Implementation of Exome Sequencing in Prenatal Diagnostics: Chances and Challenges
Abstract
1. Introduction
2. Methods
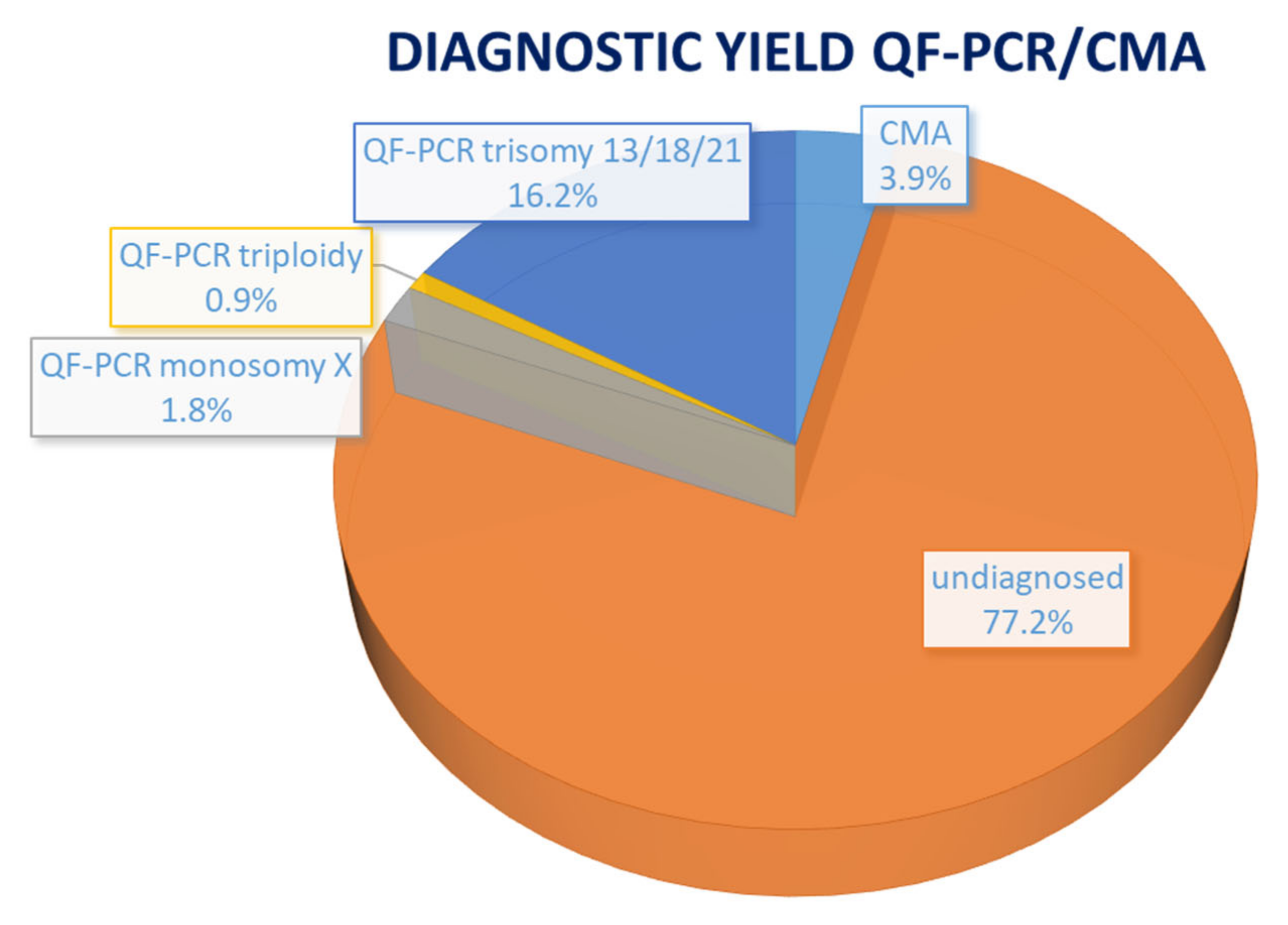
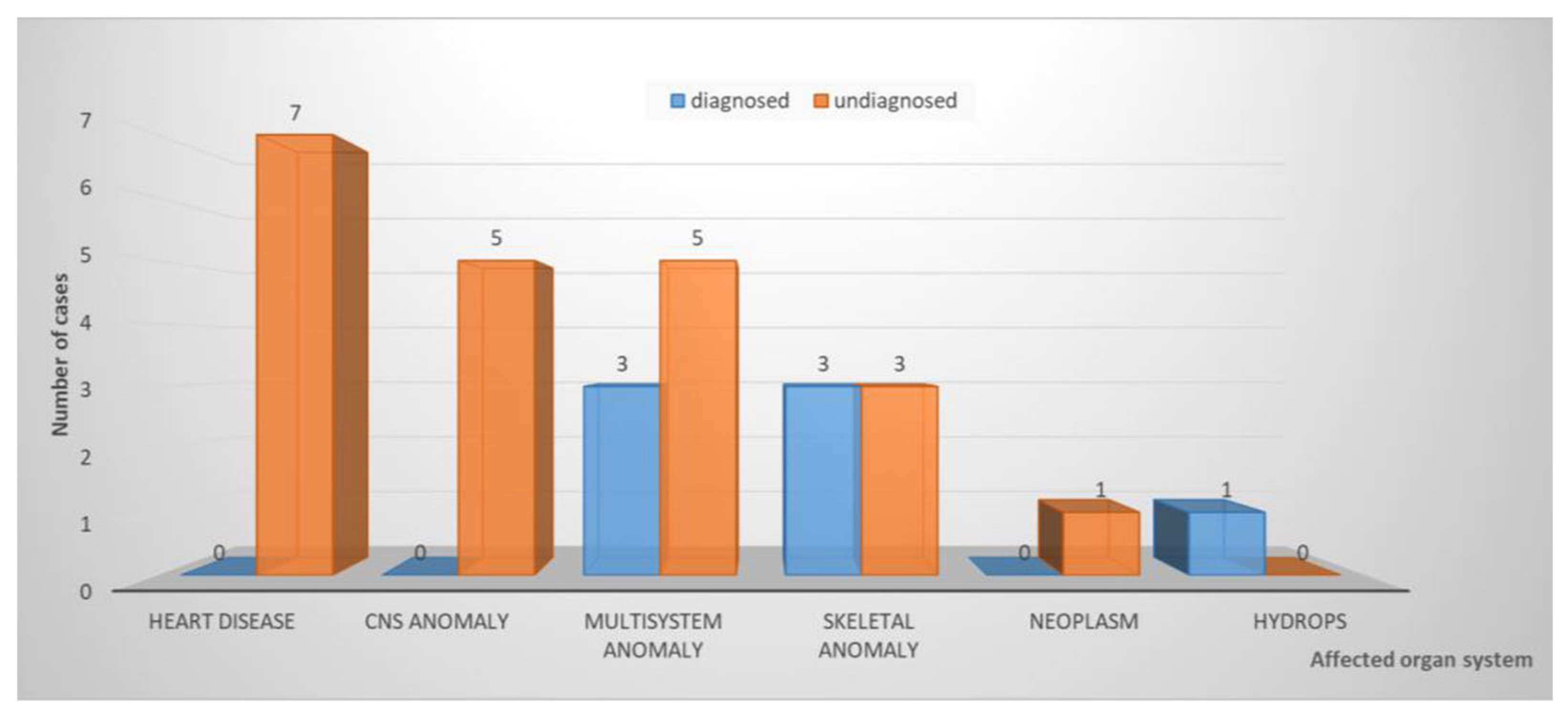
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyd, P.A.; Tonks, A.M.; Rankin, J.; Rounding, C.; Wellesley, D.; Draper, E.S. Monitoring the prenatal detection of structural fetal congenital anomalies in England and Wales: Register-based study. J. Med. Screen. 2011, 18, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, E.; Barisic, I.; Loane, M.; Morris, J.; Wellesley, D.; Dolk, H.; Addor, M.; Arriola, L.; Bianchi, F.; Neville, A.J.; et al. Epidemiology of multiple congenital anomalies in Europe: A EUROCAT population-based registry study. Birth Defects Res. Part A Clin. Mol. Teratol. 2014, 100, 270–276. [Google Scholar] [CrossRef] [PubMed]
- Wapner, R.J.; Martin, C.L.; Levy, B.; Ballif, B.C.; Eng, C.M.; Zachary, J.M.; Savage, M.; Platt, L.D.; Saltzman, D.; Grobman, W.A.; et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012, 367, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Mellis, R.; Oprych, K.; Scotchman, E.; Hill, M.; Chitty, L.S. Diagnostic yield of exome sequencing for prenatal diagnosis of fetal structural anomalies: A systematic review and meta-analysis. Prenat. Diagn. 2022, 42, 662–685. [Google Scholar] [CrossRef]
- Pauta, M.; Martinez-Portilla, R.J.; Borrell, A. Diagnostic yield of exome sequencing in fetuses with multisystem malformations: Systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2022, 59, 715–722. [Google Scholar] [CrossRef]
- Mone, F.; Mellis, R.; Gabriel, H.; Baptiste, C.; Giordano, J.; Wapner, R.; Chitty, L.S. Should we offer prenatal exome sequencing for intrauterine growth restriction or short long bones? A systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2022. online ahead of print. [Google Scholar] [CrossRef]
- Mellis, R.; Eberhardt, R.Y.; Hamilton, S.J.; PAGE Consortium; McMullan, D.J.; Kilby, M.D.; Maher, E.R.; Hurles, M.E.; Giordano, J.L.; Aggarwal, V.; et al. Fetal exome sequencing for isolated increased nuchal translucency: Should we be doing it? BJOG 2022, 129, 52–61. [Google Scholar] [CrossRef]
- Pauta, M.; Martinez-Portilla, E.J.; Borrell, A. Prenatal Exome Sequencing in Recurrent Fetal Structural Anomalies: Systematic Review and Meta-Analysis. J. Clin. Med. 2021, 10, 4739. [Google Scholar] [CrossRef]
- Van den Veyver, I.B.; Chandler, N.; Wilkins-Haug, L.E.; Wapner, R.J.; Chitty, L.S. International Society for Prenatal Diagnosis Updated Position Statement on the use of genome-wide sequencing for prenatal diagnosis. Prenat. Diagn. 2022, 42, 796–803. [Google Scholar] [CrossRef]
- Vanakker, O.; Vilain, C.; Janssens, K.; Van der Aa, N.; Smits, G.; Bandelier, C.; Blaumeiser, B.; Bulk, S.; Caberg, J.H.; De Leener, A.; et al. Implementation of genomic arrays in prenatal diagnosis: The Belgian approach to meet the challenges. Eur. J. Med. Genet. 2014, 57, 151–156. [Google Scholar] [CrossRef]
- Vandeweyer, G.; Van Laer, L.; Loeys, B.; Van den Bulcke, T.; Kooy, R.F. VariantDB: A flexible annotation and filtering portal for next generation sequencing data. Genome Med. 2014, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Vears, D.; Amor, D.J. A framework for reporting secondary and incidental findings in prenatal sequencing: When and for whom? Prenat. Diagn. 2022, 42, 697–704. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.T.; Lee, K.; Chung, W.K.; Gordon, A.S.; Herman, G.E.; Klein, T.E.; Stewart, D.R.; Amendola, L.M.; Adelman, K.; Bale, S.J.; et al. ACMG SF v3.0 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2021, 23, 1381–1390. [Google Scholar] [CrossRef]
- Winsor, E.J.; Silver, M.P.; Theve, R.; Wright, M.; Ward, B.E. Maternal cell contamination in uncultured amniotic fluid. Prenat. Diagn. 1996, 16, 49–54. [Google Scholar] [CrossRef]
- Gray, K.J.; Wilkins-Haug, L.E.; Herrig, N.J.; Vora, N.L. Fetal phenotypes emerge as genetic technologies become robust. Prenat. Diagn. 2019, 39, 811–817. [Google Scholar] [CrossRef]
- Giordano, J.L.; Wapner, R.J. The fetal sequencing consortium: The value of multidisciplinary dialog and collaboration. Prenat. Diagn. 2022, 42, 807–810. [Google Scholar] [CrossRef]
- Lamouroux, A.; Dauge, C.; Wells, C.; Mousty, E.; Pinson, L.; Cavé, H.; Capri, Y.; Faure, J.M.; Grosjean, F.; Sauvestre, F.; et al. Extending the prenatal Noonan’s phenotype by review of ultrasound and autopsy data. Prenat. Diagn. 2022, 42, 574–582. [Google Scholar] [CrossRef]
- Westenius, E.; Sahlin, E.; Conner, P.; Lindstrand, A.; Iwarsson, E. Diagnostic yield using whole-genome sequencing and in-silico panel of 281 genes associated with non-immune hydrops fetalis in clinical setting. Ultrasound Obstet. Gynecol. 2022, 60, 487–493. [Google Scholar] [CrossRef]
- Das, S.; Sharma, C.; Gothwal, M.; Tada, N. Whole exome sequencing, clinical exome or targeted gene panels: What to choose for suspected lethal skeletal dysplasia (short rib thoracic dysplasia type IV). BMJ Case Rep. 2022, 15, e251118. [Google Scholar] [CrossRef]
- Cao, J.; Chen, A.; Tian, L.; Yan, L.; Li, H.; Zhou, B. Application of whole exome sequencing in fetal cases with skeletal abnormalities. Heliyon 2022, 8, e09819. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, M.; Wang, H. Prenatal trio-based whole exome sequencing in fetuses with abnormalities of the skeletal system. Mol. Genet. Genom. 2022, 297, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- So, P.L.; Luk, H.M.; Cheung, K.W.; Hui, W.; Chung, M.Y.; Mak, A.S.; Lok, W.Y.; Yu, K.P.T.; Cheng, S.S.; Hau, E.W.; et al. Prenatal phenotype of Kabuki syndrome: A case series and literature review. Prenat. Diagn. 2021, 41, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, A.D. Fanconi anemia and its diagnosis. Mutat. Res. 2009, 668, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, M.; Rac, M.W.F.; McKinney, J. Radial Ray Malformation. Am. J. Obstet. Gynecol. 2019, 221, B16–B18. [Google Scholar] [CrossRef] [PubMed]
- Kennelly, M.M.; Moran, P. A clinical algorithm of prenatal diagnosis of Radial Ray Defects with two and three dimensional ultrasound. Prenat. Diagn. 2007, 27, 730–737. [Google Scholar] [CrossRef] [PubMed]
- Ruaud, L.; Roux, N.; Boutaud, L.; Bessières, B.; Ageorges, F.; Achaiaa, A.; Bole, C.; Nitschke, P.; Masson, C.; Vekemans, M.; et al. Biallelic THOC6 pathogenic variants: Prenatal phenotype and review of the literature. Birth Defects Res. 2022, 114, 499–504. [Google Scholar] [CrossRef]
- Tjon, J.K.; Tan-Sindhunata, G.M.; Bugiani, M.; Witbreuk, M.M.; van der Sluijs, J.A.; Weiss, M.M.; van de Pol, L.A.; van Weissenbruch, M.M.; van der Knoop, B.J.; de Vries, J.I. Fetal akinesia deformation sequence, arthrogryposis multiplex congenita, and bilateral clubfeet: Is motor assessment of additional value for in utero diagnosis? A 10-year cohort study. Prenat. Diagn. 2019, 39, 219–231. [Google Scholar] [CrossRef]
- Barros, F.; Rolo, L.; Nardozza, L. Prenatal Diagnosis of Lethal Multiple Pterygium Syndrome Using Two-and Three-Dimensional Ultrasonography. J. Clin. Imaging Sci. 2012, 2, 65. [Google Scholar] [CrossRef]
| Case No. | Phenotype | Phenotypic Group | Gene | Variant | Inheritance | Classification | Associated Syndrome | Outcome |
|---|---|---|---|---|---|---|---|---|
| 1 | Abnormal ears, bilateral talipes equinovarus | skeletal | KMT2D | c.450G > A p.(Trp150*) | AD–de novo | path | Kabuki syndrome 1 (OMIM# 147920) | TOP |
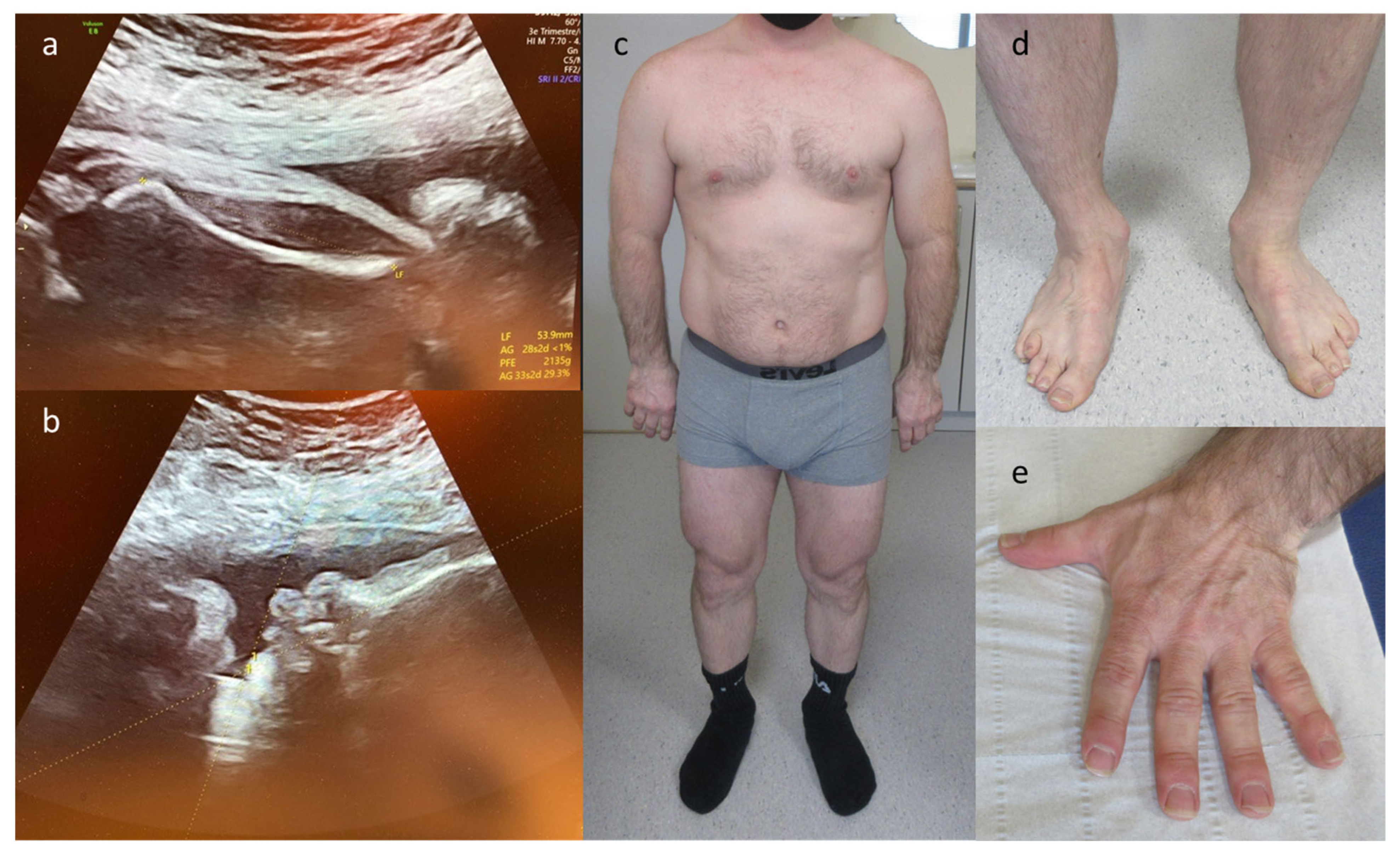
| 8 | IUGR, oligodactyly left hand, hypoplastic ray right hand | skeletal | FANCG | c.115C > T p.(Arg39*) | AR–hom | path | Fanconi anemia, complementation group G (OMIM# 614082) | TOP |
| 10 | Rhizomelic shortening and bowing of the long bones, microretrognathia and clenched hands | skeletal | COL2A1 | c.2710C > T p.(Arg904Cys) | AD–pat | path | Stickler syndrome type I (OMIM# 108300) | LB |
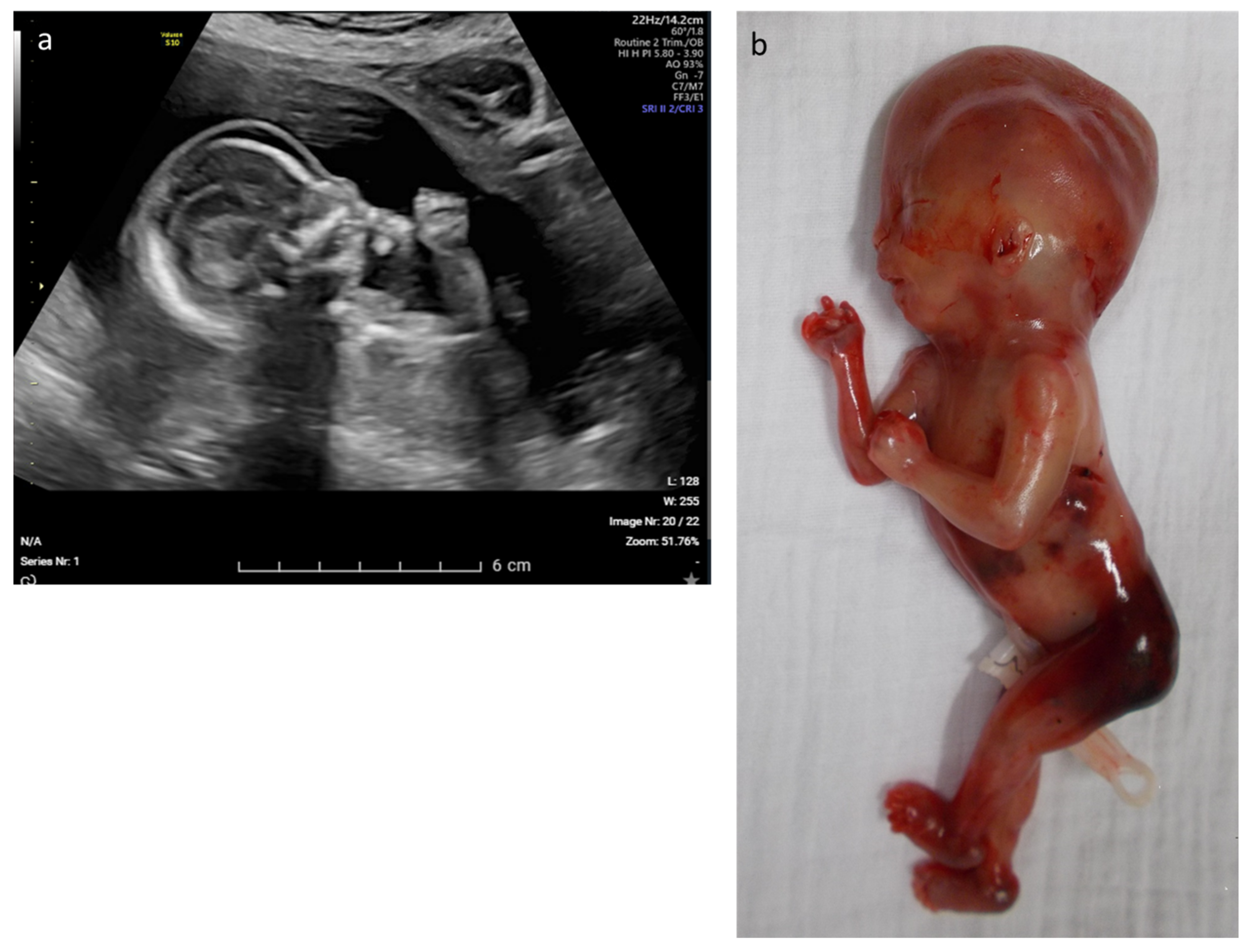
| 12 | Edema, rocker bottom foot, retrognathia, abnormal thorax and ribs, increased NT | multisystem | CHRNA1 | c.548A > G p.(Asp183Gly) | AR–hom | likely path | Multiple pterygium syndrome, lethal type (OMIM# 253290) | TOP |
| 23 | Hydrops, acites, hydrothorax | hydrops | RIT1 | c.297T > A p.(Phe99Leu) | AD–de novo | path | Noonan syndroom 8 (OMIM# 615355) | TOP |
| STXBP1 | c.875G > A p.(Arg292His) | AD–de novo | path (IF) | Developmental and epileptic encephalopathy type 4 (OMIM# 612164) | ||||
| 26 | Olivopontocerebellar hypoplasia, tetralogy of Fallot, hypospadias | multisystem | THOC6 | c.298T > A p.(Trp100Arg) | AR–het (mat) | path | Beaulieu-Boycott-Innes syndrome (OMIM# 613680) | TOP |
| THOC6 | c.700G > C p.(Val234Leu) | AR–het (mat) | path | |||||
| THOC6 | c.824G > A p.(Gly275Asp) | AR–het(mat) | path | |||||
| THOC6 | c.569G > A p.(Gly190Glu) | AR–het (pat) | path | |||||
| 28 | Fetal akinesia, hypotonia, rocker-bottom feet, hydrops, hydrothorax, ascites | multisystem | MUSK | c.2201G > T p.(Gly734Val) | AR–hom | likely path | Fetal akinesia deformation sequence 1 (OMIM# 208150) | TOP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janicki, E.; De Rademaeker, M.; Meunier, C.; Boeckx, N.; Blaumeiser, B.; Janssens, K. Implementation of Exome Sequencing in Prenatal Diagnostics: Chances and Challenges. Diagnostics 2023, 13, 860. https://doi.org/10.3390/diagnostics13050860
Janicki E, De Rademaeker M, Meunier C, Boeckx N, Blaumeiser B, Janssens K. Implementation of Exome Sequencing in Prenatal Diagnostics: Chances and Challenges. Diagnostics. 2023; 13(5):860. https://doi.org/10.3390/diagnostics13050860
Chicago/Turabian StyleJanicki, Ewa, Marjan De Rademaeker, Colombine Meunier, Nele Boeckx, Bettina Blaumeiser, and Katrien Janssens. 2023. "Implementation of Exome Sequencing in Prenatal Diagnostics: Chances and Challenges" Diagnostics 13, no. 5: 860. https://doi.org/10.3390/diagnostics13050860
APA StyleJanicki, E., De Rademaeker, M., Meunier, C., Boeckx, N., Blaumeiser, B., & Janssens, K. (2023). Implementation of Exome Sequencing in Prenatal Diagnostics: Chances and Challenges. Diagnostics, 13(5), 860. https://doi.org/10.3390/diagnostics13050860






