4 on a scale) with the image upload procedure, praising its efficiency and ease.
- Accuracy and usability: The tool’s accuracy and usability garnered positive reviews, with 72% of users being “Very Satisfied” with it (5).
- Quickness: The majority of users (76%) were “Very Satisfied” (5) with the tool’s quickness in providing diagnosis findings.
- User interface: The user interface’s aesthetic attractiveness and intuitiveness satisfied 68% of users, who were “Very Satisfied” with it (5).
- Diagnostic explanation: 74% of users were “Very Satisfied” with the diagnostic explanation (5), indicating that it was clear and easy to comprehend.
- Accuracy trust: 70% of users indicated strong trust in the tool’s diagnosis (rating 4 on a scale).
- Issues or mistakes: There were no reported incidents of users having issues or mistakes during the diagnosis procedure, which is encouraging.
- Suggestions for change: Users gave useful suggestions for improving the tool’s functionality and user experience, exhibiting involvement and a desire for change.
- Privacy and security: The picture upload and data management privacy and security safeguards were well-received, with 68% of users being “Very Satisfied” with them (5).
- Overall satisfaction: 76% of users were “Very Satisfied” (5) in terms of overall satisfaction, indicating a very good reaction.
- Recommendation: Users expressed a strong willingness to suggest the product to others, with 82% being “Very Satisfied” with it (5).
- Demographics: Data on age groups, gender, and location were gathered for demographic analysis.
7. Results and Discussion
8. Conclusions
9. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbrevations
| Acronyms | Expanded Form | Acronyms | Expanded Form |
| ABCD | Asymmetry, Border, Color, Diameter | AI | Artificial Intelligence |
| AKIEC | Actinic Keratoses and Intraepithelial Carcinoma | AUC | Area Under the Curve |
| BCC | Basal Cell Carcinoma | BKLs | Benign Keratosis-like Lesions |
| CAD | Computer-Aided Detection | CNNs | Convolutional Neural Networks |
| COCO | Common Objects in Context | DCNNs | Deep Convolutional Neural Networks |
| DF | Dermatofibroma | EIS | Electrical Impedance Spectroscopy |
| F1 | F1 Score | Gradio | Gradio (Python Package) |
| HAM | HAM10000 Dataset | ISIC | International Skin Imaging Collaboration |
| MEL | Melanoma | MSI | Multispectral Imaging |
| NDOELM | Non-destructive Online Ensemble Learning Method | NMSC | Non-Melanoma Skin Cancer |
| NV | Melanocytic Nevi | PC | Personal Computer |
| PSNR | Peak Signal-to-Noise Ratio | RCM | Reflectance Confocal Microscopy |
| ResNet | Residual Network | RGB | Red, Green, Blue |
| ROC | Receiver Operating Characteristic | SGD | Stochastic Gradient Descent |
| SSIM | Structural Similarity Index Measure | SVM | Support Vector Machine |
| VASCs | Vascular Lesions | VGG | Visual Geometry Group |
References
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Chen, W. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Jetter, N.; Chandan, N.; Wang, S.; Tsoukas, M. Field cancerization therapies for management of actinic keratosis: A narrative review. Am. J. Clin. Dermatol. 2018, 19, 543–557. [Google Scholar] [CrossRef] [PubMed]
- McConnell, N.; Miron, A.; Wang, Z.; Li, Y. Integrating residual, dense, and inception blocks into the nnunet. In Proceedings of the 2022 IEEE 35th International Symposium on Computer-Based Medical Systems (CBMS), Shenzen, China, 21–23 July 2022; pp. 217–222. [Google Scholar]
- Binder, M.; Schwarz, M.; Winkler, A.; Steiner, A.; Kaider, A.; Wolff, K.; Pehamberger, H. Epiluminescence Microscopy: A Useful Tool for the Diagnosis of Pigmented Skin Lesions for Formally Trained Dermatologists. Arch. Dermatol. 1995, 131, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Aljohani, K.; Turki, T. Automatic Classification of Melanoma Skin Cancer with Deep Convolutional Neural Networks. AI 2022, 3, 512–525. [Google Scholar] [CrossRef]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. In Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 13–15 May 2010; pp. 249–256. [Google Scholar]
- Ndipenoch, N.; Miron, A.; Wang, Z.; Li, Y. Simultaneous segmentation of layers and fluids in retinal oct images. In Proceedings of the 2022 15th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 5–7 November 2022; pp. 1–6. [Google Scholar]
- Manzoor, K.; Majeed, F.; Siddique, A.; Meraj, T.; Rauf, H.T.; El-Meligy, M.A.; Sharaf, M.; Elgawad, A.E.E.A. A lightweight approach for skin lesion detection through optimal features fusion. Comput. Mater. Contin. 2022, 70, 1617–1630. [Google Scholar] [CrossRef]
- Ferguson, M.K.; Ronay, A.; Lee, Y.-T.T.; Law, K.H. Detection and segmentation of manufacturing defects with convolutional neural networks and transfer learning. Smart Sustain. Manuf. Syst. 2018, 2, 137–164. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In International Workshop on Machine Learning in Medical Imaging; Springer: Cham, Switzerland, 2015; pp. 118–126. [Google Scholar]
- Almaraz-Damian, J.-A.; Ponomaryov, V.; Sadovnychiy, S.; Castillejos-Fernandez, H. Melanoma and nevus skin lesion classification using handcraft and deep learning feature fusion via mutual data measures. Entropy 2020, 22, 484. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, M.J.M.; Rosado, L.; Ferreira, M. A new color assessment methodology using cluster-based features for skin lesion analysis. In Proceedings of the 2015 38th International Convention on Data and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 25–29 May 2015; pp. 373–378. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Kaur, P.; Dana, K.J.; Cula, G.O.; Mack, M.C. Hybrid deep learning for reflectance confocal microscopy skin images. In Proceedings of the 2016 23rd International conference on pattern recognition (ICPR), Cancun, Mexico, 4–8 December 2016; pp. 1466–1471. [Google Scholar]
- Harangi, B. Skin lesion detection based on an ensemble of deep convolutional neural network. arXiv 2017, arXiv:1705.03360. [Google Scholar]
- Papachristou, I.; Bosanquet, N. Improving the prevention and diagnosis of melanoma on a national scale: A comparative study of performance in the United Kingdom and Australia. J. Public Health Policy 2020, 41, 28–38. [Google Scholar] [CrossRef]
- Bebis, G.; Boyle, R.; Parvin, B.; Koracin, D.; Ushizima, D.; Chai, S.; Sueda, S.; Lin, X.; Lu, A.; Thalmann, D.; et al. Advances in Visual Computing: 14th International Symposium on Visual Computing, ISVC 2019, Lake Tahoe, NV, USA, 7–9 October 2019, Proceedings, Part II Vol. 11845; Springer Nature: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- di Ruffano, L.F.; Takwoingi, Y.; Dinnes, J.; Chuchu, N.; Bayliss, S.E.; Davenport, C.; Matin, R.N.; Godfrey, K.; O'Sullivan, C.; Gulati, A.; et al. Computer-assisted diagnosis techniques (dermoscopy and spectroscopy-based) for diagnosing skin cancer in adults. Cochrane Database Syst. Rev. 2018, 2018, CD013186. [Google Scholar]
- Li, Y.; Shen, L. Skin lesion analysis towards melanoma detection using deep learning network. Sensors 2018, 18, 556. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, J.; Xie, F.; Liu, J.; Shu, C.; Wang, Y.; Zheng, Y.; Zhang, H. A convolutional neural network trained with dermoscopic images of psoriasis performed on par with 230 dermatologists. Comput. Biol. Med. 2021, 139, 104924. [Google Scholar] [CrossRef]
- Bisla, D.; Choromanska, A.; Berman, R.S.; Stein, J.A.; Polsky, D. Towards automated melanoma detection with deep learning: Data purification and augmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Long Beach, CA, USA, 16–17 June 2019. [Google Scholar]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.H.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Majtner, T.; Bajić, B.; Yildirim, S.; Hardeberg, J.Y.; Lindblad, J.; Sladoje, N. Ensemble of convolutional neural networks for dermoscopic images classification. arXiv 2018, arXiv:1808.05071. [Google Scholar]
- Mercuri, S.R.; Rizzo, N.; Bellinzona, F.; Pampena, R.; Brianti, P.; Moffa, G.; Flink, L.C.; Bearzi, P.; Longo, C.; Paolino, G. Digital ex-vivo confocal imaging for fast Mohs surgery in nonmelanoma skin cancers: An emerging technique in dermatologic surgery. Dermatol. Ther. 2019, 32, e13127. [Google Scholar] [CrossRef] [PubMed]
- Sae-Lim, W.; Wettayaprasit, W.; Aiyarak, P. Convolutional neural networks using MobileNet for skin lesion classification. In Proceedings of the 2019 16th International Joint Conference on Computer Science and Software Engineering (JCSSE), Chonburi, Thailand, 10–12 July 2019; pp. 242–247. [Google Scholar]
- Sahu, S.; Singh, A.K.; Ghrera, S.; Elhoseny, M. An approach for de-noising and contrast enhancement of retinal fundus image using CLAHE. Opt. Laser Technol. 2019, 110, 87–98. [Google Scholar]
- Maarouf, M.; Costello, C.; Gonzalez, S.; Angulo, I.; Curiel-Lewandrowski, C.; Shi, V. In vivo reflectance confocal microscopy: Emerging role in noninvasive diagnosis and monitoring of eczematous dermatoses. Actas Dermo-Sifiliográficas 2019, 110, 626–636. [Google Scholar] [CrossRef]
- Wang, L.; Chen, A.; Zhang, Y.; Wang, X.; Zhang, Y.; Shen, Q.; Xue, Y. AK-DL: A shallow neural network model for diagnosing actinic keratosis with better performance than deep neural networks. Diagnostics 2020, 10, 217. [Google Scholar] [CrossRef]
- Dodo, B.I.; Li, Y.; Eltayef, K.; Liu, X. Min-cut segmentation of retinal oct images. In Biomedical Engineering Systems and Technologies: 11th International Joint Conference, BIOSTEC 2018, Funchal, Madeira, Portugal, January 19–21, 2018, Revised Selected Papers 11; Springer: Cham, Switzerland, 2019; pp. 86–99. [Google Scholar]
- Dodo, B.I.; Li, Y.; Eltayef, K.; Liu, X. Automatic annotation of retinal layers in optical coherence tomography images. J. Med. Syst. 2019, 43, 336. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Van Dyk, D.A.; Meng, X.-L. The art of data augmentation. J. Comput. Graph. Stat. 2001, 10, 1–50. [Google Scholar] [CrossRef]
- Karmaoui, A.; Yoganandan, G.; Sereno, D.; Shaukat, K.; Jaafari, S.E.; Hajji, L. Global network analysis of links between business, climate change, and sustainability and setting up the interconnections framework. Environ. Dev. Sustain. 2023, 1–25. [Google Scholar] [CrossRef]
- Abid, A.; Abdalla, A.; Abid, A.; Khan, D.; Alfozan, A.; Zou, J. Gradio: Hassle-free sharing and testing of ml models in the wild. arXiv 2019, arXiv:1906.02569. [Google Scholar]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef] [PubMed]
- Acosta, M.F.J.; Tovar, L.Y.C.; Garcia-Zapirain, M.B.; Percybrooks, W.S. Melanoma diagnosis using deep learning techniques on dermatoscopic images. BMC Med. Imaging 2021, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Fan, H.; Li, Y.; Jiang, Z.; Meng, R.; Bovik, A. Melanoma classification on dermoscopy images using a neural network ensemble model. IEEE Trans. Med. Imaging 2016, 36, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, K.; Luo, S.; Varadharajan, V. A novel machine learning approach for detecting first-time-appeared malware. Eng. Appl. Artif. Intell. 2024, 131, 107801. [Google Scholar] [CrossRef]
- Didona, D.; Paolino, G.; Bottoni, U.; Cantisani, C. Non melanoma skin cancer pathogenesis overview. Biomedicines 2018, 6, 6. [Google Scholar] [CrossRef]
- Ali, K.; Shaikh, Z.A.; Khan, A.A.; Laghari, A.A. Multiclass skin cancer classification using EfficientNets–a first step towards preventing skin cancer. Neurosci. Inform. 2022, 2, 100034. [Google Scholar] [CrossRef]
- Venugopal, V.; Raj, N.I.; Nath, M.K.; Stephen, N. A deep neural network using modified EfficientNet for skin cancer detection in dermoscopic images. Decis. Anal. J. 2023, 8, 100278. [Google Scholar] [CrossRef]
- Shaukat, K.; Luo, S.; Varadharajan, V. A novel deep learning-based approach for malware detection. Eng. Appl. Artif. Intell. 2023, 122, 106030. [Google Scholar] [CrossRef]
- Akter, M.S.; Shahriar, H.; Sneha, S.; Cuzzocrea, A. Multi-class skin cancer classification architecture based on deep convolutional neural network. In Proceedings of the 2022 IEEE International Conference on Big Data (Big Data), Osaka, Japan, 17–20 December 2022; pp. 5404–5413. [Google Scholar]
- Arshed, M.A.; Mumtaz, S.; Ibrahim, M.; Ahmed, S.; Tahir, M.; Shafi, M. Multi-Class Skin Cancer Classification Using Vision Transformer Networks and Convolutional Neural Network-Based Pre-Trained Models. Data 2023, 14, 415. [Google Scholar] [CrossRef]
- Khan, M.A.; Akram, T.; Zhang, Y.D.; Alhaisoni, M.; Al Hejaili, A.; Shaban, K.A.; Tariq, U.; Zayyan, M.H. SkinNet-ENDO: Multiclass skin lesion recognition using deep neural network and Entropy-Normal distribution optimization algorithm with ELM. Int. J. Imaging Syst. Technol. 2023, 33, 1275–1292. [Google Scholar] [CrossRef]

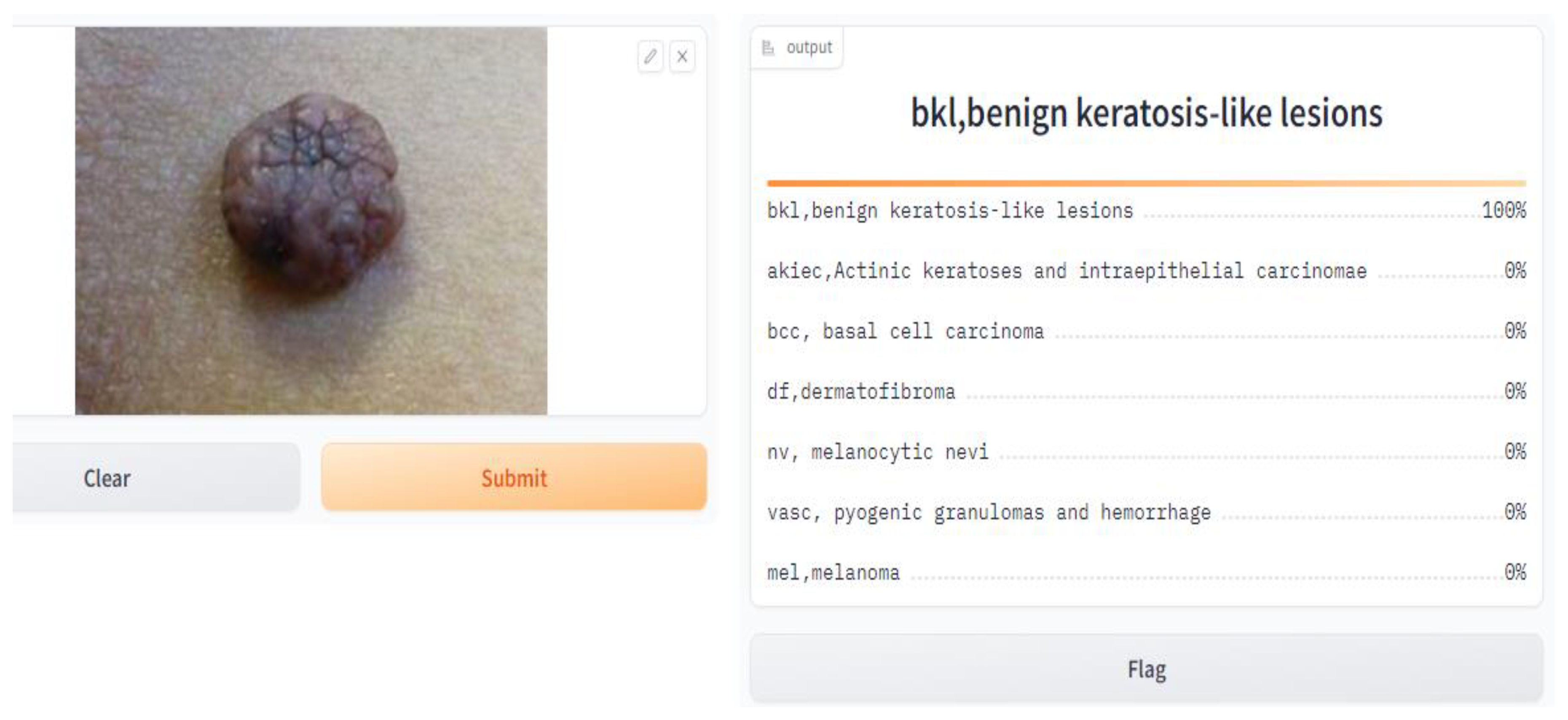

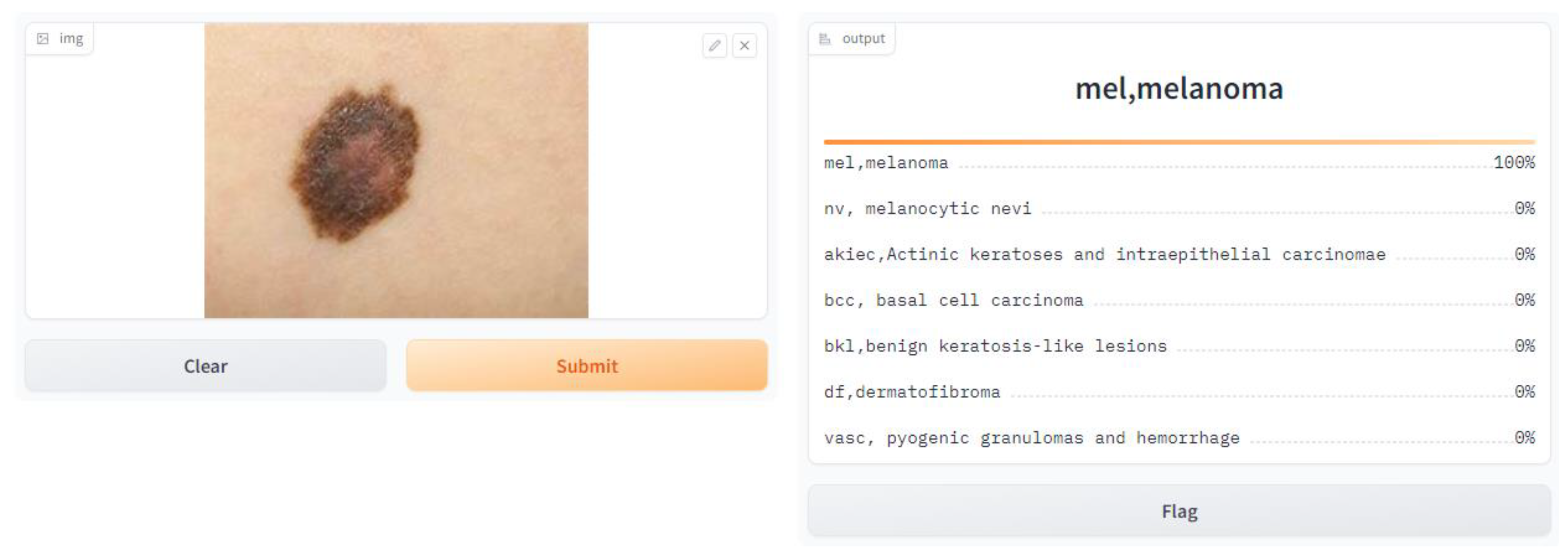
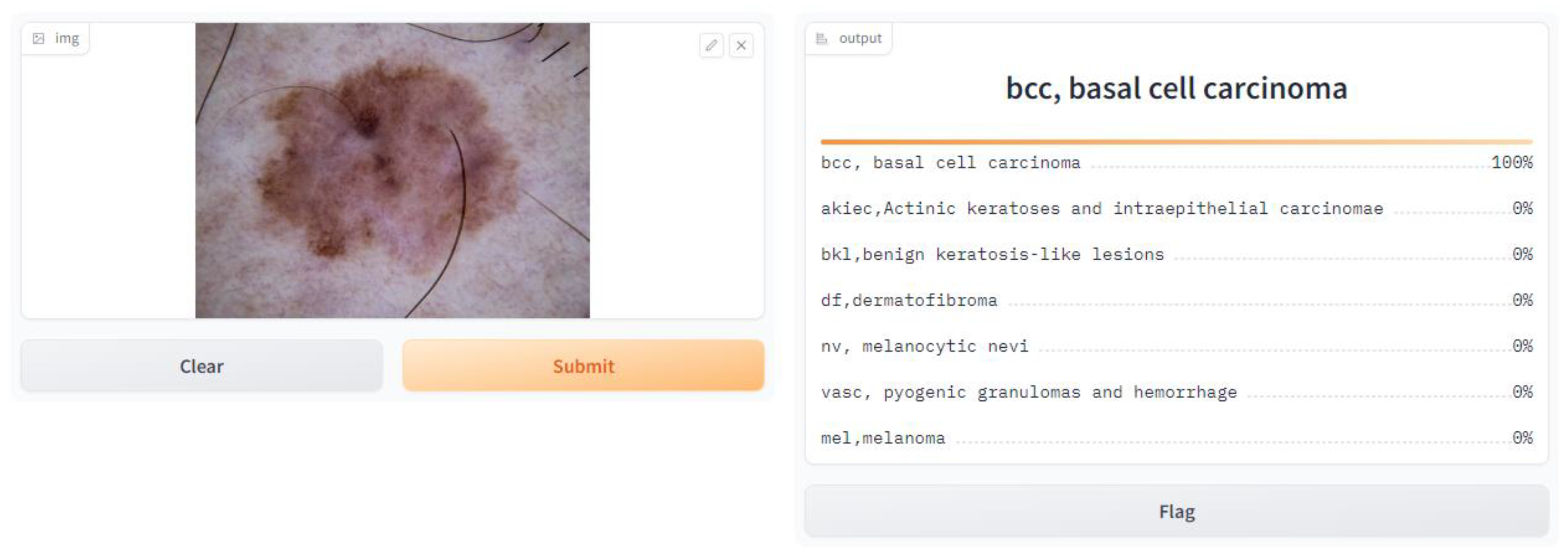

| User Interface Web Browser | Gradio App |
|---|---|
| Predictions are displayed based on user input - Captures or uploads photos from the user’s camera - Local storage of images |
| - Resizes and preprocesses the incoming picture and converts photos to a model-compatible format |
| - Neural network model trained on image data - Accepts preprocessed images as inputs - Outputs predictions for each class |
| - Generates human-readable labels and confidence scores from model predictions |
| - Displays the predicted class labels and their corresponding confidence scores |
| - Renders the predictions on the user interface - Shows the class labels and confidence scores |
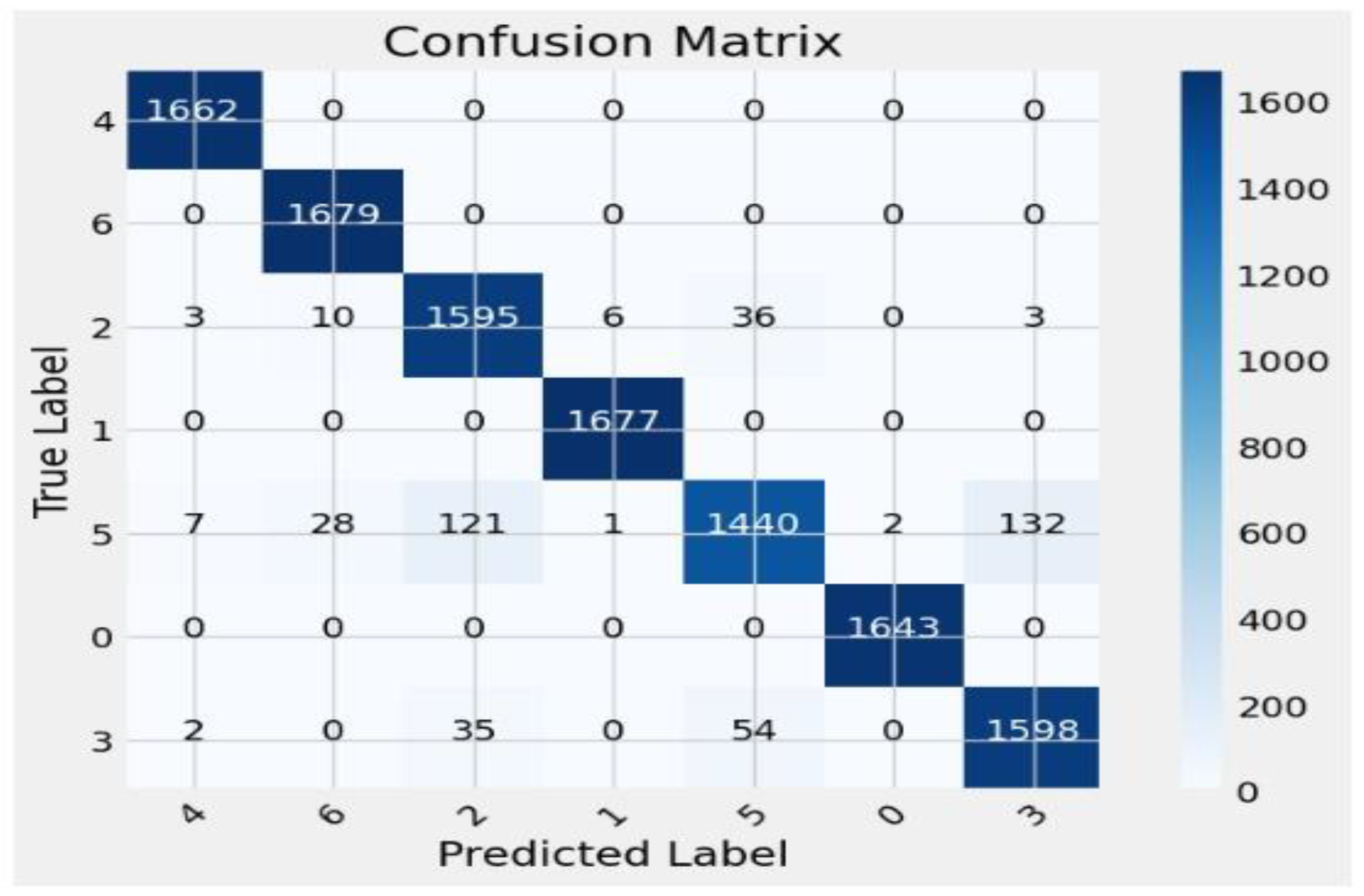
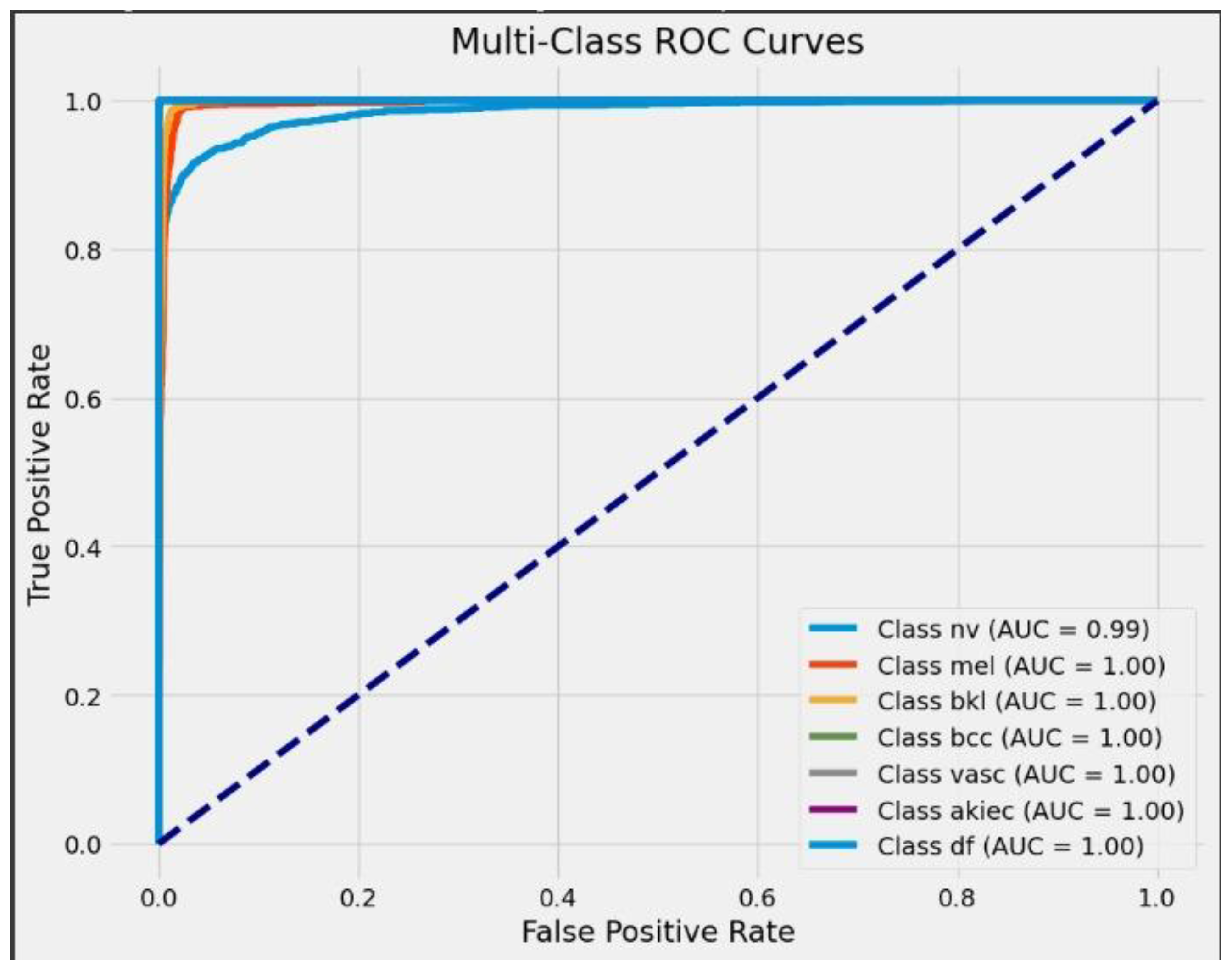
| Classes | Precision | Recall | F1 Score | Sensitivity | Specificity | AUC | ROC | Support |
|---|---|---|---|---|---|---|---|---|
| 0 | 0.99 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.99 | 1662 |
| 1 | 0.98 | 1.00 | 0.99 | 1.00 | 1.00 | 0.99 | 1.00 | 1679 |
| 2 | 0.91 | 0.96 | 0.94 | 1.00 | 1.00 | 0.99 | 1.00 | 1653 |
| 3 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1677 |
| 4 | 0.94 | 0.83 | 0.88 | 1.00 | 1.00 | 0.99 | 1.00 | 1731 |
| 5 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 1.00 | 1643 |
| 6 | 0.92 | 0.95 | 0.93 | 1.00 | 1.00 | 0.99 | 1.00 | 1689 |
| Accuracy | 96.25% | 11734 | ||||||
| Macro avg | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 11734 |
| Weighted avg | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 0.96 | 11734 |
| Training accuracy | 98. 26% |
| Article | Dataset | Preprocessing | Model | Accuracy | Classes | Image Type | Precision | Recall | F1 Score | AUC and ROC | Sensitivity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| [41] | HAM10000 | Yes | CNN transfer learning | 87.9% | 7 | RGB | 86% | 86% | 85% | -- | -- |
| [42] | HAM10000 | yes | Modified EfficientNet | 95.95 | 7 | RGB | 0.83 | 0.94 | 0.88 | -- | -- |
| [39] | HAM10000 | Yes | VGG 19 | 86 | 7 | RGB | 71 | 74 | 72 | -- | -- |
| [43] | HAM10000 | Yes | ResNet-50 + VGG-16 | 94.14%. | 7 | RGB | No | No | No | -- | -- |
| [44] | HAM10000 | Yes | Inception v3 | 0.90 | 7 | RGB | 0.90 | 0.90 | 0.90 | -- | -- |
| [45] | HAM10000 | Yes | Vision transformers (RGB images) | 92.14% | 7 | RGB | 92.61% | 92.14% | 92.17% | -- | -- |
| [46] | HAM10000 | Yes | Entropy-NDOELM algorithm | 95.7% | 7 | RGB | No | No | No | -- | -- |
| Proposed | HAM10000 | Yes | Sequential convolutional neural network | 96.25 | 7 | RGB | 96.27 | 96.23 | 96.23 | 0.96 | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdullah; Siddique, A.; Shaukat, K.; Jan, T. An Intelligent Mechanism to Detect Multi-Factor Skin Cancer. Diagnostics 2024, 14, 1359. https://doi.org/10.3390/diagnostics14131359
Abdullah, Siddique A, Shaukat K, Jan T. An Intelligent Mechanism to Detect Multi-Factor Skin Cancer. Diagnostics. 2024; 14(13):1359. https://doi.org/10.3390/diagnostics14131359
Chicago/Turabian StyleAbdullah, Ansar Siddique, Kamran Shaukat, and Tony Jan. 2024. "An Intelligent Mechanism to Detect Multi-Factor Skin Cancer" Diagnostics 14, no. 13: 1359. https://doi.org/10.3390/diagnostics14131359
APA StyleAbdullah, Siddique, A., Shaukat, K., & Jan, T. (2024). An Intelligent Mechanism to Detect Multi-Factor Skin Cancer. Diagnostics, 14(13), 1359. https://doi.org/10.3390/diagnostics14131359







