Optimal Skin Cancer Detection Model Using Transfer Learning and Dynamic-Opposite Hunger Games Search
Abstract
1. Introduction
- A pre-trained deep learning is used to learn and extract new representations for skin cancer images.
- A novel FS algorithm is proposed to reduce the dimensionality of extracted features and improve the overall performance by determining the relevant features.
- Two real-world datasets are used to validate and compare the proposed method to well-known methods.
- A more general framework is suggested to integrate the proposed method into the system.
2. Related Works
2.1. Deep Learning-Based Medical Images
2.2. Medical Images Classification Using FS Optimizers
3. Background
3.1. Efficient Neural Networks
3.2. Hunger Games Search
| Algorithm 1 Steps of HGS |
|
3.3. Particle Swarm Optimization
| Algorithm 2 Algorithm of PSO |
|
3.4. Dynamic-Opposite Learning
4. Proposed Model
4.1. Deep Learning for Feature Extraction
- (1)
- Replacing the two last output layers in MobileNetV3 with dense connected blocks including two convolutions for feature extraction and classification, respectively;
- (2)
- Fine-tuning the modified MobileNetV3 on the skin cancer dataset;
- (3)
- Extracting the corresponding feature vector of each image from the convolution layer added to the MobileNetV3 model; where the extracted features for each image are flattened into a vector of size 128.
- (4)
- Later, the extracted features for each image are fed to the feature selection part in our framework.
4.2. Steps of DOLHGS Feature Selection Algorithm
4.3. Framework of the Developed Skin Cancer Detection
5. Experiments and Results
5.1. Description of Datasets
5.2. Performance Measures
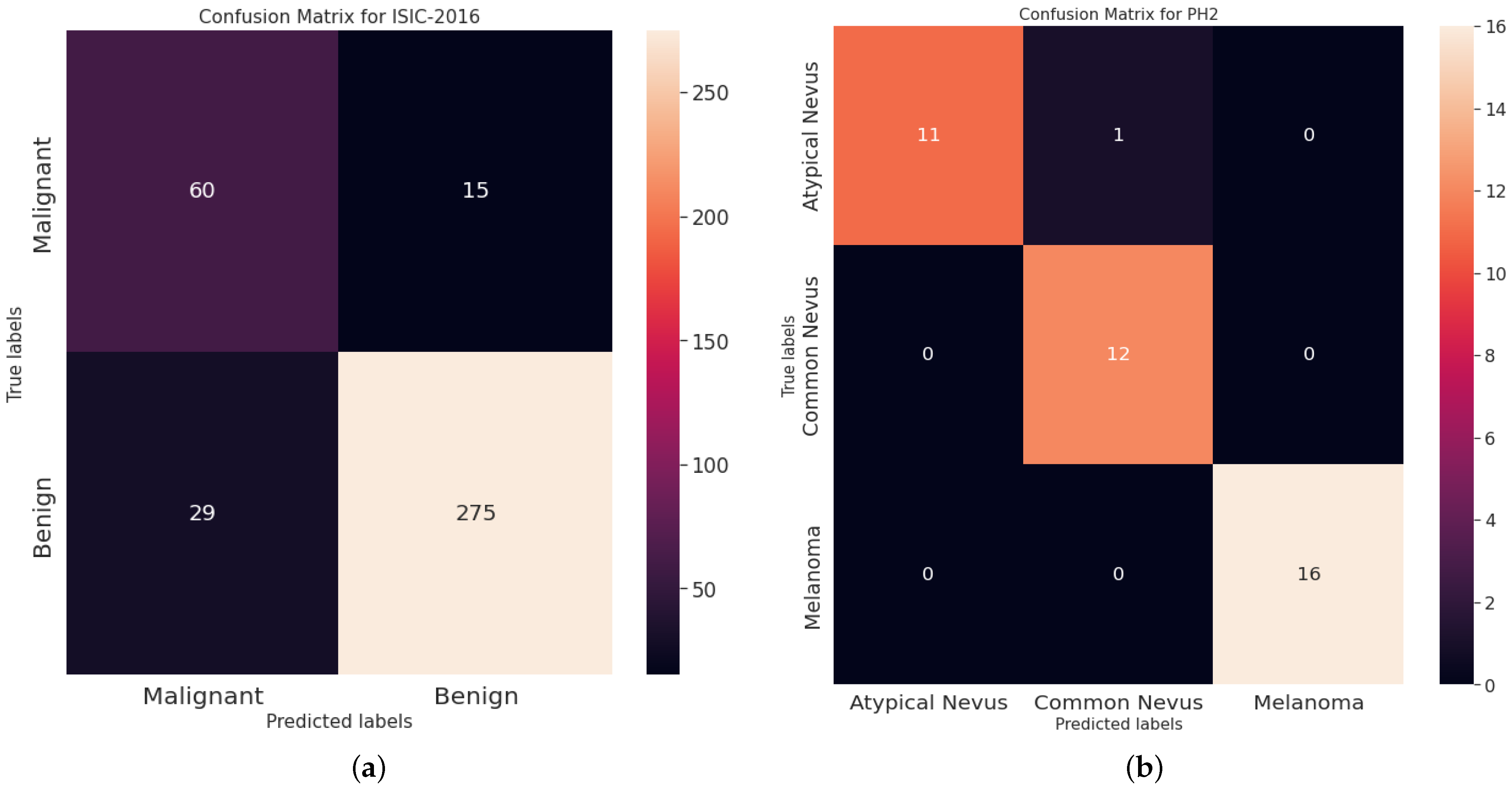
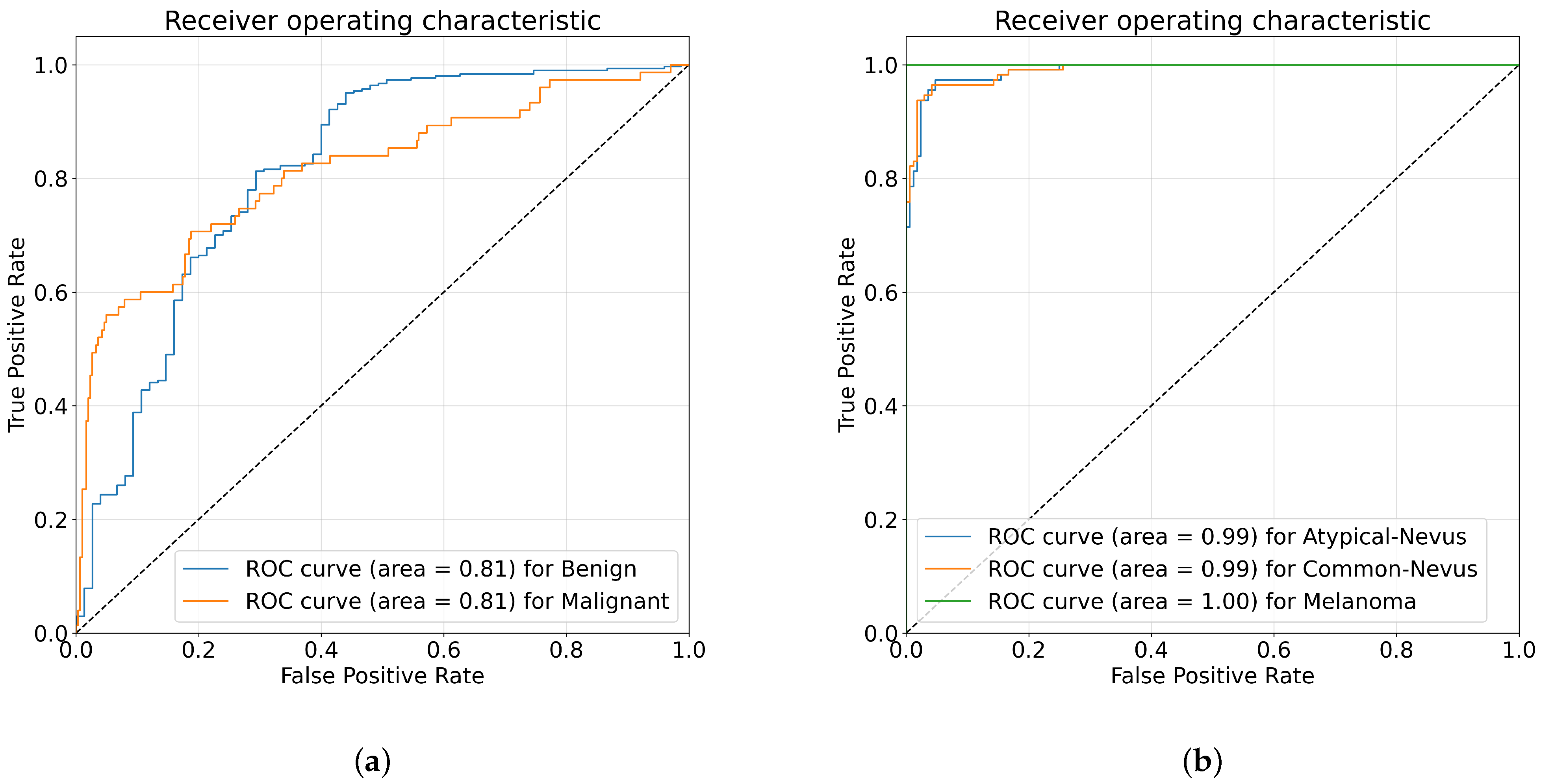
5.3. Results and Discussion
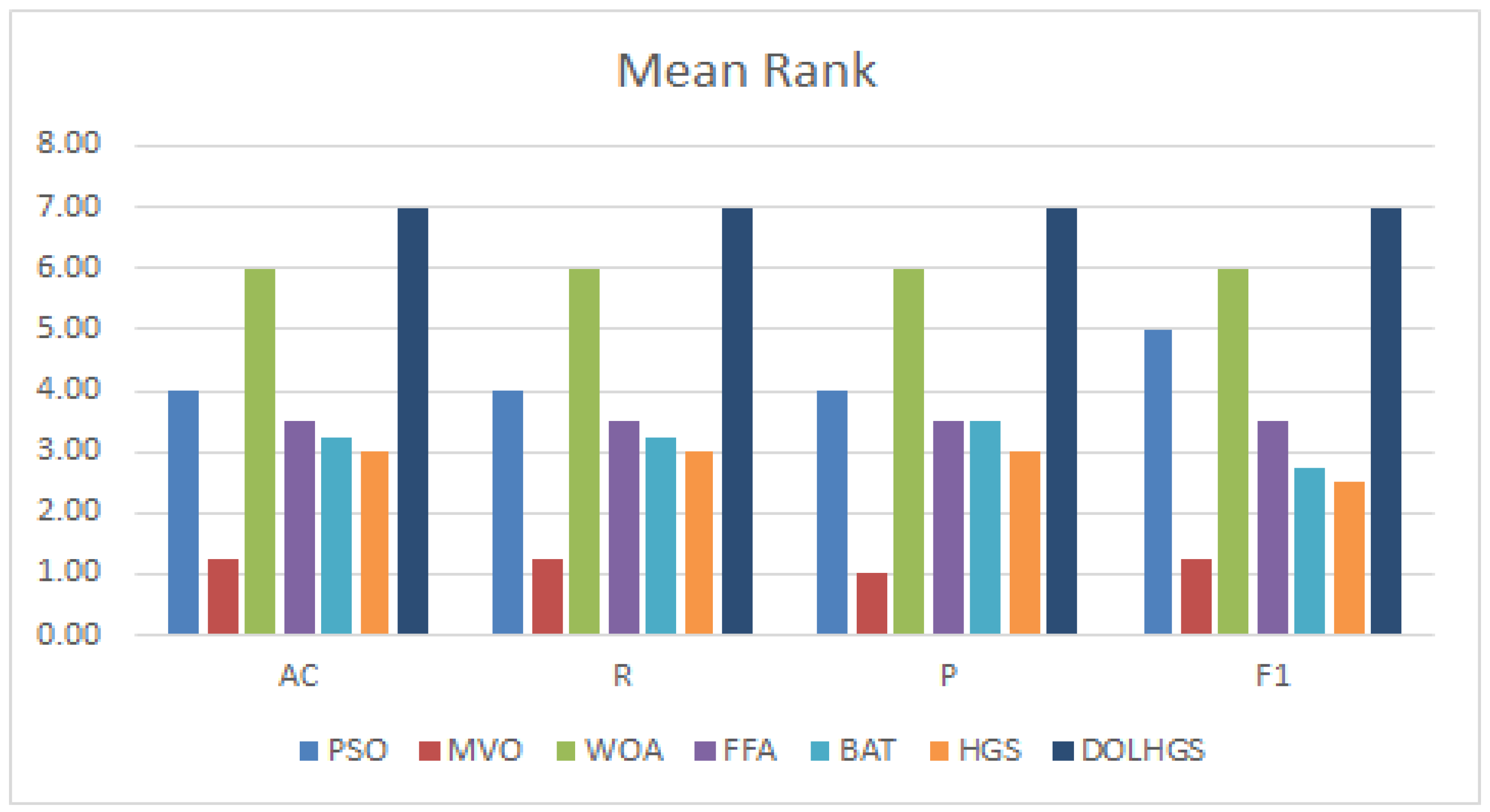
5.3.1. Comparison with FS Methods
5.3.2. Comparison with Previous Works
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Le, P.T.; Chang, C.C.; Li, Y.H.; Hsu, Y.C.; Wang, J.C. Antialiasing Attention Spatial Convolution Model for Skin Lesion Segmentation with Applications in the Medical IoT. Wirel. Commun. Mob. Comput. 2022, 2022, 1278515. [Google Scholar] [CrossRef]
- Thomas, S.M.; Lefevre, J.G.; Baxter, G.; Hamilton, N.A. Interpretable deep learning systems for multi-class segmentation and classification of non-melanoma skin cancer. Med. Image Anal. 2021, 68, 101915. [Google Scholar] [CrossRef] [PubMed]
- Kassani, S.H.; Kassani, P.H. A comparative study of deep learning architectures on melanoma detection. Tissue Cell 2019, 58, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Ding, K.; Hu, H. Automatic skin cancer detection in dermoscopy images based on ensemble lightweight deep learning network. IEEE Access 2020, 8, 99633–99647. [Google Scholar] [CrossRef]
- Hammad, M.; Iliyasu, A.M.; Subasi, A.; Ho, E.S.; Abd El-Latif, A.A. A multitier deep learning model for arrhythmia detection. IEEE Trans. Instrum. Meas. 2020, 70, 1–9. [Google Scholar] [CrossRef]
- Rodrigues, D.D.A.; Ivo, R.F.; Satapathy, S.C.; Wang, S.; Hemanth, J.; Reboucas Filho, P.P. A new approach for classification skin lesion based on transfer learning, deep learning, and IoT system. Pattern Recognit. Lett. 2020, 136, 8–15. [Google Scholar] [CrossRef]
- Abbas, F.; Yasmin, M.; Fayyaz, M.; Elaziz, M.A.; Lu, S.; El-Latif, A.A.A. Gender Classification Using Proposed CNN-Based Model and Ant Colony Optimization. Mathematics 2021, 9, 2499. [Google Scholar] [CrossRef]
- Jing, H.; He, X.; Han, Q.; Abd El-Latif, A.A.; Niu, X. Saliency detection based on integrated features. Neurocomputing 2014, 129, 114–121. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, X.; Zhou, F.; Qin, J.; Ni, D.; Chen, S.; Lei, B.; Wang, T. Melanoma recognition in dermoscopy images via aggregated deep convolutional features. IEEE Trans. Biomed. Eng. 2018, 66, 1006–1016. [Google Scholar] [CrossRef]
- Zhang, J.; Xie, Y.; Wu, Q.; Xia, Y. Medical image classification using synergic deep learning. Med. Image Anal. 2019, 54, 10–19. [Google Scholar] [CrossRef]
- Mabrouk, A.; Dahou, A.; Elaziz, M.A.; Díaz Redondo, R.P.; Kayed, M. Medical Image Classification Using Transfer Learning and Chaos Game Optimization on the Internet of Medical Things. Comput. Intell. Neurosci. 2022, 2022, 9112634. [Google Scholar] [CrossRef] [PubMed]
- Elaziz, M.A.; Dahou, A.; El-Sappagh, S.; Mabrouk, A.; Gaber, M.M. AHA-AO: Artificial Hummingbird Algorithm with Aquila Optimization for Efficient Feature Selection in Medical Image Classification. Appl. Sci. 2022, 12, 9710. [Google Scholar] [CrossRef]
- Adel, H.; Dahou, A.; Mabrouk, A.; Abd Elaziz, M.; Kayed, M.; El-Henawy, I.M.; Alshathri, S.; Amin Ali, A. Improving Crisis Events Detection Using DistilBERT with Hunger Games Search Algorithm. Mathematics 2022, 10, 447. [Google Scholar] [CrossRef]
- Mabrouk, A.; Redondo, R.P.D.; Kayed, M. Deep learning-based sentiment classification: A comparative survey. IEEE Access 2020, 8, 85616–85638. [Google Scholar] [CrossRef]
- Mabrouk, A.; Redondo, R.P.D.; Kayed, M. SEOpinion: Summarization and Exploration of Opinion from E-Commerce Websites. Sensors 2021, 21, 636. [Google Scholar] [CrossRef]
- Kadry, S.; Taniar, D.; Damaševičius, R.; Rajinikanth, V.; Lawal, I.A. Extraction of abnormal skin lesion from dermoscopy image using VGG-SegNet. In Proceedings of the 2021 Seventh International Conference on Bio Signals, Images, and Instrumentation (ICBSII), Chennai, India, 25–27 March 2021; pp. 1–5. [Google Scholar]
- Nawaz, M.; Nazir, T.; Masood, M.; Ali, F.; Khan, M.A.; Tariq, U.; Sahar, N.; Damaševičius, R. Melanoma segmentation: A framework of improved DenseNet77 and UNET convolutional neural network. Int. J. Imaging Syst. Technol. 2022, 32, 2137–2153. [Google Scholar] [CrossRef]
- Abd Elaziz, M.; Mabrouk, A.; Dahou, A.; Chelloug, S.A. Medical Image Classification Utilizing Ensemble Learning and Levy Flight-Based Honey Badger Algorithm on 6G-Enabled Internet of Things. Comput. Intell. Neurosci. 2022, 2022, 5830766. [Google Scholar] [CrossRef]
- Abayomi-Alli, O.O.; Damasevicius, R.; Misra, S.; Maskeliunas, R.; Abayomi-Alli, A. Malignant skin melanoma detection using image augmentation by oversamplingin nonlinear lower-dimensional embedding manifold. Turk. J. Electr. Eng. Comput. Sci. 2021, 29, 2600–2614. [Google Scholar] [CrossRef]
- Mabrouk, A.; Díaz Redondo, R.P.; Dahou, A.; Abd Elaziz, M.; Kayed, M. Pneumonia Detection on Chest X-ray Images Using Ensemble of Deep Convolutional Neural Networks. Appl. Sci. 2022, 12, 6448. [Google Scholar] [CrossRef]
- Niu, S.; Liu, Y.; Wang, J.; Song, H. A decade survey of transfer learning (2010–2020). IEEE Trans. Artif. Intell. 2020, 1, 151–166. [Google Scholar] [CrossRef]
- Niu, S.; Wang, J.; Liu, Y.; Song, H. Transfer learning based data-efficient machine learning enabled classification. In Proceedings of the 2020 IEEE International Conference on Dependable, Autonomic and Secure Computing, International Conference on Pervasive Intelligence and Computing, International Conference on Cloud and Big Data Computing, International Conference on Cyber Science and Technology Congress (DASC/PiCom/CBDCom/CyberSciTech), Calgary, AB, Canada, 17–22 August 2020; pp. 620–626. [Google Scholar]
- Niu, S.; Liu, M.; Liu, Y.; Wang, J.; Song, H. Distant domain transfer learning for medical imaging. IEEE J. Biomed. Health Inform. 2021, 25, 3784–3793. [Google Scholar] [CrossRef]
- Niu, S.; Hu, Y.; Wang, J.; Liu, Y.; Song, H. Feature-based distant domain transfer learning. In Proceedings of the 2020 IEEE International Conference on Big Data (Big Data), Atlanta, GA, USA, 10–13 December 2020; pp. 5164–5171. [Google Scholar]
- Rashid, J.; Ishfaq, M.; Ali, G.; Saeed, M.R.; Hussain, M.; Alkhalifah, T.; Alturise, F.; Samand, N. Skin Cancer Disease Detection using Transfer Learning Technique. Appl. Sci. 2022, 12, 5714. [Google Scholar] [CrossRef]
- Lopez, A.R.; Giro-i Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017; pp. 49–54. [Google Scholar]
- Ayan, E.; Ünver, H.M. Data augmentation importance for classification of skin lesions via deep learning. In Proceedings of the 2018 Electric Electronics, Computer Science, Biomedical Engineerings’ Meeting (EBBT), Istanbul, Turkey, 18–19 April 2018; pp. 1–4. [Google Scholar]
- Tang, P.; Yan, X.; Nan, Y.; Xiang, S.; Krammer, S.; Lasser, T. FusionM4Net: A multi-stage multi-modal learning algorithm for multi-label skin lesion classification. Med. Image Anal. 2022, 76, 102307. [Google Scholar] [CrossRef] [PubMed]
- Manickam, P.; Mariappan, S.A.; Murugesan, S.M.; Hansda, S.; Kaushik, A.; Shinde, R.; Thipperudraswamy, S. Artificial Intelligence (AI) and Internet of Medical Things (IoMT) Assisted Biomedical Systems for Intelligent Healthcare. Biosensors 2022, 12, 562. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.W.; Chiang, M.C.; Ksentini, A.; Chen, M. Metaheuristic algorithms for healthcare: Open issues and challenges. Comput. Electr. Eng. 2016, 53, 421–434. [Google Scholar] [CrossRef]
- Kang, L.; Chen, R.S.; Cao, W.; Chen, Y.C.; Hu, Y.X. Mechanism analysis of non-inertial particle swarm optimization for Internet of Things in edge computing. Eng. Appl. Artif. Intell. 2020, 94, 103803. [Google Scholar] [CrossRef]
- Stephen, V.K.; Sharma, S.; Manalang, A.R.; Al-Harthy, F.R.A. A Multi-hop Energy-Efficient Cluster-Based Routing Using Multi-verse Optimizer in IoT. In Computer Networks and Inventive Communication Technologies; Springer: Berlin/Heidelberg, Germany, 2021; pp. 1–14. [Google Scholar]
- Alharbi, A.; Alosaimi, W.; Alyami, H.; Rauf, H.T.; Damaševičius, R. Botnet Attack Detection Using Local Global Best Bat Algorithm for Industrial Internet of Things. Electronics 2021, 10, 1341. [Google Scholar] [CrossRef]
- El-Shafeiy, E.; Sallam, K.M.; Chakrabortty, R.K.; Abohany, A.A. A clustering based Swarm Intelligence optimization technique for the Internet of Medical Things. Expert Syst. Appl. 2021, 173, 114648. [Google Scholar] [CrossRef]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning transferable architectures for scalable image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.C.; Chen, B.; Tan, M.; Wang, W.; Zhu, Y.; Pang, R.; Vasudevan, V.; et al. Searching for mobilenetv3. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; pp. 1314–1324. [Google Scholar]
- Tan, M.; Chen, B.; Pang, R.; Vasudevan, V.; Sandler, M.; Howard, A.; Le, Q.V. Mnasnet: Platform-aware neural architecture search for mobile. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 2820–2828. [Google Scholar]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. Shufflenet: An extremely efficient convolutional neural network for mobile devices. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning. PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 6105–6114. [Google Scholar]
- Ji, J.; Krishna, R.; Fei-Fei, L.; Niebles, J.C. Action genome: Actions as compositions of spatio-temporal scene graphs. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10236–10247. [Google Scholar]
- Liu, J.; Inkawhich, N.; Nina, O.; Timofte, R. NTIRE 2021 multi-modal aerial view object classification challenge. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 588–595. [Google Scholar]
- Ignatov, A.; Romero, A.; Kim, H.; Timofte, R. Real-time video super-resolution on smartphones with deep learning, mobile AI 2021 challenge: Report. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 2535–2544. [Google Scholar]
- Ramachandran, P.; Zoph, B.; Le, Q.V. Searching for activation functions. arXiv 2017, arXiv:1710.05941. [Google Scholar]
- Elfwing, S.; Uchibe, E.; Doya, K. Sigmoid-weighted linear units for neural network function approximation in reinforcement learning. Neural Netw. 2018, 107, 3–11. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Yang, Y.; Chen, H.; Heidari, A.A.; Gandomi, A.H. Hunger games search: Visions, conception, implementation, deep analysis, perspectives, and towards performance shifts. Expert Syst. Appl. 2021, 177, 114864. [Google Scholar] [CrossRef]
- Eberhart, R.; Kennedy, J. A new optimizer using particle swarm theory. In Proceedings of the Sixth International Symposium on Micro Machine and Human Science, Nagoya, Japan, 4–6 October 1995; pp. 39–43. [Google Scholar]
- Noman, S.; Shamsuddin, S.M.; Hassanien, A.E. Hybrid learning enhancement of RBF network with particle swarm optimization. In Foundations of Computational, Intelligence; Springer: Berlin/Heidelberg, Germany, 2009; Volume 1, pp. 381–397. [Google Scholar]
- Niknam, T.; Amiri, B. An efficient hybrid approach based on PSO, ACO and k-means for cluster analysis. Appl. Soft Comput. 2010, 10, 183–197. [Google Scholar] [CrossRef]
- Tizhoosh, H.R. Opposition-based learning: A new scheme for machine intelligence. In Proceedings of the International Conference on Computational Intelligence for Modelling, Control and Automation and International Conference on Intelligent Agents, Web Technologies and Internet Commerce (CIMCA-IAWTIC’06), Vienna, Austria, 28–30 November 2005; Volume 1, pp. 695–701. [Google Scholar]
- Ewees, A.A.; Abd Elaziz, M.; Houssein, E.H. Improved grasshopper optimization algorithm using opposition-based learning. Expert Syst. Appl. 2018, 112, 156–172. [Google Scholar] [CrossRef]
- Ibrahim, R.A.; Ewees, A.A.; Oliva, D.; Abd Elaziz, M.; Lu, S. Improved salp swarm algorithm based on particle swarm optimization for feature selection. J. Ambient. Intell. Humaniz. Comput. 2019, 10, 3155–3169. [Google Scholar] [CrossRef]
- Yu, Z.; Jiang, F.; Zhou, F.; He, X.; Ni, D.; Chen, S.; Wang, T.; Lei, B. Convolutional descriptors aggregation via cross-net for skin lesion recognition. Appl. Soft Comput. 2020, 92, 106281. [Google Scholar] [CrossRef]
- Gutman, D.; Codella, N.C.; Celebi, E.; Helba, B.; Marchetti, M.; Mishra, N.; Halpern, A. Skin lesion analysis toward melanoma detection: A challenge at the international symposium on biomedical imaging (ISBI) 2016, hosted by the international skin imaging collaboration (ISIC). arXiv 2016, arXiv:1605.01397. [Google Scholar]
- Mendonça, T.; Ferreira, P.M.; Marques, J.S.; Marcal, A.R.; Rozeira, J. PH 2-A dermoscopic image database for research and benchmarking. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5437–5440. [Google Scholar]
- Mirjalili, S.; Mirjalili, S.M.; Hatamlou, A. Multi-verse optimizer: A nature-inspired algorithm for global optimization. Neural Comput. Appl. 2016, 27, 495–513. [Google Scholar] [CrossRef]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Yang, X.S. A new metaheuristic bat-inspired algorithm. In Nature Inspired Cooperative Strategies for Optimization (NICSO 2010); Springer: Berlin/Heidelberg, Germany, 2010; pp. 65–74. [Google Scholar]
- Yang, X.S. Firefly algorithm, Levy flights and global optimization. In Research and Development in Intelligent Systems XXVI; Springer: Berlin/Heidelberg, Germany, 2010; pp. 209–218. [Google Scholar]
- Derrac, J.; García, S.; Molina, D.; Herrera, F. A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm Evol. Comput. 2011, 1, 3–18. [Google Scholar] [CrossRef]
- Yu, L.; Chen, H.; Dou, Q.; Qin, J.; Heng, P.A. Automated melanoma recognition in dermoscopy images via very deep residual networks. IEEE Trans. Med. Imaging 2016, 36, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Demyanov, S.; Bozorgtabar, B.; Abedini, M.; Chakravorty, R.; Bowling, A.; Garnavi, R. Exploiting local and generic features for accurate skin lesions classification using clinical and dermoscopy imaging. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, Australia, 18–21 April 2017; pp. 986–990. [Google Scholar]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P. Automated detection of melanocytes related pigmented skin lesions: A clinical framework. Biomed. Signal Process. Control. 2019, 51, 59–72. [Google Scholar] [CrossRef]
- Ozkan, I.A.; Koklu, M. Skin lesion classification using machine learning algorithms. Int. J. Intell. Syst. Appl. Eng. 2017, 5, 285–289. [Google Scholar] [CrossRef]
- Moradi, N.; Mahdavi-Amiri, N. Kernel sparse representation based model for skin lesions segmentation and classification. Comput. Methods Programs Biomed. 2019, 182, 105038. [Google Scholar] [CrossRef]
- Al Nazi, Z.; Abir, T.A. Automatic skin lesion segmentation and melanoma detection: Transfer learning approach with u-net and dcnn-svm. In Proceedings of the International Joint Conference on Computational Intelligence, Dhaka, Bangladesh, 20–21 November 2020; pp. 371–381. [Google Scholar]
- Afza, F.; Sharif, M.; Mittal, M.; Khan, M.A.; Hemanth, D.J. A hierarchical three-step superpixels and deep learning framework for skin lesion classification. Methods 2021, 202, 88–102. [Google Scholar] [CrossRef]



| Input | Operator | Output | SE | NL | Stride |
|---|---|---|---|---|---|
| 2d-Conv | 16 | FALSE | HS | 2 | |
| 16 | FALSE | RE | 1 | ||
| 24 | FALSE | RE | 2 | ||
| 24 | FALSE | RE | 1 | ||
| 40 | TRUE | RE | 2 | ||
| 40 | TRUE | RE | 1 | ||
| 40 | TRUE | RE | 1 | ||
| 80 | FALSE | HS | 2 | ||
| 80 | FALSE | HS | 1 | ||
| 80 | FALSE | HS | 1 | ||
| 80 | FALSE | HS | 1 | ||
| 112 | TRUE | HS | 1 | ||
| 112 | TRUE | HS | 1 | ||
| 160 | TRUE | HS | 2 | ||
| 160 | TRUE | HS | 1 | ||
| 160 | TRUE | HS | 1 | ||
| 2d-Conv | 960 | FALSE | HS | 1 | |
| Adaptive average pooling | 960 | FALSE | - | 1 | |
| Image embedding | 128 | FALSE | HS | 1 |
| Dataset | Skin Disease | # Training Images | # Testing Images | Total Images per Category |
|---|---|---|---|---|
| ISIC-2016 | Malignant | 173 | 75 | 248 |
| Benign | 727 | 304 | 1031 | |
| Total images | 900 | 379 | 1279 | |
| Common Nevus | 68 | 12 | 80 | |
| Atypical Nevus | 68 | 12 | 80 | |
| Melanoma | 34 | 6 | 40 | |
| Total images | 170 | 30 | 200 |
| Algorithm | Value of the Parameters |
|---|---|
| DOLHGS | EPSILON = 10 × 10, MIN-PROB = 0, MAX-PROB = −1 |
| WOA | a = 2 to 0, a2 = −1 to −2 |
| BAT | QMin = 0, QMax = 2 |
| MVO | WEPMax = 1, WEPMin = 0.2 |
| PSO | VMax = 6, WMax = 0.9, WMin = 0.2 |
| FFA | Alpha = 0.5, BetaMin = 0.2, Gamma = 1 |
| HGS | EPSILON = 10 × 10, POS = 0, F IT = 1 |
| ISIC | PH2 | |||||||
|---|---|---|---|---|---|---|---|---|
| AC | R | P | F1 | AC | R | P | F1 | |
| PSO | 0.865699 | 0.865699 | 0.856919 | 0.852251 | 0.956429 | 0.956429 | 0.956949 | 0.956522 |
| MVO | 0.863325 | 0.863325 | 0.853915 | 0.849824 | 0.956071 | 0.956071 | 0.956575 | 0.956165 |
| WOA | 0.86781 | 0.86781 | 0.860512 | 0.853141 | 0.957143 | 0.957143 | 0.957592 | 0.957233 |
| FFA | 0.865435 | 0.865435 | 0.857003 | 0.85143 | 0.956429 | 0.956429 | 0.956918 | 0.956521 |
| BAT | 0.867018 | 0.867018 | 0.860102 | 0.851955 | 0.956071 | 0.956071 | 0.956581 | 0.956165 |
| HGS | 0.864908 | 0.864908 | 0.85652 | 0.850973 | 0.956429 | 0.956429 | 0.95694 | 0.956513 |
| DOLHGS | 0.88185 | 0.87517 | 0.87633 | 0.87575 | 0.96429 | 0.97429 | 0.97699 | 0.97563 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dahou, A.; Aseeri, A.O.; Mabrouk, A.; Ibrahim, R.A.; Al-Betar, M.A.; Elaziz, M.A. Optimal Skin Cancer Detection Model Using Transfer Learning and Dynamic-Opposite Hunger Games Search. Diagnostics 2023, 13, 1579. https://doi.org/10.3390/diagnostics13091579
Dahou A, Aseeri AO, Mabrouk A, Ibrahim RA, Al-Betar MA, Elaziz MA. Optimal Skin Cancer Detection Model Using Transfer Learning and Dynamic-Opposite Hunger Games Search. Diagnostics. 2023; 13(9):1579. https://doi.org/10.3390/diagnostics13091579
Chicago/Turabian StyleDahou, Abdelghani, Ahmad O. Aseeri, Alhassan Mabrouk, Rehab Ali Ibrahim, Mohammed Azmi Al-Betar, and Mohamed Abd Elaziz. 2023. "Optimal Skin Cancer Detection Model Using Transfer Learning and Dynamic-Opposite Hunger Games Search" Diagnostics 13, no. 9: 1579. https://doi.org/10.3390/diagnostics13091579
APA StyleDahou, A., Aseeri, A. O., Mabrouk, A., Ibrahim, R. A., Al-Betar, M. A., & Elaziz, M. A. (2023). Optimal Skin Cancer Detection Model Using Transfer Learning and Dynamic-Opposite Hunger Games Search. Diagnostics, 13(9), 1579. https://doi.org/10.3390/diagnostics13091579








