Liver Transplant in Patients with Hepatocarcinoma: Imaging Guidelines and Future Perspectives Using Artificial Intelligence
Abstract
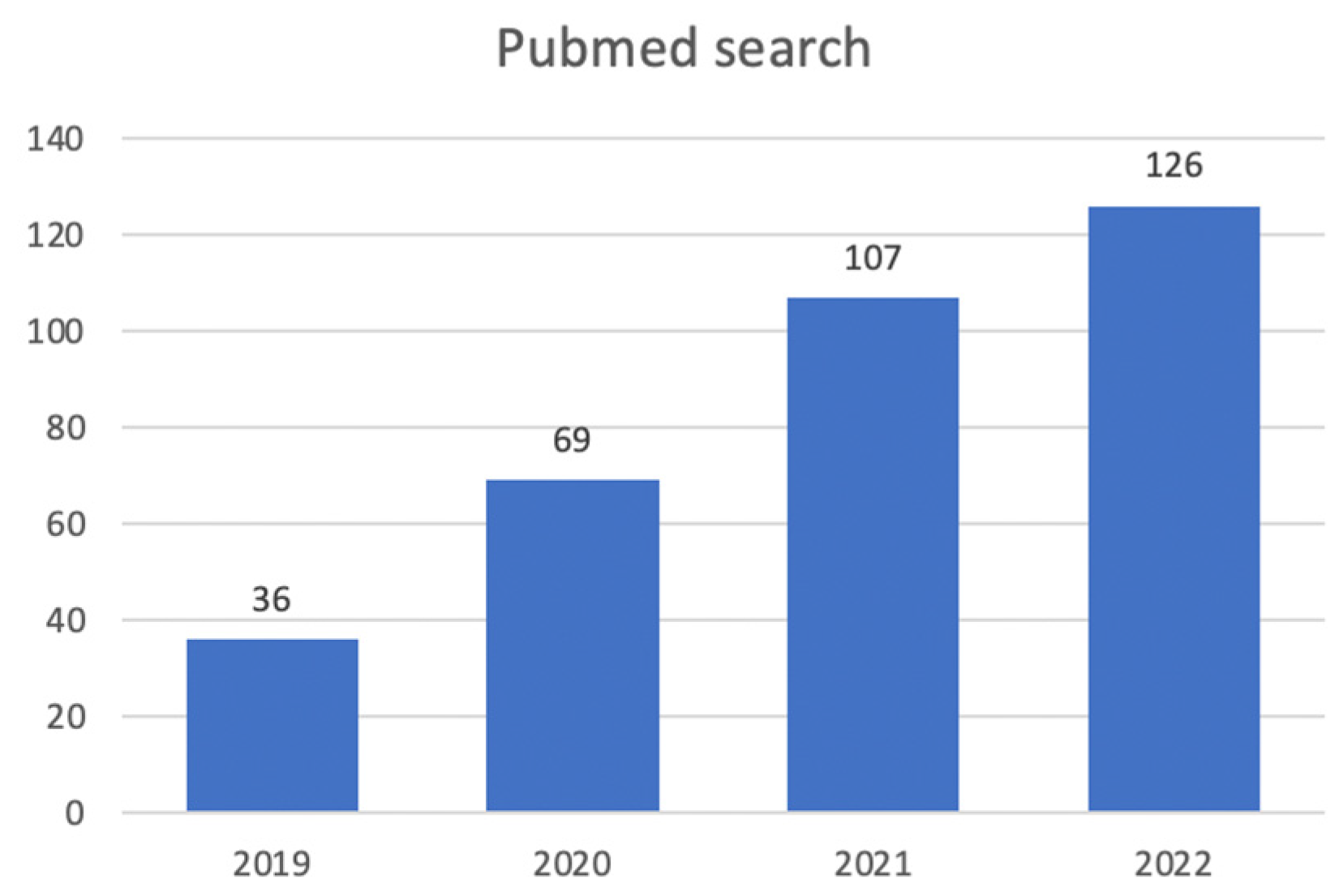
1. Introduction
2. Liver Transplant in HCC
3. Extending Milan
4. AI-Aided Evaluations in Candidates for LT with HCC
4.1. Detection
| Author | Year | Modality | AI-Method | Sensitivity |
|---|---|---|---|---|
| Tiyarattanachai et al. [50] | 2022 | US | DL (RetinaNet CNN) | 89.8% |
| Lee et al. [51] | 2019 | CECT | DL (CNN) | 93.8% |
| Kim et al. [52] | 2021 | CECT | DL (Mask R-CNN) | 84.8% |
| Kim et al. [53] | 2020 | MRI | DL (CNN) | 87% |
| Fabijańska et al. [54] | 2018 | MRI | DL (U-Net CNN) | 90.8% |
4.2. Segmentation
| Author | Year | Scope | Modality | AI-Method | DICE Score |
|---|---|---|---|---|---|
| Tian et al. [63] | 2019 | Couinaud segmentation | CECT | DL (GLC-UNet CNN) | 92.46% |
| Wang et al. [64] | 2022 | Couinaud segmentation | CECT | DL (ARH-CNet CNN) | 84% |
| Han et al. [65] | 2022 | Couinaud segmentation | MRI | DL (U-Net CNN) | 90.2% |
| Jimenez-Pastor et al. [69] | 2021 | Liver segmentation, fat, and iron quantification | MRI | DL (CNN) | 93% |
| Bousabarah et al. [70] | 2021 | Liver and HCC segmentation | MRI | DL (U-Net CNN) | 91% for liver 68% for HCC |
| Durand et al. [74] | 2020 | Sarcopenia evaluation | CT | DL (U-Net CNN) | 97% |
4.3. Classification
4.3.1. Microvascular Invasion
4.3.2. HCC Grading Prediction
4.3.3. Molecular Evaluation
| Author | Year | Scope | Data | AI-Method | AUC |
|---|---|---|---|---|---|
| Gu et al. [90] | 2020 | GPC3 prediction | DCE-MRI (Gd- DTPA) + Clinical | Radiomics | 0.914 |
| Chong et al. [91] | 2023 | GPC3 prediction | DCE-MRI (Gd-EOB-DTPA) + Clinical | Radiomics | 0.943 |
| Zhang et al. [93] | 2022 | CK19 prediction | US + Clinical | Radiomics | 0.867 |
| Yang et al. [94] | 2021 | CK19 prediction | DCE-MRI (Gd-EOB-DTPA) | Radiomics | 0.79 |
| Chen et al. [95] | 2021 | CK19 prediction | DCE-MRI (Gd-EOB-DTPA) + Clinical | Radiomics | 0.833 |
5. Discussion and Limitations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Hepatol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Akinyemiju, T.; Abera, S.; Ahmed, M.; Alam, N.; Alemayohu, M.A.; Allen, C.; Al-Raddadi, R.; Alvis-Guzman, N.; Amoako, Y.; Artaman, A.; et al. Global Burden of Disease Liver Cancer Collaboration. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: Results from the global burden of disease study 2015. JAMA Oncol. 2017, 3, 1683–1691. [Google Scholar] [PubMed]
- Anstee, Q.M.; Reeves, H.L.; Kotsiliti, E.; Govaere, O.; Heikenwalder, M. From NASH to HCC: Current concepts and future challenges. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 411–428. [Google Scholar] [CrossRef]
- Araújo, A.R.; Rosso, N.; Bedogni, G.; Tiribelli, C.; Bellentani, S. Global epidemiology of non-alcoholic fatty liver disease/non-alcoholic steatohepatitis: What we need in the future. Liver Int. 2018, 38, 47–51. [Google Scholar] [CrossRef]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef]
- Kim, W.R.; Lake, J.R.; Smith, J.M.; Schladt, D.P.; Skeans, M.A.; Harper, A.M.; Wainright, J.L.; Snyder, J.J.; Israni, A.K.; Kasiske, B.L. OPTN/SRTR 2016 Annual Data Report: Liver. Am. J. Transpl. 2018, 18 (Suppl. S1), 172–253. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- Mehta, N.; Dodge, J.L.; Hirose, R.; Roberts, J.P.; Yao, F.Y. Predictors of low risk for dropout from the liver transplant waiting list for hepatocellular carcinoma in long wait time regions: Implications for organ allocation. Am. J. Transpl. 2019, 19, 2210–2218. [Google Scholar] [CrossRef]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- Do, S.; Song, K.D.; Chung, J.W. Basics of deep learning: A radiologist’s guide to understanding published radiology articles on deep learning. Korean J. Radiol. 2020, 21, 33–41. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Into Imaging 2020, 11, 93. [Google Scholar] [CrossRef]
- Min, S.; Lee, B.; Yoon, S. Deep learning in bioinformatics. Brief. Bioinform. 2017, 18, 851–869. [Google Scholar] [CrossRef]
- Ueda, D.; Shimazaki, A.; Miki, Y. Technical and clinical overview of deep learning in radiology. Jpn. J. Radiol. 2019, 37, 15–33. [Google Scholar] [CrossRef]
- Minor, L.B. Harnessing the Power of Data in Health. Stanf. Med. Health Trends Rep. 2017. Available online: https://med.stanford.edu/content/dam/sm/sm-news/documents/StanfordMedicineHealthTrendsWhitePaper2017.pdf (accessed on 7 March 2023).
- Briceño, J.; Cruz-Ramírez, M.; Prieto, M.; Navasa, M.; De Urbina, J.O.; Orti, R.; Gómez-Bravo, M.Á.; Otero, A.; Varo, E.; Tomé, S.; et al. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: Results from a multicenter Spanish study. J. Hepatol. 2014, 61, 1020–1028. [Google Scholar] [CrossRef]
- Briceño, J.; Ayllón, M.D.; Ciria, R. Machine-learning algorithms for predicting results in liver transplantation: The problem of donor–recipient matching. Curr. Opin. Organ Transplant. 2020, 25, 406–411. [Google Scholar] [CrossRef]
- Bertsimas, D.; Kung, J.; Trichakis, N.; Wang, Y.; Hirose, R.; Vagefi, P.A. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. Am. J. Transplant. 2019, 19, 1109–1118. [Google Scholar] [CrossRef]
- Wingfield, L.R.; Ceresa, C.; Thorogood, S.; Fleuriot, J.; Knight, S. Using artificial intelligence for predicting survival of individual grafts in liver transplantation: A systematic review. Liver Transplant. 2020, 26, 922–934. [Google Scholar] [CrossRef]
- Survarachakan, S.; Prasad, P.J.R.; Naseem, R.; de Frutos, J.P.; Kumar, R.P.; Langø, T.; Cheikh, F.A.; Elle, O.J.; Lindseth, F. Deep learning for image-based liver analysis—A comprehensive review focusing on malignant lesions. Artif. Intell. Med. 2022, 130, 102331. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, H.; Huang, Y.; Yan, B.; Chang, Z.; Liu, Z.; Zhao, M.; Cui, L.; Song, J.; Li, F. Trends in the application of deep learning networks in medical image analysis: Evolution between 2012 and 2020. Eur. J. Radiol. 2022, 146, 110069. [Google Scholar] [CrossRef] [PubMed]
- Nam, D.; Chapiro, J.; Paradis, V.; Seraphin, T.P.; Kather, J.N. Artificial intelligence in liver diseases: Improving diagnostics, prognostics and response prediction. JHEP Rep. 2022, 4, 100443. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, K.G.; Schalekamp, S.; Rutten, M.J.; van Ginneken, B.; de Rooij, M. Artificial intelligence in radiology: 100 commercially available products and their scientific evidence. Eur. Radiol. 2021, 31, 3797–3804. [Google Scholar] [CrossRef] [PubMed]
- Mazzaferro, V.; Regalia, E.; Doci, R.; Andreola, S.; Pulvirenti, A.; Bozzetti, F.; Montalto, F.; Ammatuna, M.; Morabito, A.; Gennari, L. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. New Engl. J. Med. 1996, 334, 693–700. [Google Scholar] [CrossRef]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.L.; Schirmacher, P.; Vilgrain, V. EASL clinical practice guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef]
- Vogel, A.; Cervantes, A.; Chau, I.; Daniele, B.; Llovet, J.M.; Meyer, T.; Nault, J.C.; Neumann, U.; Ricke, J.; Sangro, B.; et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv238–iv255. [Google Scholar] [CrossRef]
- Benson, A.B.; D’Angelica, M.I.; Abbott, D.E.; Anaya, D.A.; Anders, R.; Are, C.; Bachini, M.; Borad, M.; Brown, D.; Burgoyne, A.; et al. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 541–565. Available online: https://jnccn.org/view/journals/jnccn/19/5/article-p541.xml (accessed on 7 March 2023). [CrossRef]
- Heimbach, J.K.; Kulik, L.M.; Finn, R.S.; Sirlin, C.B.; Abecassis, M.M.; Roberts, L.R.; Zhu, A.X.; Murad, M.H.; Marrero, J.A. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018, 67, 358–380. [Google Scholar] [CrossRef]
- Available online: https://www.eurotransplant.org (accessed on 7 March 2023).
- Murali, A.R.; Patil, S.; Phillips, K.T.; Voigt, M.D. Locoregional therapy with curative intent versus primary liver transplant for hepatocellular carcinoma: Systematic review and meta-analysis. Transplantation 2017, 101, e249–e257. [Google Scholar] [CrossRef]
- Yao, F.Y.; Ferrell, L.; Bass, N.M.; Watson, J.J.; Bacchetti, P.; Venook, A.; Ascher, N.L.; Roberts, J.P. Liver transplantation for hepatocellular carcinoma: Expansion of the tumor size limits does not adversely impact survival. Hepatology 2001, 33, 1394–1403. [Google Scholar] [CrossRef]
- Mazzaferro, V.; Llovet, J.M.; Miceli, R.; Bhoori, S.; Schiavo, M.; Mariani, L.; Camerini, T.; Roayaie, S.; Schwartz, M.E.; Grazi, G.L.; et al. Predicting survival after liver transplantation in patients with hepatocellular carcinoma beyond the Milan criteria: A retrospective, exploratory analysis. Lancet Oncol. 2009, 10, 35–43. [Google Scholar] [CrossRef]
- Mazzaferro, V.; Sposito, C.; Zhou, J.; Pinna, A.D.; De Carlis, L.; Fan, J.; Cescon, M.; Di Sandro, S.; Yi-Feng, H.; Lauterio, A.; et al. Metroticket 2.0 model for analysis of competing risks of death after liver transplantation for hepatocellular carcinoma. Gastroenterology 2018, 154, 128–139. [Google Scholar] [CrossRef]
- Duvoux, C.; Roudot-Thoraval, F.; Decaens, T.; Pessione, F.; Badran, H.; Piardi, T.; Francoz, C.; Compagnon, P.; Vanlemmens, C.; Dumortier, J.; et al. Liver Transplantation French Study Group. Liver transplantation for hepatocellular carcinoma: A model including α-fetoprotein improves the performance of Milan criteria. Gastroenterology 2012, 143, 986–994. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. OPTN/UNOS Liver and Intestinal Organ Transplantation Committee. Available online: https://optn.transplant.hrsa.gov/media/1922/liver_hcc_criteria_for_auto_approval_20160815.pdf (accessed on 13 December 2022).
- DuBay, D.; Sandroussi, C.; Sandhu, L.; Cleary, S.; Guba, M.; Cattral, M.S.; McGilvray, I.; Ghanekar, A.; Selzner, M.; Greig, P.D.; et al. Liver transplantation for advanced hepatocellular carcinoma using poor tumor differentiation on biopsy as an exclusion criterion. Ann. Surg. 2011, 253, 166–172. [Google Scholar] [CrossRef]
- Toso, C.; Trotter, J.; Wei, A.; Bigam, D.L.; Shah, S.; Lancaster, J.; Grant, D.R.; Greig, P.D.; Shapiro, A.J.; Kneteman, N.M. Total tumor volume predicts risk of recurrence following liver transplantation in patients with hepatocellular carcinoma. Liver Transplant. 2008, 14, 1107–1115. [Google Scholar] [CrossRef]
- Zheng, S.S.; Xu, X.; Wu, J.; Chen, J.; Wang, W.L.; Zhang, M.; Liang, T.B.; Wu, L.M. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation 2008, 85, 1726–1732. [Google Scholar] [CrossRef]
- Lai, Q.; Nicolini, D.; Inostroza Nunez, M.; Iesari, S.; Goffette, P.; Agostini, A.; Giovagnoni, A.; Vivarelli, M.; Lerut, J. A Novel Prognostic Index in Patients With Hepatocellular Cancer Waiting for Liver Transplantation. Ann. Surg. 2016, 264, 787–796. [Google Scholar] [CrossRef]
- Hameed, B.; Mehta, N.; Sapisochin, G.; Roberts, J.P.; Yao, F.Y. Alpha-fetoprotein level > 1000 ng/mL as an exclusion criterion for liver transplantation in patients with hepatocellular carcinoma meeting the Milan criteria. Liver Transplant. 2014, 20, 945–951. [Google Scholar] [CrossRef]
- Sapisochin, G.; Goldaracena, N.; Laurence, J.M.; Dib, M.; Barbas, A.; Ghanekar, A.; Cleary, S.P.; Lilly, L.; Cattral, M.S.; Marquez, M.; et al. The extended Toronto criteria for liver transplantation in patients with hepatocellular carcinoma: A prospective validation study. Hepatology 2016, 64, 2077–2088. [Google Scholar] [CrossRef]
- Shimamura, T.; Goto, R.; Watanabe, M.; Kawamura, N.; Takada, Y. Liver transplantation for Hepatocellular Carcinoma: How Should We Improve the Thresholds? Cancers 2022, 14, 419. [Google Scholar] [CrossRef] [PubMed]
- Thuluvath, P.J.; To, C.; Amjad, W. Role of locoregional therapies in patients with hepatocellular cancer awaiting liver transplantation. Off. J. Am. Coll. Gastroenterol. ACG 2021, 116, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Mehta, N.; Bhangui, P.; Yao, F.Y.; Mazzaferro, V.; Toso, C.; Akamatsu, N.; Durand, F.; Ijzermans, J.; Polak, W.; Zheng, S.; et al. Liver transplantation for hepatocellular carcinoma. Working group report from the ILTS transplant oncology consensus conference. Transplantation 2020, 104, 1136–1142. [Google Scholar] [CrossRef] [PubMed]
- Tzartzeva, K.; Obi, J.; Rich, N.E.; Parikh, N.D.; Marrero, J.A.; Yopp, A.; Waljee, A.K.; Singal, A.G. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: A meta-analysis. Gastroenterology 2018, 154, 1706–1718. [Google Scholar] [CrossRef]
- Roberts, L.R.; Sirlin, C.B.; Zaiem, F.; Almasri, J.; Prokop, L.J.; Heimbach, J.K.; Murad, M.H.; Mohammed, K. Imaging for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. Hepatology 2018, 67, 401–421. [Google Scholar] [CrossRef]
- Goldberg, D.S.; Olthoff, K.M. Standardizing MELD exceptions: Current challenges and future directions. Curr. Transplant. Rep. 2014, 1, 232–237. [Google Scholar] [CrossRef]
- Song, K.D. Current status of deep learning applications in abdominal ultrasonography. Ultrasonography 2021, 40, 177. [Google Scholar] [CrossRef]
- Tiyarattanachai, T.; Apiparakoon, T.; Marukatat, S.; Sukcharoen, S.; Yimsawad, S.; Chaichuen, O.; Bhumiwat, S.; Tanpowpong, N.; Pinjaroen, N.; Rerknimitr, R.; et al. The feasibility to use artificial intelligence to aid detecting focal liver lesions in real-time ultrasound: A preliminary study based on videos. Sci. Rep. 2022, 12, 7749. [Google Scholar] [CrossRef]
- Lee, G.; Kim, J.; Lee, J.G.; Ahn, G.; Park, S.H.; Kim, S.Y.; Kim, K.W.; Lee, S.S.; Kim, N. Automatic hepatocellular carcinoma lesion detection with dynamic enhancement characteristic from multi-phase CT images. In International Forum on Medical Imaging in Asia 2019; SPIE: Bellingham, DC, USA, 2019; Volume 11050, pp. 203–208. [Google Scholar]
- Kim, D.W.; Lee, G.; Kim, S.Y.; Ahn, G.; Lee, J.G.; Lee, S.S.; Kim, K.W.; Park, S.H.; Lee, Y.J.; Kim, N. Deep learning–based algorithm to detect primary hepatic malignancy in multiphase CT of patients at high risk for HCC. Eur. Radiol. 2021, 31, 7047–7057. [Google Scholar] [CrossRef]
- Kim, J.; Min, J.H.; Kim, S.K.; Shin, S.Y.; Lee, M.W. Detection of hepatocellular carcinoma in contrast-enhanced magnetic resonance imaging using deep learning classifier: A multi-center retrospective study. Sci. Rep. 2020, 10, 9458. [Google Scholar] [CrossRef]
- Fabijańska, A.; Vacavant, A.; Lebre, M.A.; Pavan, A.L.; de Pina, D.R.; Abergel, A.; Chabrot, P.; Magnin, B. U-CatcHCC: An accurate HCC detector in hepatic DCE-MRI sequences based on an U-Net framework. In Proceedings of the International Conference on Computer Vision, ICCVG 2018, Warsaw, Poland, 17–19 September 2018; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 319–328. [Google Scholar]
- Gotra, A.; Sivakumaran, L.; Chartrand, G.; Vu, K.N.; Vandenbroucke-Menu, F.; Kauffmann, C.; Kadoury, S.; Gallix, B.; de Guise, J.A.; Tang, A. Liver segmentation: Indications, techniques and future directions. Insights Into Imaging 2017, 8, 377–392. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Heimann, T.; Styner, M. 3D Segmentation in the Clinic: A Grand Challenge. In MICCAI Workshop on 3D Segmentation in the Clinic: A Grand Challenge; 2007; Volume 1, pp. 7–15. Available online: http://rumc-gcorg-p-public.s3.amazonaws.com/f/challenge/8/5db8e512-75f2-4878-a6a1-b426f43104a5/p7.pdf (accessed on 7 March 2023).
- Deng, X.; Du, G. 3D Segmentation in the Clinic: A Grand Challenge II-Liver Tumor Segmentation. In MICCAI Workshop; 2008. Available online: https://www.researchgate.net/profile/Guangwei-Du-2/publication/267699971_Editorial_3D_Segmentation_in_the_Clinic_A_Grand_Challenge_II_-Liver_Tumor_Segmentation/links/549a28f40cf2b8037135913b/Editorial-3D-Segmentation-in-the-Clinic-A-Grand-Challenge-II-Liver-Tumor-Segmentation.pdf (accessed on 7 March 2023).
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.; Han, X.; Heng, P.; Hesser, J.W.; et al. The Liver Tumor Segmentation Benchmark (LiTS). Med. Image Anal. 2019, 84, 102680. [Google Scholar] [CrossRef]
- Singh, H.R.; Rabi, S. Study of morphological variations of liver in human. Transl. Res. Anat. 2019, 14, 1–5. [Google Scholar] [CrossRef]
- Vernuccio, F.; Whitney, S.A.; Ravindra, K.; Marin, D. CT and MR imaging evaluation of living liver donors. Abdom. Radiol. 2021, 46, 17–28. [Google Scholar] [CrossRef]
- Lim, M.C.; Tan, C.H.; Cai, J.; Zheng, J.; Kow, A.W.C. CT volumetry of the liver: Where does it stand in clinical practice? Clin. Radiol. 2014, 69, 887–895. [Google Scholar] [CrossRef]
- Couinaud, C. Le Foie: Études Anatomiques et Chirurgicales; Masson: Paris, France, 1957. [Google Scholar]
- Tian, J.; Liu, L.; Shi, Z.; Xu, F. Automatic couinaud segmentation from CT volumes on liver using GLC-UNet. In Proceedings of the 10th International Workshop, MLMI 2019, Shenzhen, China, 13 October 2019; pp. 274–282. [Google Scholar]
- Wang, M.; Jin, R.; Lu, J.; Song, E.; Ma, G. Automatic CT liver Couinaud segmentation based on key bifurcation detection with attentive residual hourglass-based cascaded network. Comput. Biol. Med. 2022, 144, 105363. [Google Scholar] [CrossRef]
- Han, X.; Wu, X.; Wang, S.; Xu, L.; Xu, H.; Zheng, D.; Yu, N.; Hong, Y.; Yu, Z.; Yang, D.; et al. Automated segmentation of liver segment on portal venous phase MR images using a 3D convolutional neural network. Insights Into Imaging 2022, 13, 26. [Google Scholar] [CrossRef]
- Hamar, M.; Selzner, M. Steatotic donor livers: Where is the risk-benefit maximized? Liver Transplant. 2017, 23, S34–S39. [Google Scholar] [CrossRef]
- Yersiz, H.; Lee, C.; Kaldas, F.M.; Hong, J.C.; Rana, A.; Schnickel, G.T.; Wertheim, J.A.; Zarrinpar, A.; Agopian, V.G.; Gornbein, J.; et al. Assessment of hepatic steatosis by transplant surgeon and expert pathologist: A prospective, double-blind evaluation of 201 donor livers. Liver Transplant. 2013, 19, 437–449. [Google Scholar] [CrossRef]
- Gu, J.; Liu, S.; Du, S.; Zhang, Q.; Xiao, J.; Dong, Q.; Xin, Y. Diagnostic value of MRI-PDFF for hepatic steatosis in patients with non-alcoholic fatty liver disease: A meta-analysis. Eur. Radiol. 2019, 29, 3564–3573. [Google Scholar] [CrossRef]
- Jimenez-Pastor, A.; Alberich-Bayarri, A.; Lopez-Gonzalez, R.; Marti-Aguado, D.; França, M.; Bachmann, R.S.M.; Mazzucco, J.; Marti-Bonmati, L. Precise whole liver automatic segmentation and quantification of PDFF and R2* on MR images. Eur. Radiol. 2021, 31, 7876–7887. [Google Scholar] [CrossRef] [PubMed]
- Bousabarah, K.; Letzen, B.; Tefera, J.; Savic, L.; Schobert, I.; Schlachter, T.; Staib, L.H.; Kocher, M.; Chapiro, J.; Lin, M. Automated detection and delineation of hepatocellular carcinoma on multiphasic contrast-enhanced MRI using deep learning. Abdom. Radiol. 2021, 46, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Van Vugt, J.L.A.; Levolger, S.; De Bruin, R.W.F.; van Rosmalen, J.; Metselaar, H.J.; IJzermans, J.N.M. Systematic review and meta-analysis of the impact of computed tomography–assessed skeletal muscle mass on outcome in patients awaiting or undergoing liver transplantation. Am. J. Transplant. 2016, 16, 2277–2292. [Google Scholar] [CrossRef] [PubMed]
- Shafaat, O.; Liu, Y.; Jackson, K.R.; Motter, J.D.; Boyarsky, B.J.; Latif, M.A.; Yuan, F.; Khalil, A.; King, E.A.; Zaheer, A.; et al. Association between abdominal CT measurements of body composition before deceased donor liver transplant with posttransplant outcomes. Radiology 2022, 306, 212403. [Google Scholar] [CrossRef] [PubMed]
- Rozynek, M.; Kucybała, I.; Urbanik, A.; Wojciechowski, W. Use of artificial intelligence in the imaging of sarcopenia: A narrative review of current status and perspectives. Nutrition 2021, 89, 111227. [Google Scholar] [CrossRef]
- Blanc-Durand, P.; Schiratti, J.B.; Schutte, K.; Jehanno, P.; Herent, P.; Pigneur, F.; Lucidarme, O.; Benaceur, Y.; Sadate, A.; Luciani, A.; et al. Abdominal musculature segmentation and surface prediction from CT using deep learning for sarcopenia assessment. Diagn. Interv. Imaging 2020, 101, 789–794. [Google Scholar] [CrossRef]
- Roayaie, S.; Blume, I.N.; Thung, S.N.; Guido, M.; Fiel, M.I.; Hiotis, S.; Labow, D.M.; Llovet, J.M.; Schwartz, M.E. A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology 2009, 137, 850–855. [Google Scholar] [CrossRef]
- Chen, Y.D.; Zhang, L.; Zhou, Z.P.; Lin, B.; Jiang, Z.J.; Tang, C.; Dang, Y.W.; Xia, Y.W.; Song, B.; Long, L.L. Radiomics and nomogram of magnetic resonance imaging for preoperative prediction of microvascular invasion in small hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 4399. [Google Scholar] [CrossRef]
- Jiang, Y.Q.; Cao, S.E.; Cao, S.; Chen, J.N.; Wang, G.Y.; Shi, W.Q.; Deng, Y.N.; Cheng, N.; Ma, K.; Zeng, K.N.; et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J. Cancer Res. Clin. Oncol. 2021, 147, 821–833. [Google Scholar] [CrossRef]
- Sun, B.Y.; Gu, P.Y.; Guan, R.Y.; Zhou, C.; Lu, J.W.; Yang, Z.F.; Pan, C.; Zhou, P.Y.; Zhu, Y.P.; Li, J.R.; et al. Deep-learning-based analysis of preoperative MRI predicts microvascular invasion and outcome in hepatocellular carcinoma. World J. Surg. Oncol. 2022, 20, 189. [Google Scholar] [CrossRef]
- Zhou, W.; Jian, W.; Cen, X.; Zhang, L.; Guo, H.; Liu, Z.; Liang, C.; Wang, G. Prediction of microvascular invasion of hepatocellular carcinoma based on contrast-enhanced MR and 3D convolutional neural networks. Front. Oncol. 2021, 11, 189. [Google Scholar] [CrossRef]
- Tamura, S.; Kato, T.; Berho, M.; Misiakos, E.P.; O’Brien, C.; Reddy, K.R.; Nery, J.R.; Burke, G.W.; Schiff, E.R.; Miller, J.; et al. Impact of histological grade of hepatocellular carcinoma on the outcome of liver transplantation. Arch. Surg. 2001, 136, 25–30. [Google Scholar] [CrossRef]
- Martins-Filho, S.N.; Paiva, C.; Azevedo, R.S.; Alves, V.A.F. Histological grading of hepatocellular carcinoma—A systematic review of literature. Front. Med. 2017, 4, 193. [Google Scholar] [CrossRef]
- Mao, B.; Zhang, L.; Ning, P.; Ding, F.; Wu, F.; Lu, G.; Geng, Y.; Ma, J. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur. Radiol. 2020, 30, 6924–6932. [Google Scholar] [CrossRef]
- Wu, M.; Tan, H.; Gao, F.; Hai, J.; Ning, P.; Chen, J.; Zhu, S.; Wang, M.; Dou, S.; Shi, D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur. Radiol. 2019, 29, 2802–2811. [Google Scholar] [CrossRef]
- Han, Y.E.; Cho, Y.; Kim, M.J.; Park, B.J.; Sung, D.J.; Han, N.Y.; Sim, K.C.; Park, Y.S.; Park, B.N. Hepatocellular carcinoma pathologic grade prediction using radiomics and machine learning models of gadoxetic acid-enhanced MRI: A two-center study. Abdom. Radiol. 2023, 48, 244–256. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, G.; Xie, G.; Zhang, L. Grading of hepatocellular carcinoma based on diffusion weighted images with multiple b-values using convolutional neural networks. Med. Phys. 2019, 46, 3951–3960. [Google Scholar] [CrossRef]
- Zhou, Q.; Zhou, Z.; Chen, C.; Fan, G.; Chen, G.; Heng, H.; Ji, J.; Dai, Y. Grading of hepatocellular carcinoma using 3D SE-DenseNet in dynamic enhanced MR images. Comput. Biol. Med. 2019, 107, 47–57. [Google Scholar] [CrossRef]
- Xiao, W.K.; Qi, C.Y.; Chen, D.; Li, S.Q.; Fu, S.J.; Peng, B.G.; Liang, L.J. Prognostic significance of glypican-3 in hepatocellular carcinoma: A meta-analysis. BMC Cancer 2014, 14, e9702. [Google Scholar] [CrossRef]
- Wang, Y.L.; Zhu, Z.J.; Teng, D.H.; Yao, Z.; Gao, W.; Shen, Z.Y. Glypican-3 expression and its relationship with recurrence of HCC after liver transplantation. World J. Gastroenterol. WJG 2012, 18, 2408. [Google Scholar] [CrossRef]
- Cui, X.; Li, Z.; Gao, P.J.; Gao, J.; Zhu, J.Y. Prognostic value of glypican-3 in patients with HBV-associated hepatocellular carcinoma after liver transplantation. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 157–163. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Xie, Y.; Wei, J.; Li, W.; Ye, Z.; Zhu, Z.; Tian, J.; Li, X. MRI-based radiomics signature: A potential biomarker for identifying glypican 3-positive hepatocellular carcinoma. J. Magn. Reson. Imaging 2020, 52, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.; Gong, Y.; Zhang, Y.; Dai, Y.; Sheng, R.; Zeng, M. Radiomics on Gadoxetate Disodium-enhanced MRI: Non-invasively Identifying Glypican 3-Positive Hepatocellular Carcinoma and Postoperative Recurrence. Acad. Radiol. 2023, 30, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Miltiadous, O.; Sia, D.; Hoshida, Y.; Fiel, M.I.; Harrington, A.N.; Thung, S.N.; Tan, P.S.; Dong, H.; Revill, K.; Chang, C.Y.; et al. Progenitor cell markers predict outcome of patients with hepatocellular carcinoma beyond Milan criteria undergoing liver transplantation. J. Hepatol. 2015, 63, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qi, Q.; Li, Q.; Ren, S.; Liu, S.; Mao, B.; Li, X.; Wu, Y.; Yang, L.; Liu, L.; et al. Ultrasomics prediction for cytokeratin 19 expression in hepatocellular carcinoma: A multicenter study. Front. Oncol. 2022, 12, 994456. [Google Scholar] [CrossRef]
- Yang, F.; Wan, Y.; Xu, L.; Wu, Y.; Shen, X.; Wang, J.; Lu, D.; Shao, C.; Zheng, S.; Niu, T.; et al. MRI-Radiomics prediction for cytokeratin 19-Positive hepatocellular carcinoma: A multicenter study. Front. Oncol. 2021, 11, 672126. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Zhang, Y.; Lin, Z.; Wang, M.; Huang, L.; Huang, M.; Tang, M.; Zhou, X.; Peng, Z.; et al. Preoperative Prediction of Cytokeratin 19 Expression for Hepatocellular Carcinoma with Deep Learning Radiomics Based on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging. J. Hepatocell. Carcinoma 2021, 8, 795–808. [Google Scholar] [CrossRef]
- Court, C.M.; Harlander-Locke, M.P.; Markovic, D.; French, S.W.; Naini, B.V.; Lu, D.S.; Raman, S.S.; Kaldas, F.M.; Zarrinpar, A.; Farmer, D.G.; et al. Determination of hepatocellular carcinoma grade by needle biopsy is unreliable for liver transplant candidate selection. Liver Transplant. 2017, 23, 1123–1132. [Google Scholar] [CrossRef]
- Hong, S.B.; Choi, S.H.; Kim, S.Y.; Shim, J.H.; Lee, S.S.; Byun, J.H.; Park, S.H.; Kim, K.W.; Kim, S.; Lee, N.K. MRI features for predicting microvascular invasion of hepatocellular carcinoma: A systematic review and meta-analysis. Liver Cancer 2021, 10, 94–106. [Google Scholar] [CrossRef]
- Choi, S.Y.; Kim, S.H.; Park, C.K.; Min, J.H.; Lee, J.E.; Choi, Y.H.; Lee, B.R. Imaging features of gadoxetic acid–enhanced and diffusion-weighted MR imaging for identifying cytokeratin 19-positive hepatocellular carcinoma: A Retrospective Observational Study. Radiology 2018, 286, 897–908. [Google Scholar] [CrossRef]
- Cho, E.S.; Choi, J.Y. MRI features of hepatocellular carcinoma related to biologic behavior. Korean J. Radiol. 2015, 16, 449–464. [Google Scholar] [CrossRef]
- Johnson, P.; Zhou, Q.; Dao, D.Y.; Lo, Y.D. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 670–681. [Google Scholar] [CrossRef]
- Soler, L.; Hostettler, A.; Agnus, V.; Charnoz, A.; Fasquel, J.-B.; Moreau, J.; Osswald, A.-B.; Bouhadjar, M.; Marescaux, J. 3D Image Reconstruction for Comparison of Algorithm Database: A Patient Specific Anatomical and Medical Image Database; Technical Report; IRCAD: Strasbourg, France, 2010. [Google Scholar]
- Kavur, A.E.; Selver, M.A.; Dicle, O.; Barıs, M.; Gezer, N.S. CHAOS-combined (CT-MR) healthy abdominal organ segmentation challenge data. Med. Image. Anal. 2019, 69, 101950. [Google Scholar] [CrossRef]
- Cardobi, N.; Dal Palù, A.; Pedrini, F.; Beleù, A.; Nocini, R.; De Robertis, R.; Ruzzenente, A.; Salvia, R.; Montemezzi, S.; D’Onofrio, M. An Overview of Artificial Intelligence Applications in Liver and Pancreatic Imaging. Cancers 2021, 13, 2162. [Google Scholar] [CrossRef]
- Zhang, J.; Huang, S.; Xu, Y.; Wu, J. Diagnostic accuracy of artificial intelligence based on imaging data for preoperative prediction of microvascular invasion in hepatocellular carcinoma: A systematic review and meta-analysis. Front. Oncol. 2022, 12, 763842. [Google Scholar] [CrossRef]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable deep learning models in medical image analysis. J. Imaging 2020, 6, 52. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E., Jr. Checklist for artificial intelligence in medical imaging (CLAIM): A guide for authors and reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef]

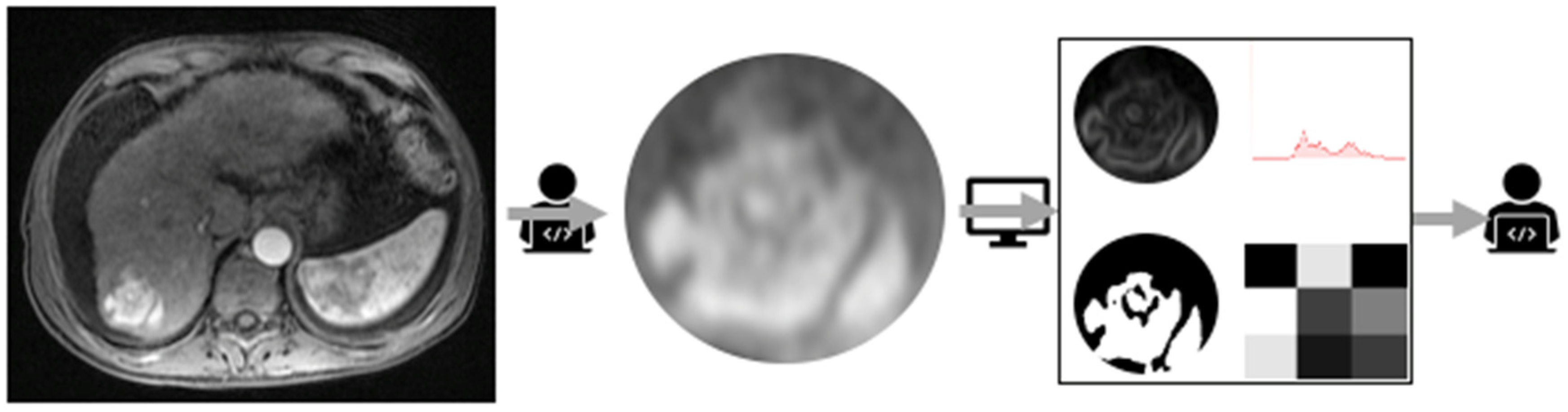





| CRITERIA | REPORT |
|---|---|
| MILAN [25] | One lesion ≤5 cm or a maximum of 3 lesions each ≤3 cm |
| University of California, San Francisco (UCSF) [32] | One lesion ≤6.5 cm or a maximum of 3 lesions with the largest tumor diameter ≤4.5 cm and a total tumor diameter ≤8 |
| Up-to-7 [33] | The sum of the number of lesions and the diameter of the largest lesion ≤7 |
| Updated Up-to-7/Metroticket V2.0 [34] | A combination of the sum of the number of lesions, the largest lesion diameter and AFP |
| AFP model [35] | A score based on the largest tumour diameter, number of nodules and AFP; A result of ≤2 is an indication of a transplant |
| UNOS criteria [36] | One lesion ≥2 cm and ≤5 cm or maximum 3 lesions each ≥1 cm and ≤3; AFP ≤1000 ng/dl |
| Extended Toronto [37] | No tumour size and number limit; Biopsy needed beyond Milan to exclude poorly differentiated |
| Total tumor volume (TTV) [38] | TTV of less than 115 cm3 |
| Hangzhou criteria [39] | Total tumor diameter ≤8 cm or >8 cm with histopathologic grade 1 or 2 and a preoperative AFP value of ≤400 |
| TRAIN score [40] | mRECIST response; AFP slope; Neutrophil-to-lymphocyte ratio (NLR) and platelet-to-lymphocyte ratio (PLR); Waitlist time |
| Author | Year | Scope | Data | AI-Method | AUC |
|---|---|---|---|---|---|
| Chen et al. [76] | 2022 | MVI prediction | DCE-MRI (Gd-EOB-DTPA) + Clinical | Radiomics | 0.971 |
| Jiang et al. [77] | 2021 | MVI prediction | CECT | DL (CNN) | 0.906 |
| Sun et al. [78] | 2022 | MVI prediction | DCE-MRI (Gd-EOB-DTPA) + Clinical | DL (ResNet CNN) | 0.824 |
| Zhou et al. [79] | 2021 | MVI prediction | DCE-MRI (Gd- DTPA) | DL (CNN) | 0.926 |
| Author | Year | Scope | Data | AI-Method | AUC |
|---|---|---|---|---|---|
| Mao et al. [82] | 2020 | Grading prediction | CECT + Clinical | Radiomics | 0.801 |
| Wu et al. [83] | 2019 | Grading prediction | MRI + Clinical | Radiomics | 0.8 |
| Han et al. [84] | 2023 | Grading prediction | DCE MRI (Gd-EOB-DTPA) | Radiomics | 0.8 |
| Zhou et al. [85] | 2019 | Grading prediction | MRI | DL (CNN) | 0.83 |
| Zhou et al. [86] | 2019 | Grading prediction | DCE MRI (Gd- DTPA) | DL (DenseNet CNN) | 0.83 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pomohaci, M.D.; Grasu, M.C.; Dumitru, R.L.; Toma, M.; Lupescu, I.G. Liver Transplant in Patients with Hepatocarcinoma: Imaging Guidelines and Future Perspectives Using Artificial Intelligence. Diagnostics 2023, 13, 1663. https://doi.org/10.3390/diagnostics13091663
Pomohaci MD, Grasu MC, Dumitru RL, Toma M, Lupescu IG. Liver Transplant in Patients with Hepatocarcinoma: Imaging Guidelines and Future Perspectives Using Artificial Intelligence. Diagnostics. 2023; 13(9):1663. https://doi.org/10.3390/diagnostics13091663
Chicago/Turabian StylePomohaci, Mihai Dan, Mugur Cristian Grasu, Radu Lucian Dumitru, Mihai Toma, and Ioana Gabriela Lupescu. 2023. "Liver Transplant in Patients with Hepatocarcinoma: Imaging Guidelines and Future Perspectives Using Artificial Intelligence" Diagnostics 13, no. 9: 1663. https://doi.org/10.3390/diagnostics13091663
APA StylePomohaci, M. D., Grasu, M. C., Dumitru, R. L., Toma, M., & Lupescu, I. G. (2023). Liver Transplant in Patients with Hepatocarcinoma: Imaging Guidelines and Future Perspectives Using Artificial Intelligence. Diagnostics, 13(9), 1663. https://doi.org/10.3390/diagnostics13091663








