Retrospective Analysis to Optimize the Detection of MET Exon 14 Skipping Mutations in Non-Small Cell Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Validation of Oncomine™ Focus Assay Results
3.2. Validation of the Pan Lung Cancer PCR Panel Results
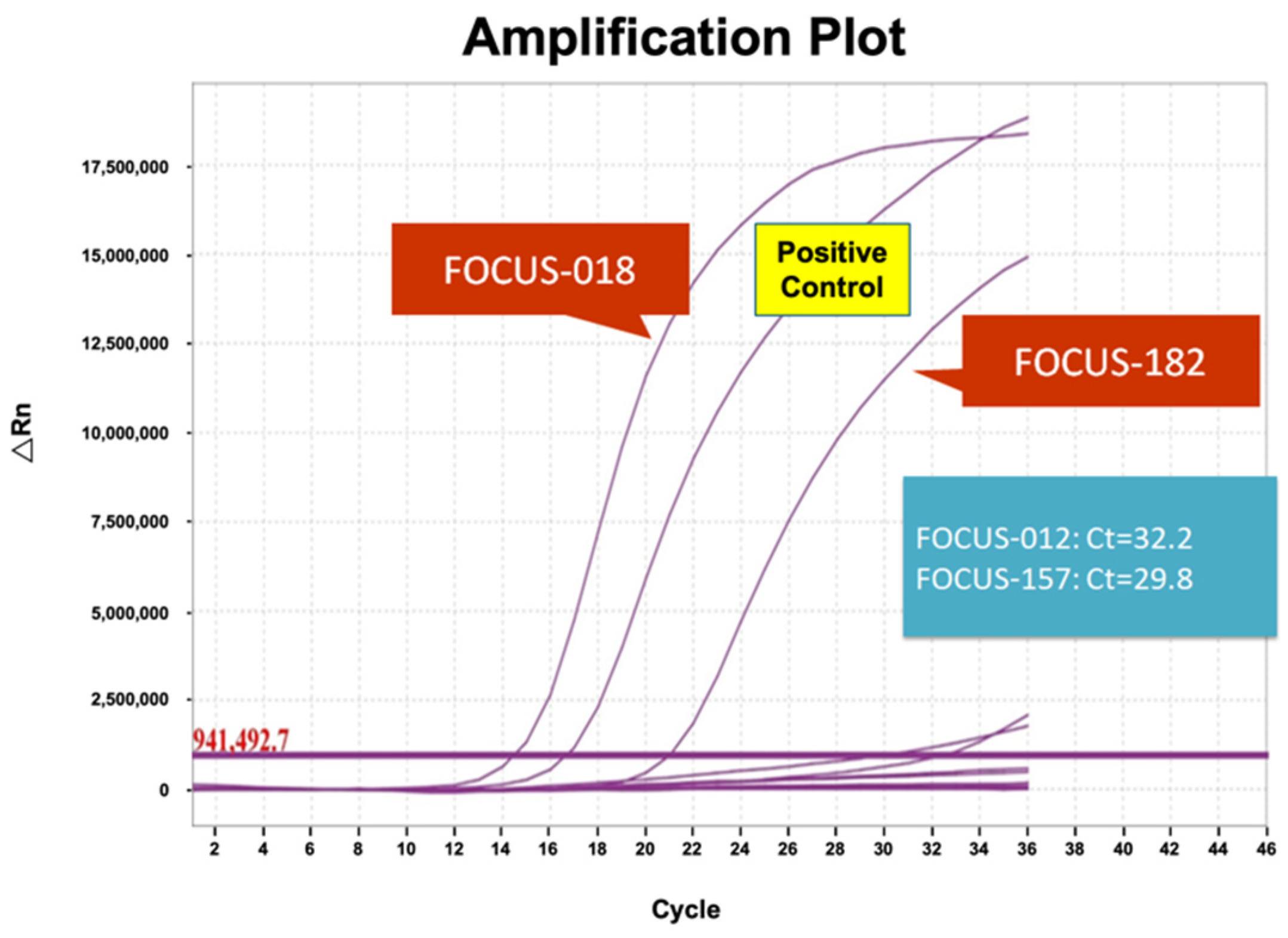
3.3. Validation of RT-PCR Results
3.4. Comparative Analysis of Three Platforms Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gherardi, E.; Birchmeier, W.; Birchmeier, C.; Vande Woude, G. Targeting MET in cancer: Rationale and progress. Nat. Rev. Cancer 2012, 12, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Socinski, M.A.; Pennell, N.A.; Davies, K.D. MET Exon 14 Skipping Mutations in Non-Small-Cell Lung Cancer: An Overview of Biology, Clinical Outcomes, and Testing Considerations. JCO Precis. Oncol. 2021, 5, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.H.; Yeung, S.F.; Chan, A.W.; Chung, L.Y.; Chau, S.L.; Lung, R.W.; Tong, C.Y.; Chow, C.; Tin, E.K.; Yu, Y.H.; et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. 2016, 22, 3048–3056. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.; Qiu, Y.; Zhang, J.; Wei, W.; Gao, H.; Yuan, Y.; Wang, X. Clinicopathological characteristics of Non-Small Cell Lung Cancer (NSCLC) patients with c-MET exon 14 skipping mutation, MET overexpression and amplification. BMC Pulm. Med. 2023, 23, 240. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, M.D.; Ciuleanu, T.E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef] [PubMed]
- Yatabe, Y.; Sunami, K.; Goto, K.; Nishio, K.; Aragane, N.; Ikeda, S.; Inoue, A.; Kinoshita, I.; Kimura, H.; Sakamoto, T.; et al. Multiplex gene-panel testing for lung cancer patients. Pathol. Int. 2020, 70, 921–931. [Google Scholar] [CrossRef] [PubMed]
- Teishikata, T.; Shiraishi, K.; Shinno, Y.; Kobayashi, Y.; Kashima, J.; Ishiyama, T.; Yoshida, T.; Mori, T.; Yatabe, Y. An Alert to Possible False Positives with a Commercial Assay for MET Exon 14 Skipping. J. Thorac. Oncol. 2021, 16, 2133–2138. [Google Scholar] [CrossRef] [PubMed]
- Onodera, Y.; Sekine, A.; Hagiwara, E.; Yamada, S.; Ikeda, S.; Tabata, E.; Kitamura, H.; Baba, T.; Komatsu, S.; Okudela, K.; et al. Successful tepotinib treatment of adenocarcinoma with MET exon 14 skipping and discordant results between Oncomine Dx target test and ArcherMET: A case report. Mol. Clin. Oncol. 2023, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Lee, S.H.; Jung, C.K.; Park, G.; Lee, K.Y.; Choi, H.J.; Min, K.O.; Kim, T.J.; Lee, E.J.; Lee, Y.S. Use of the Ion AmpliSeq Cancer Hotspot Panel in clinical molecular pathology laboratories for analysis of solid tumours: With emphasis on validation with relevant single molecular pathology tests and the Oncomine Focus Assay. Pathol. Res. Pract. 2018, 214, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.L.; Walsh, K.; Diamond, A.; Oniscu, A.; Deans, Z.C. Validation of the Oncomine() focus panel for next-generation sequencing of clinical tumour samples. Virchows Arch. 2018, 473, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Balasubramaniam, S.; Zhang, H.; Berman, T.; Narayan, P.; Suzman, D.; Bloomquist, E.; Tang, S.; Gong, Y.; Sridhara, R.; et al. FDA Approval Summary: Olaparib Monotherapy or in Combination with Bevacizumab for the Maintenance Treatment of Patients with Advanced Ovarian Cancer. Oncologist 2021, 26, e164–e172. [Google Scholar] [CrossRef] [PubMed]
- Kunimasa, K.; Matsumoto, S.; Kawamura, T.; Inoue, T.; Tamiya, M.; Kanzaki, R.; Maniwa, T.; Okami, J.; Honma, K.; Goto, K.; et al. Clinical application of the AMOY 9-in-1 panel to lung cancer patients. Lung Cancer 2023, 179, 107190. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Madison, R.; Classon, A.; Gjoerup, O.; Rosenzweig, M.; Frampton, G.M.; Alexander, B.M.; Oxnard, G.R.; Venstrom, J.M.; Awad, M.M.; et al. Characterization of Non-Small-Cell Lung Cancers with MET Exon 14 Skipping Alterations Detected in Tissue or Liquid: Clinicogenomics and Real-World Treatment Patterns. JCO Precis. Oncol. 2021, 5, 1354–1376. [Google Scholar] [CrossRef] [PubMed]
- Mazieres, J.; Vioix, H.; Pfeiffer, B.M.; Campden, R.I.; Chen, Z.; Heeg, B.; Cortot, A.B. MET Exon 14 Skipping in NSCLC: A Systematic Literature Review of Epidemiology, Clinical Characteristics, and Outcomes. Clin. Lung Cancer 2023, 24, P483–P497. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chen, M.; Lin, J.; Chen, X.; Yu, X.; Chen, Z.; Jin, L. Identifying a wide range of actionable variants using capture-based ultra-deep targeted sequencing in treatment-naive patients with primary lung adenocarcinoma. Int. J. Clin. Exp. Pathol. 2020, 13, 525–535. [Google Scholar] [PubMed]
- Kato, K.; Okami, J.; Nakamura, H.; Honma, K.; Sato, Y.; Nakamura, S.; Kukita, Y.; Nakatsuka, S.I.; Higashiyama, M. Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics. Diagnostics 2023, 13, 1476. [Google Scholar] [CrossRef] [PubMed]


| Sample | Oncomine Focus Assay | AmoyDx (Ct < 28) | RT-PCR | ||
|---|---|---|---|---|---|
| Read Count (Cut Off >120) | Total Mapped Fusion Reads | Fusion Reads Count/Total Mapped Fusion Reads | |||
| FOCUS-004 | 302 | 221,438 | 0.13% | - | - |
| FOCUS-012 | 612 | 280,337 | 0.22% | - | - |
| FOCUS-018 | 10,177 | 218,892 | 4.65% | + * | + * |
| FOCUS-090 | 261 | 318,617 | 0.08% | - | - |
| FOCUS-128 | 154 | 282,856 | 0.05% | - | - |
| FOCUS-157 | 201 | 153,230 | 0.13% | - | - |
| FOCUS-159 | 179 | 476,881 | 0.04% | - | - |
| FOCUS-160 | 319 | 581,357 | 0.05% | - | - |
| FOCUS-182 | 2540 | 197,851 | 1.28% | + * | + * |
| FOCUS-199 | 333 | 133,050 | 0.25% | - | - |
| FOCUS-202 | 212 | 122,505 | 0.17% | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.-J.; Tsai, S.-H.; Lin, L.-C.; Chiueh, T.-S. Retrospective Analysis to Optimize the Detection of MET Exon 14 Skipping Mutations in Non-Small Cell Lung Cancer. Diagnostics 2024, 14, 1110. https://doi.org/10.3390/diagnostics14111110
Lu J-J, Tsai S-H, Lin L-C, Chiueh T-S. Retrospective Analysis to Optimize the Detection of MET Exon 14 Skipping Mutations in Non-Small Cell Lung Cancer. Diagnostics. 2024; 14(11):1110. https://doi.org/10.3390/diagnostics14111110
Chicago/Turabian StyleLu, Jang-Jih, Shu-Hui Tsai, Lee-Chung Lin, and Tzong-Shi Chiueh. 2024. "Retrospective Analysis to Optimize the Detection of MET Exon 14 Skipping Mutations in Non-Small Cell Lung Cancer" Diagnostics 14, no. 11: 1110. https://doi.org/10.3390/diagnostics14111110
APA StyleLu, J.-J., Tsai, S.-H., Lin, L.-C., & Chiueh, T.-S. (2024). Retrospective Analysis to Optimize the Detection of MET Exon 14 Skipping Mutations in Non-Small Cell Lung Cancer. Diagnostics, 14(11), 1110. https://doi.org/10.3390/diagnostics14111110






