Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA and RNA Samples
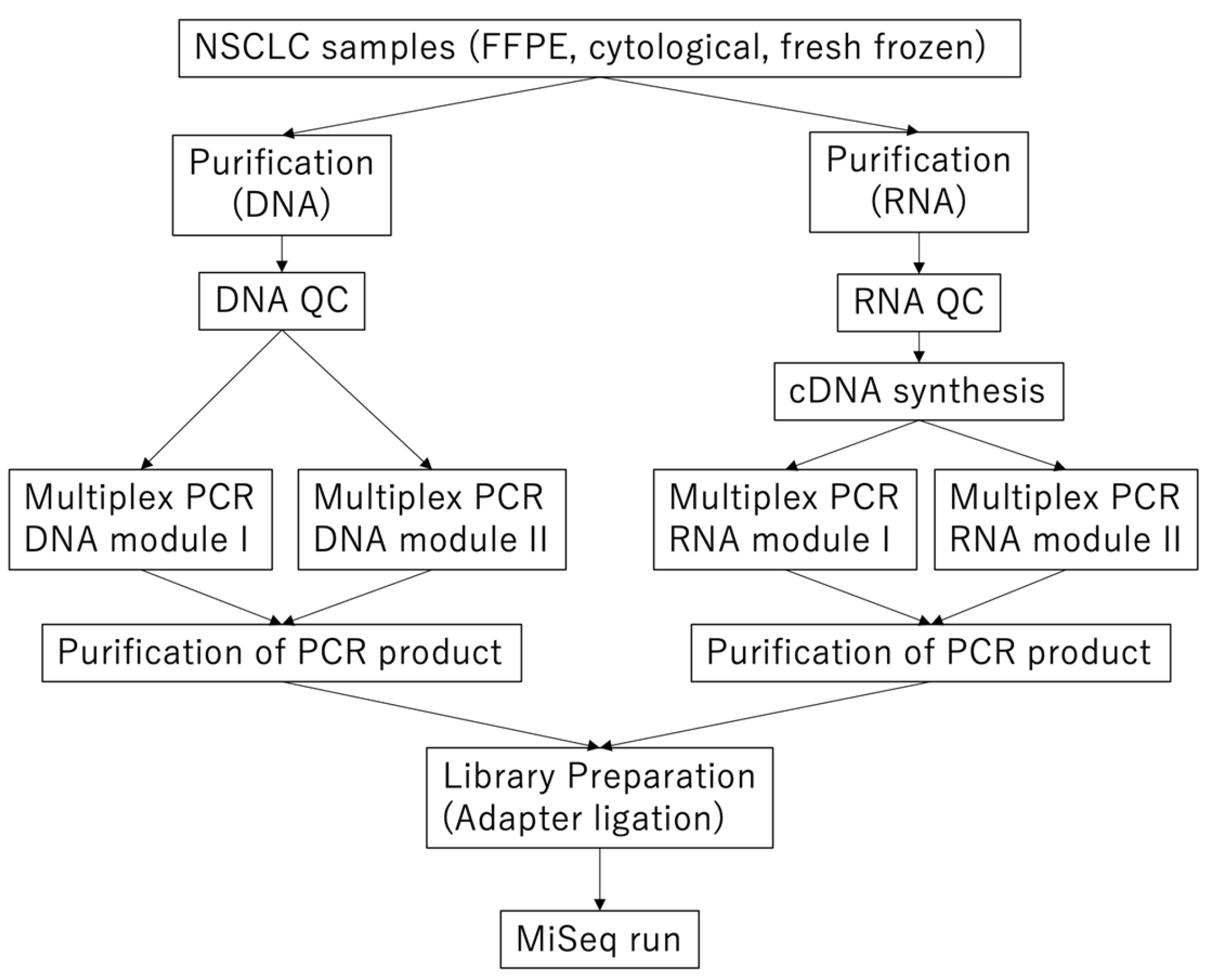
2.2. Library Preparation and Sequencing of DNA Modules
2.3. Library Preparation and Sequencing of RNA Modules
2.4. Sequence Data Analysis
2.5. Analysis of Discordant Samples
2.6. Preparation of Artificial Samples
3. Results
3.1. Design of the Compact Panel
3.2. Sensitivity of Mutation Detection Using DNA as a Template
3.3. Sensitivity of Fusion Detection Using RNA as a Template
3.4. Quantification of Mutation Detection
3.5. Concordance with Conventional Diagnostic Tests
3.6. Incidence of Mutations and Fusions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.; Camidge, D.R.; Shaw, A.T.; Gadgeel, S.; Ahn, J.S.; Kim, D.W.; Ou, S.I.; Pérol, M.; Dziadziuszko, R.; Rosell, R.; et al. Alectinib versus Crizotinib in Untreated ALK-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Riely, G.J.; Bang, Y.J.; Kim, D.W.; Camidge, D.R.; Solomon, B.J.; Varella-Garcia, M.; Iafrate, A.J.; Shapiro, G.I.; Usari, T.; et al. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): Updated results, including overall survival, from PROFILE 1001. Ann. Oncol. 2019, 30, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Smit, E.F.; Groen, H.J.M.; Mazieres, J.; Besse, B.; Helland, Å.; Giannone, V.; D’Amelio, A.M., Jr.; Zhang, P.; Mookerjee, B.; et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: An open-label, phase 2 trial. Lancet Oncol. 2017, 18, 1307–1316. [Google Scholar] [CrossRef]
- Paik, P.K.; Felip, E.; Veillon, R.; Sakai, H.; Cortot, A.B.; Garassino, M.C.; Mazieres, J.; Viteri, S.; Senellart, H.; Van Meerbeeck, J.; et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020, 383, 931–943. [Google Scholar] [CrossRef]
- Drilon, A.; Oxnard, G.R.; Tan, D.S.W.; Loong, H.H.F.; Johnson, M.; Gainor, J.; McCoach, C.E.; Gautschi, O.; Besse, B.; Cho, B.C.; et al. Efficacy of Selpercatinib in RET Fusion-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 813–824. [Google Scholar] [CrossRef]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Singh, N.; Temin, S.; Baker, S., Jr.; Blanchard, E.; Brahmer, J.R.; Celano, P.; Duma, N.; Ellis, P.M.; Elkins, I.B.; Haddad, R.Y.; et al. Therapy for Stage IV Non-Small-Cell Lung Cancer with Driver Alterations: ASCO Living Guideline. J. Clin. Oncol. 2022, 40, 3310–3322. [Google Scholar] [CrossRef]
- Zhang, S.S.; Zhu, V.W. Spotlight on Mobocertinib (TAK-788) in NSCLC with EGFR Exon 20 Insertion Mutations. Lung Cancer 2021, 12, 61–65. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Kukita, Y.; Matoba, R.; Uchida, J.; Hamakawa, T.; Doki, Y.; Imamura, F.; Kato, K. High-fidelity target sequencing of individual molecules identified using barcode sequences: De novo detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. DNA Res. 2015, 22, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Attili, I.; Passaro, A.; Pisapia, P.; Malapelle, U.; de Marinis, F. Uncommon EGFR Compound Mutations in Non-Small Cell Lung Cancer (NSCLC): A Systematic Review of Available Evidence. Curr. Oncol. 2022, 29, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Kukita, Y.; Uchida, J.; Oba, S.; Nishino, K.; Kumagai, T.; Taniguchi, K.; Okuyama, T.; Imamura, F.; Kato, K. Quantitative identification of mutant alleles derived from lung cancer in plasma cell-free DNA via anomaly detection using deep sequencing data. PLoS ONE 2013, 8, e81468. [Google Scholar] [CrossRef]
- Russell, H.J.; Richardson, T.T.; Emptage, K.; Connolly, B.A. The 3′-5′ proofreading exonuclease of archaeal family-B DNA polymerase hinders the copying of template strand deaminated bases. Nucleic Acids Res. 2009, 37, 7603–7611. [Google Scholar] [CrossRef]
- Seto, T.; Matsumoto, S.; Yoh, K.; Fujiwara, Y.; Yokoyama, T.; Nishino, K.; Kato, T.; Sugawara, S.; Shingoji, M.; Kodani, M.; et al. Contribution of nationwide genome screening in Japan (LC-SCRUM-Japan) to the development of precision medicine for non-small cell lung cancer. J. Clin. Oncol. 2018, 36, 9085. [Google Scholar] [CrossRef]
- Singh, N.; Temin, S.; Baker, S., Jr.; Blanchard, E.; Brahmer, J.R.; Celano, P.; Duma, N.; Ellis, P.M.; Elkins, I.B.; Haddad, R.Y.; et al. Therapy for Stage IV Non-Small-Cell Lung Cancer without Driver Alterations: ASCO Living Guideline. J. Clin. Oncol. 2022, 40, 3323–3343. [Google Scholar] [CrossRef]
- Robert, N.J.; Nwokeji, E.D.; Espirito, J.L.; Chen, L.; Karhade, M.; Evangelist, M.C.; Spira, A.I.; Neubauer, M.A.; Bullock, S.A.; Coleman, R.L. Biomarker tissue journey among patients (pts) with untreated metastatic non-small cell lung cancer (mNSCLC) in the U.S. Oncology Network community practices. J. Clin. Oncol. 2021, 39, 9004. [Google Scholar] [CrossRef]
- Melosky, B.; Kambartel, K.; Hantschel, M.; Bennetts, M.; Nickens, D.J.; Brinkmann, J.; Kayser, A.; Moran, M.; Cappuzzo, F. Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis. Mol. Diagn. Ther. 2022, 26, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.M.; Morrison, C.; Gold, E.J.; Tradonsky, A.; Layton, A.J. Multiple Biomarker Testing Tissue Consumption and Completion Rates with Single-gene Tests and Investigational Use of Oncomine Dx Target Test for Advanced Non-Small-cell Lung Cancer: A Single-center Analysis. Clin. Lung Cancer 2019, 20, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Ariyasu, R.; Uchibori, K.; Ninomiya, H.; Ogusu, S.; Tsugitomi, R.; Manabe, R.; Sakamaoto, H.; Tozuka, T.; Yoshida, H.; Amino, Y.; et al. Feasibility of next-generation sequencing test for patients with advanced NSCLC in clinical practice. Thorac. Cancer 2021, 12, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, D.; Yokose, T.; Katayama, K.; Murakami, S.; Kato, T.; Saito, H.; Suzuki, M.; Eriguchi, D.; Samejima, J.; Nagashima, T.; et al. Tissue surface area and tumor cell count affect the success rate of the Oncomine Dx Target Test in the analysis of biopsy tissue samples. Thorac. Cancer 2021, 12, 194–200. [Google Scholar] [CrossRef]
- Takeyasu, Y.; Yoshida, T.; Motoi, N.; Teishikata, T.; Tanaka, M.; Matsumoto, Y.; Shinno, Y.; Okuma, Y.; Goto, Y.; Horinouchi, H.; et al. Feasibility of next-generation sequencing (Oncomine™ DX Target Test) for the screening of oncogenic mutations in advanced non-small-cell lung cancer patients. Jpn. J. Clin. Oncol. 2021, 51, 1114–1122. [Google Scholar] [CrossRef]
- Minami, D.; Takigawa, N.; Nakajima, Y.; Miyahara, N.; Mizumori, Y.; Ueda, M.; Nakamura, S.; Suzuki, F.; Sato, Y.; Morikawa, K.; et al. Use of a highly sensitive lung cancer compact panel to detect KRAS G12D in the wash fluid from a lung tumor: A case report. Thorac. Cancer 2022, 13, 1735–1738. [Google Scholar] [CrossRef]
- Morikawa, K.; Kida, H.; Handa, H.; Inoue, T.; Saji, H.; Koike, J.; Nakamura, S.; Sato, Y.; Ueda, Y.; Suzuki, F.; et al. A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens. Cancers 2022, 14, 3784. [Google Scholar] [CrossRef]
- Morikawa, K.; Kinoshita, K.; Kida, H.; Inoue, T.; Mineshita, M. Preliminary Results of NGS Gene Panel Test Using NSCLC Sputum Cytology and Therapeutic Effect Using Corresponding Molecular-Targeted Drugs. Genes 2022, 13, 812. [Google Scholar] [CrossRef]
- Uchida, J.; Kato, K.; Kukita, Y.; Kumagai, T.; Nishino, K.; Daga, H.; Nagatomo, I.; Inoue, T.; Kimura, M.; Oba, S.; et al. Diagnostic Accuracy of Noninvasive Genotyping of EGFR in Lung Cancer Patients by Deep Sequencing of Plasma Cell-Free DNA. Clin. Chem. 2015, 61, 1191–1196. [Google Scholar] [CrossRef]
- Li, B.T.; Smit, E.F.; Goto, Y.; Nakagawa, K.; Udagawa, H.; Mazieres, J.; Nagasaka, M.; Bazhenova, L.; Saltos, A.N.; Felip, E.; et al. Trastuzumab Deruxtecan in HER2-Mutant Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2022, 386, 241–251. [Google Scholar] [CrossRef]
- Grafstrom, R.C.; Fornace, A.J., Jr.; Autrup, H.; Lechner, J.F.; Harris, C.C. Formaldehyde damage to DNA and inhibition of DNA repair in human bronchial cells. Science 1983, 220, 216–218. [Google Scholar] [CrossRef] [PubMed]
- Florell, S.R.; Coffin, C.M.; Holden, J.A.; Zimmermann, J.W.; Gerwels, J.W.; Summers, B.K.; Jones, D.A.; Leachman, S.A. Preservation of RNA for functional genomic studies: A multidisciplinary tumor bank protocol. Mod. Pathol. 2001, 14, 116–128. [Google Scholar] [CrossRef] [PubMed]

| Gene | Target Mutations/Fusion Variants | |
|---|---|---|
| DNA module I | EGFR | Exon 19 deletion, L858R, T790M, L861Q, L861R |
| BRAF | V600E | |
| KRAS | G12C | |
| DNA module II | EGFR | G719X, E709X, S768I, exon 20 insertion |
| HER2 | Exon 20 mutations | |
| MET | Exon 14 skipping | |
| RNA module I | ALK | EML4, 22 variants; KIF5, 3; TFG, 1; HIP, 3; KLC1, 1 |
| MET | Exon 14 skipping | |
| RNA module II | ROS1 | CD74, 5 variants; SLC34A2, 7; EZR, 1; GOPC, 2; SDC4, 4; LRIG, 1; TPM3, 1; CCDC6, 1; KDELR2, 1 |
| RET | KIF5B, 7 variants; CCDC6, 1; NCOA4, 1 |
| Threshold of Detection DNA, % Allele Frequency; RNA, TM Score | Sensitivity Test | ||
|---|---|---|---|
| False Negative | False Positive | ||
| DNA module | |||
| EGFR exon 19 del | 0.14 | 0 | 0 |
| EGFR L858R | 0.2 | 0 | 0 |
| EGFR T790M | 0.48 | 0 | 0 |
| BRAF V600E | 0.24 | 0 | 0 |
| KRAS G12C | 0.2 | 0 | 0 |
| RNA module | |||
| ALK fusion | 188 | 0 | 1 |
| ROS1 fusion | 32 | 0 | 1 |
| RET fusion | 18 | 0 | 0 |
| MET exon 14 skipping | 28 | 0 | 0 |
| Compact Panel Positive | Compact Panel Negative | Positive Identity Rate | Negative Identity Rate | Reference Test | ||
|---|---|---|---|---|---|---|
| EGFR | Reference test positive | 73 | 0 | 100 (95.5–100) | - | Cobas EGFR Mutation Test v2 |
| Reference test negative | 7 | 70 | - | 90.9 (82.2–96.3) | ||
| BRAF V600E | Reference test positive | 7 | 0 | 100 (59.0–100) | - | Oncomine Dx Target Test |
| Reference test negative | 0 | 70 | - | 100 (94.9–100) | ||
| KRAS G12C | Reference test positive | 49 | 0 | 100 (92.7–100) | - | therascreen KRAS RGQ PCR Kit |
| Reference test negative | 0 | 51 | - | 100 (93.0–100) | ||
| ALK | Reference test positive | 29 | 1 | 96.7 (83.8–99.9) | - | Histofine ALK iAEP kit/Vysis ALK Break Apart FISH Probe Kit |
| Reference test negative | 11 | 692 | - | 98.4 (97.2–99.2) | ||
| ROS1 | Reference test positive | 9 | 0 | 100 (66.4–100) | - | OncoGuide AmoyDx ROS1 |
| Reference test negative | 1 | 99 | - | 99.0 (94.6–100) | ||
| MET | Reference test positive | 48 | 1 | 98.0 (89.0–99.9) | - | ArcherMET |
| Reference test negative | 0 | 50 | - | 100 (92.8–100) | ||
| RET | Reference test positive | 15 | 1 | 93.8 (69.8–100) | - | Oncomine Dx Target Test |
| Reference test negative | 0 | 70 | - | 100 (94.9–100) |
| Compact Panel Positive | Compact Panel Negative | Positive Identity Rate | Reference Test | ||
|---|---|---|---|---|---|
| BRAF V600E | Reference test positive | 77 | 0 | 100 (95.3–100) | Oncomine Dx Target Test |
| Reference test no call | 3 | 0 | - | ||
| ROS1 | Reference test positive | 99 | 1 | 99.0 (94.6–100) | OncoGuide AmoyDx ROS1 |
| RET | Reference test positive | 79 | 1 | 98.8 (93.2–100) | Oncomine Dx Target Test |
| Osaka International Cancer Institute | Scrum-Japan | |||
|---|---|---|---|---|
| Assay system | Compact panel | Oncomine comprehensive assay | ||
| Total number of samples | 827 | 3919 | ||
| Gene | Number of variant-positive samples | Percent frequency | Number of variant-positive samples | Percent frequency |
| KRAS | 202 | 24.4 (21.5–27.5) | 382 | 9.7 (8.8–10.7) |
| BRAF | 36 | 4.4 (3.1–6.0) | 97 | 2.5 (2.0–3.0) |
| ALK | 41 | 5 (3.6–6.7) | 97 | 2.5 (2.0–3.0) |
| ROS1 | 10 | 1.2 (0.6–2.2) | 142 | 3.6 (3.1–4.3) |
| MET | 56 | 6.8 (5.2–8.7) | 98 | 2.5 (2.0–3.0) |
| RET | 18 | 2.2 (1.3–3.4) | 100 | 2.6 (2.1–3.1) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, K.; Okami, J.; Nakamura, H.; Honma, K.; Sato, Y.; Nakamura, S.; Kukita, Y.; Nakatsuka, S.-i.; Higashiyama, M. Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics. Diagnostics 2023, 13, 1476. https://doi.org/10.3390/diagnostics13081476
Kato K, Okami J, Nakamura H, Honma K, Sato Y, Nakamura S, Kukita Y, Nakatsuka S-i, Higashiyama M. Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics. Diagnostics. 2023; 13(8):1476. https://doi.org/10.3390/diagnostics13081476
Chicago/Turabian StyleKato, Kikuya, Jiro Okami, Harumi Nakamura, Keiichiro Honma, Yoshiharu Sato, Seiji Nakamura, Yoji Kukita, Shin-ichi Nakatsuka, and Masahiko Higashiyama. 2023. "Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics" Diagnostics 13, no. 8: 1476. https://doi.org/10.3390/diagnostics13081476
APA StyleKato, K., Okami, J., Nakamura, H., Honma, K., Sato, Y., Nakamura, S., Kukita, Y., Nakatsuka, S.-i., & Higashiyama, M. (2023). Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics. Diagnostics, 13(8), 1476. https://doi.org/10.3390/diagnostics13081476







