A New Approach for Gastrointestinal Tract Findings Detection and Classification: Deep Learning-Based Hybrid Stacking Ensemble Models
Abstract
1. Introduction
2. Related Works
2.1. Literature Gaps
2.2. Contributions
- It is the most detailed and large-scale study in the literature. It can simultaneously detect and classify pathological findings, anatomical points, therapeutic interventions, and mucosal images on two datasets with balanced and unbalanced sample distribution.
- It presents three new CNN models with high performance and low hyperparameter sensitivity to the literature.
- An innovative hybrid approach to learning that efficiently evaluates CNN features and enhances deep learning models’ performance is proposed.
- It is the first study in the literature to provide reliable and objective results in which performance results are supported by applying statistical tests other than metrics. This study is a good precedent for the statistical analysis of artificial intelligence methods.
- The performance of the proposed approach is higher than other state-of-the-art methods proposed in the literature.
3. Materials and Methods
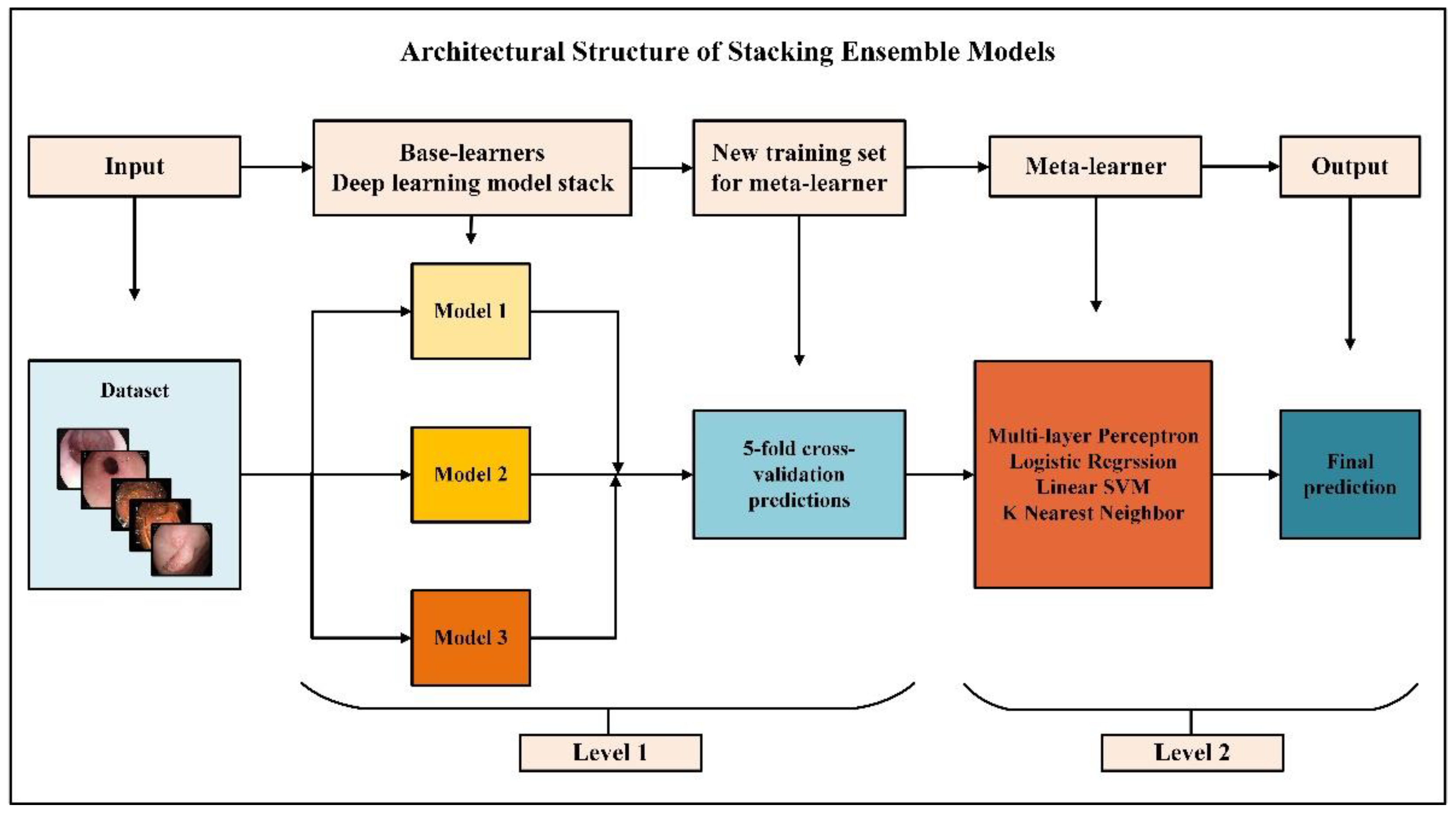
3.1. Proposed Approach
- The dataset was split into 5 using the stratified shuffle split cross-validation strategy;
- Three independent base learners are trained to the other folds, keeping one of the folds, and predictions are obtained;
- The above three steps were repeated five times to obtain out-of-sample predictions for all five folds;
- All out-of-sample predictions were used as training data for meta-learners;
- The final output was estimated with meta-learners.
3.2. Background
3.3. Datasets
3.4. Details of Proposed Approach
4. Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- PAHO. The Burden of Digestive Diseases in the Region of the Americas, 2000–2019; Pan American Health Organization: Washington, DC, USA, 2021. [Google Scholar]
- Kurumi, H.; Kanda, T.; Ikebuchi, Y.; Yoshida, A.; Kawaguchi, K.; Yashima, K.; Isomoto, H. Current Status of Photodynamic Diagnosis for Gastric Tumors. Diagnostics 2021, 11, 1967. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Shimodaira, Y.; Matsuhashi, T.; Tsuji, T.; Fukuda, S.; Sugawara, K.; Saruta, Y.; Watanabe, K.; Iijima, K. Nature and Clinical Outcomes of Acute Hemorrhagic Rectal Ulcer. Diagnostics 2022, 12, 2487. [Google Scholar] [CrossRef] [PubMed]
- Divya, V.; Sendil Kumar, S.; Gokula Krishnan, V.; Kumar, M. Signal Conducting System with Effective Optimization Using Deep Learning for Schizophrenia Classification. Comput. Syst. Sci. Eng. 2022, 45, 1869–1886. [Google Scholar] [CrossRef]
- Thapliyal, M.; Ahuja, N.J.; Shankar, A.; Cheng, X.; Kumar, M. A differentiated learning environment in domain model for learning disabled learners. J. Comput. High. Educ. 2022, 34, 60–82. [Google Scholar] [CrossRef]
- Raheja, S.; Kasturia, S.; Cheng, X.; Kumar, M. Machine learning-based diffusion model for prediction of coronavirus-19 outbreak. Neural Comput. Appl. 2021, 1, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Oka, A.; Ishimura, N.; Ishihara, S. A New Dawn for the Use of Artificial Intelligence in Gastroenterology, Hepatology and Pancreatology. Diagnostics 2021, 11, 1719. [Google Scholar] [CrossRef]
- Patel, M.; Gulati, S.; O’Neil, S.; Wilson, N.; Williams, S.; Charles-Nurse, S.; Chung-Faye, G.; Srirajaskanthan, R.; Haji, A.; Hayee, B. Artificial intelligence increases adenoma detection even in ‘high-detector’ colonoscopy: Early evidence for human: Machine interaction. In Proceedings of the Posters; BMJ Publishing Group Ltd. and British Society of Gastroenterology: London, UK, 2021; Volume 70, pp. A70–A71. [Google Scholar]
- Lee, T.J.W.; Nickerson, C.; Rees, C.J.; Patnick, J.; Rutter, M.D. Comparison of colonoscopy quality indicators between surgeons, physicians and nurse endoscopists in the NHS bowel cancer screening programme: Analysis of the national database. Gut 2012, 61, A384. [Google Scholar] [CrossRef]
- Wang, P.; Berzin, T.M.; Glissen Brown, J.R.; Bharadwaj, S.; Becq, A.; Xiao, X.; Liu, P.; Li, L.; Song, Y.; Zhang, D.; et al. Real-time automatic detection system increases colonoscopic polyp and adenoma detection rates: A prospective randomised controlled study. Gut 2019, 68, 1813–1819. [Google Scholar] [CrossRef]
- Billah, M.; Waheed, S. Gastrointestinal polyp detection in endoscopic images using an improved feature extraction method. Biomed. Eng. Lett. 2018, 8, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Chao, W.L.; Manickavasagan, H.; Krishna, S.G. Application of Artificial Intelligence in the Detection and Differentiation of Colon Polyps: A Technical Review for Physicians. Diagnostics 2019, 9, 99. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Meng, M.Q.H. Automatic polyp detection for wireless capsule endoscopy images. Expert Syst. Appl. 2012, 39, 10952–10958. [Google Scholar] [CrossRef]
- Guo, L.; Gong, H.; Wang, Q.; Zhang, Q.; Tong, H.; Li, J.; Lei, X.; Xiao, X.; Li, C.; Jiang, J.; et al. Detection of multiple lesions of gastrointestinal tract for endoscopy using artificial intelligence model: A pilot study. Surg. Endosc. 2021, 35, 6532–6538. [Google Scholar] [CrossRef]
- Charfi, S.; El Ansari, M.; Balasingham, I. Computer-aided diagnosis system for ulcer detection in wireless capsule endoscopy images. IET Image Process. 2019, 13, 1023–1030. [Google Scholar] [CrossRef]
- Wang, X.; Qian, H.; Ciaccio, E.J.; Lewis, S.K.; Bhagat, G.; Green, P.H.; Xu, S.; Huang, L.; Gao, R.; Liu, Y. Celiac disease diagnosis from videocapsule endoscopy images with residual learning and deep feature extraction. Comput. Methods Programs Biomed. 2020, 187, 105236. [Google Scholar] [CrossRef]
- Renna, F.; Martins, M.; Neto, A.; Cunha, A.; Libânio, D.; Dinis-Ribeiro, M.; Coimbra, M. Artificial Intelligence for Upper Gastrointestinal Endoscopy: A Roadmap from Technology Development to Clinical Practice. Diagnostics 2022, 12, 1278. [Google Scholar] [CrossRef]
- Liedlgruber, M.; Uhl, A. Computer-aided decision support systems for endoscopy in the gastrointestinal tract: A review. IEEE Rev. Biomed. Eng. 2011, 4, 73–88. [Google Scholar] [CrossRef]
- Naz, J.; Sharif, M.; Yasmin, M.; Raza, M.; Khan, M.A. Detection and Classification of Gastrointestinal Diseases using Machine Learning. Curr. Med. Imaging Former. Curr. Med. Imaging Rev. 2020, 17, 479–490. [Google Scholar] [CrossRef]
- Pogorelov, K.; Randel, K.R.; Griwodz, C.; Eskeland, S.L.; De Lange, T.; Johansen, D.; Spampinato, C.; Dang-Nguyen, D.T.; Lux, M.; Schmidt, P.T.; et al. Kvasir: A multi-class image dataset for computer aided gastrointestinal disease detection. In Proceedings of the 8th ACM on Multimedia Systems Conference, Taipei, Taiwan, 20–23 June 2017; pp. 164–169. [Google Scholar] [CrossRef]
- Borgli, H.; Thambawita, V.; Smedsrud, P.H.; Hicks, S.; Jha, D.; Eskeland, S.L.; Randel, K.R.; Pogorelov, K.; Lux, M.; Nguyen, D.T.D.; et al. HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci. Data 2020, 7, 283. [Google Scholar] [CrossRef]
- Dheir, I.M.; Abu-Naser, S.S. Classification of Anomalies in Gastrointestinal Tract Using Deep Learning. Int. J. Acad. Eng. Res. 2022, 6, 15–28. [Google Scholar]
- Hmoud Al-Adhaileh, M.; Mohammed Senan, E.; Alsaade, F.W.; Aldhyani, T.H.H.; Alsharif, N.; Abdullah Alqarni, A.; Uddin, M.I.; Alzahrani, M.Y.; Alzain, E.D.; Jadhav, M.E. Deep Learning Algorithms for Detection and Classification of Gastrointestinal Diseases. Complexity 2021, 2021, 6170416. [Google Scholar] [CrossRef]
- Yogapriya, J.; Chandran, V.; Sumithra, M.G.; Anitha, P.; Jenopaul, P.; Suresh Gnana Dhas, C. Gastrointestinal Tract Disease Classification from Wireless Endoscopy Images Using Pretrained Deep Learning Model. Comput. Math. Methods Med. 2021, 2021, 5940433. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, Ş.; Özkaya, U. Gastrointestinal tract classification using improved LSTM based CNN. Multimed. Tools Appl. 2020, 79, 28825–28840. [Google Scholar] [CrossRef]
- Öztürk, Ş.; Özkaya, U. Residual LSTM layered CNN for classification of gastrointestinal tract diseases. J. Biomed. Inform. 2021, 113, 103638. [Google Scholar] [CrossRef]
- Dutta, A.; Bhattacharjee, R.K.; Barbhuiya, F.A. Efficient Detection of Lesions During Endoscopy. In ICPR International Workshops and Challenges; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12668, pp. 315–322. [Google Scholar] [CrossRef]
- Ramamurthy, K.; George, T.T.; Shah, Y.; Sasidhar, P. A Novel Multi-Feature Fusion Method for Classification of Gastrointestinal Diseases Using Endoscopy Images. Diagnostics 2022, 12, 2316. [Google Scholar] [CrossRef]
- Khan, M.A.; Sahar, N.; Khan, W.Z.; Alhaisoni, M.; Tariq, U.; Zayyan, M.H.; Kim, Y.J.; Chang, B. GestroNet: A Framework of Saliency Estimation and Optimal Deep Learning Features Based Gastrointestinal Diseases Detection and Classification. Diagnostics 2022, 12, 2718. [Google Scholar] [CrossRef]
- Khan, M.A.; Muhammad, K.; Wang, S.H.; Alsubai, S.; Binbusayyis, A.; Alqahtani, A.; Majumdar, A.; Thinnukool, O. Gastrointestinal Diseases Recognition: A Framework of Deep Neural Network and Improved Moth-Crow Optimization with DCCA Fusion. Hum.-Cent. Comput. Inf. Sci. 2022, 12, 25. [Google Scholar] [CrossRef]
- Mohapatra, S.; Nayak, J.; Mishra, M.; Pati, G.K.; Naik, B.; Swarnkar, T. Wavelet Transform and Deep Convolutional Neural Network-Based Smart Healthcare System for Gastrointestinal Disease Detection. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 212–228. [Google Scholar] [CrossRef]
- Mohapatra, S.; Kumar Pati, G.; Mishra, M.; Swarnkar, T. Gastrointestinal abnormality detection and classification using empirical wavelet transform and deep convolutional neural network from endoscopic images. Ain Shams Eng. J. 2022, 14, 101942. [Google Scholar] [CrossRef]
- Afriyie, Y.; Weyori, B.A.; Opoku, A.A. Gastrointestinal tract disease recognition based on denoising capsule network. Cogent Eng. 2022, 9, 2142072. [Google Scholar] [CrossRef]
- Wang, W.; Yang, X.; Li, X.; Tang, J. Convolutional-capsule network for gastrointestinal endoscopy image classification. Int. J. Intell. Syst. 2022, 37, 5796–5815. [Google Scholar] [CrossRef]
- Ganaie, M.A.; Hu, M.; Malik, A.K.; Tanveer, M.; Suganthan, P.N. Ensemble deep learning: A review. Eng. Appl. Artif. Intell. 2021, 115, 105151. [Google Scholar] [CrossRef]
- Mohammed, M.; Mwambi, H.; Mboya, I.B.; Elbashir, M.K.; Omolo, B. A stacking ensemble deep learning approach to cancer type classification based on TCGA data. Sci. Rep. 2021, 11, 15626. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Sharma, S.; Athaiya, A. Activation Functions in Neural Networks. Int. J. Eng. Appl. Sci. Technol. 2020, 4, 310–316. [Google Scholar] [CrossRef]
- Desai, M.; Shah, M. An anatomization on breast cancer detection and diagnosis employing multi-layer perceptron neural network (MLP) and Convolutional neural network (CNN). Clin. eHealth 2021, 4, 1–11. [Google Scholar] [CrossRef]
- Boateng, E.Y.; Abaye, D.A.; Boateng, E.Y.; Abaye, D.A. A Review of the Logistic Regression Model with Emphasis on Medical Research. J. Data Anal. Inf. Process. 2019, 7, 190–207. [Google Scholar] [CrossRef]
- Chauhan, V.K.; Dahiya, K.; Sharma, A. Problem formulations and solvers in linear SVM: A review. Artif. Intell. Rev. 2019, 52, 803–855. [Google Scholar] [CrossRef]
- Mohammed, S.N.; Serdar Guzel, M.; Bostanci, E. Classification and Success Investigation of Biomedical Data Sets Using Supervised Machine Learning Models. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Hendrycks, D.; Gimpel, K. Gaussian Error Linear Units (GELUs). arXiv 2016. [Google Scholar] [CrossRef]
- Agarap, A.F. Deep Learning using Rectified Linear Units (ReLU). arXiv 2018. [Google Scholar] [CrossRef]
- Chollet, F. Deep Learning with Python; Simon and Schuster: New York, NY, USA, 2017; ISBN 9781617294433. [Google Scholar]
- Pedregosa, F.; Weiss, R.; Brucher, M.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2015; pp. 1–15. [Google Scholar]
- Hanke, M. Regularizing properties of a truncated Newton-CG algorithm for nonlinear inverse problems. Numer. Funct. Anal. Optim. 1997, 18, 971–993. [Google Scholar] [CrossRef]
- Crammer, K.; Singer, Y. On The Algorithmic Implementation of Multiclass Kernel-based Vector Machines. J. Mach. Learn. Res. 2001, 2, 265–292. [Google Scholar]
- Bostanci, B.; Bostanci, E. An evaluation of classification algorithms using Mc Nemar’s test. In Proceedings of Seventh International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA 2012); Advances in Intelligent Systems and Computing; Springer: New Delhi, India, 2013; Volume 201, pp. 15–26. [Google Scholar] [CrossRef]




| Model 1 | Model 2 | Model 3 |
|---|---|---|
| 14 weight layers | 15 weight layers | 16 weight layers |
| input (150, 150, 3) | ||
| 3 × 3 conv—64 + gelu (150, 150, 64) 3 × 3 conv—64 + gelu (150, 150, 64) | ||
| 2 × 2 max-pooling, stride 2 (75, 75, 64) | ||
| batch_normalization | ||
| 3 × 3 conv—128 + gelu (75, 75, 128) 3 × 3 conv—128+ gelu (75, 75, 128) | ||
| 2 × 2 max-pooling, stride 2 (37, 37, 128) | ||
| batch_normalization | ||
| 3 × 3 conv—256 + gelu (37, 37, 256) 3 × 3 conv—256 + gelu (37, 37, 256) 3 × 3 conv—256 + gelu (37, 37, 256) | ||
| 2 × 2 max-pooling, stride 2 (18, 18, 256) | ||
| batch_normalization | ||
| 3 × 3 conv—512 + gelu (18, 18, 512) 3 × 3 conv—512 + gelu (18, 18, 512) 3 × 3 conv—512 + gelu (18, 18, 512) | ||
| 2 × 2 max-pooling, stride 2 (9, 9, 512) | ||
| batch_normalization | ||
| 3 × 3 conv—512 + gelu (9, 9, 512) 3 × 3 conv—512 + gelu (9, 9, 512) 3 × 3 conv—512 + gelu (9, 9, 512) | ||
| 2 × 2 max-pooling, stride 2 (4, 4, 512) | ||
| global-average-pooling—512 | ||
| dense—1024 + gelu dropout—0.5 batch_normalization | dense—2048 + gelu dropout—0.5 batch_normalization | dense—4096 + gelu dropout—0.5 batch_normalization |
| dense—8/12 + softmax | dense—1024 + gelu dropout—0.5 batch_normalization | dense—2048 + gelu dropout—0.5 batch_normalization |
| dense—8/12 + softmax | dense—1024 + gelu dropout—0.5 batch_normalization | |
| dense—8/12 + softmax | ||
| Base Learners Hyperparameter | Value |
|---|---|
| Batch size | 32 |
| Learning rate | 2 × 10−3 |
| Epoch | 50 |
| Loss function | sparse_categorical_crossentropy |
| Optimization algorithm | Adam [48] |
| Meta-Learners Hyperparameter | Value |
| Logistic regression | |
| C | 10 |
| Solver | newton-cg [49] |
| max_iter | 100 |
| multi_class | ovr |
| Support vector machine | |
| C | 0.1 |
| Kernel | linear |
| Loss | squared_hinge |
| max_iter | 100 |
| multi_class | crammer_singer [50] |
| Multi-layer Perceptron | |
| hidden_layer_sizes | 128, 64 |
| Activation | ReLU |
| Solver | Adam |
| learning_rate_init | 1 × 10−3 |
| max_iter | 100 |
| K-nearest neighbors | |
| n_neighbors | 15 |
| P | 1 |
| Metric | manhattan_distance |
| Metrics | Description |
|---|---|
| True Positive (TP) | A situation where the actual and predicted data point class is correct. |
| True Negative (TN) | A situation where the actual and predicted data point class is incorrect. |
| False Positive (FP) | A situation where the actual data point class is false and the predicted data point class is true. |
| False Negative (FN) | A situation where the actual data point class is true and the predicted data point class is false. |
| ROC-AUC | The AUC (area under the ROC curve) value measures how well two classes can be distinguished, and a value of 1 is desired for perfect discrimination. |
| Accuracy (ACC) | It is the number of correct guesses divided by the total number of guesses and is required to be 1 for perfect discrimination. |
| Precision | A Type I error occurs when a true null hypothesis () is rejected. A Type I error is the definition of a class as normal that would be sick. That is why the precision metric is critical in medical applications. |
| Recall | A Type II error occurs when a false null hypothesis () is accepted. Type II error is when a class that would be defined as normal is defined as sick. |
| Score | It is the harmonic mean of precision and recall and is required to be 1 for perfect discrimination. It does not explicitly report whether a model has Type I or Type II errors but provides an objective measure of unbalanced classification problems. |
| Jaccard similarity coefficient (JSC) | It is the ratio of the intersection of two sets of labels, actual and predicted, to the union and is required to be 1 for perfect discrimination. |
| Matthews correlation coefficient (MCC) | In the case of multiple classification, the minimum value is between −1 and 0, and the maximum value is +1. The and ACC metrics are positive class-dependent, but MCC produces positive and negative class-dependent results. Many studies consider it the most reliable metric in unbalanced classification problems. |
| Classifier | ACC (%) | Precision (%) | Recall (%) | Score (%) | ROC-AUC (%) | JSC (%) | MCC (%) |
|---|---|---|---|---|---|---|---|
| KvasirV2 | |||||||
| Model 1 | 95.33 | 95.44 | 95.33 | 95.35 | 99.69 | 91.15 | 94.68 |
| Model 2 | 96.33 | 96.38 | 96.33 | 96.34 | 99.73 | 92.98 | 95.81 |
| Model 3 | 92.75 | 93.21 | 92.75 | 92.77 | 99.60 | 86.59 | 91.77 |
| Stacking Ensemble Model 1 | 97.17 | 97.22 | 97.17 | 97.18 | 99.82 | 94.54 | 96.77 |
| Stacking Ensemble Model 2 | 97.58 | 97.59 | 97.58 | 97.58 | 99.80 | 95.30 | 97.24 |
| Stacking Ensemble Model 3 | 98.42 | 98.42 | 98.42 | 98.42 | 99.84 | 96.89 | 98.19 |
| Stacking Ensemble Model 4 | 95.25 | 95.28 | 95.25 | 95.25 | 98.87 | 90.98 | 94.57 |
| HyperKvasir | |||||||
| Model 1 | 95.91 | 95.87 | 95.63 | 95.71 | 99.79 | 91.86 | 95.51 |
| Model 2 | 97.06 | 96.55 | 96.87 | 96.69 | 99.86 | 93.69 | 96.77 |
| Model 3 | 93.60 | 93.43 | 93.25 | 93.20 | 99.64 | 87.44 | 92.99 |
| Stacking Ensemble Model 1 | 98.53 | 98.45 | 98.48 | 98.46 | 99.95 | 96.98 | 98.39 |
| Stacking Ensemble Model 2 | 97.25 | 97.11 | 97.23 | 97.14 | 99.83 | 94.49 | 96.98 |
| Stacking Ensemble Model 3 | 97.76 | 97.66 | 97.65 | 97.65 | 99.90 | 95.43 | 97.54 |
| Stacking Ensemble Model 4 | 96.29 | 96.19 | 96.19 | 96.15 | 98.62 | 92.63 | 95.93 |
| KvasirV2 | ||||||
|---|---|---|---|---|---|---|
| Model 2 | Model 3 | SEM 1 | SEM 2 | SEM 3 | SEM 4 | |
| Model 1 | ↑1.33 | ←2.82 | ↑2.71 | ↑2.82 | ↑4.16 | 0 |
| Model 2 | ←4.22 | ↑1.27 | ↑1.66 | ↑3.07 | ←1.23 | |
| Model 3 | ↑5.45 | ↑5.39 | ↑6.57 | ↑2.49 | ||
| SEM 1 | ↑0.51 | ↑1.92 | ←2.33 | |||
| SEM 2 | ↑1.42 | ←3.74 | ||||
| SEM 3 | ←4.62 | |||||
| HyperKvasir | ||||||
| Model 2 | Model 3 | SEM 1 | SEM 2 | SEM 3 | SEM 4 | |
| Model 1 | ↑1.88 | ←3.4 | ↑5.04 | ↑2.52 | ↑3.37 | ↑0.58 |
| Model 2 | ←5.3 | ↑3.02 | ↑0.25 | ↑1.35 | ←1.26 | |
| Model 3 | ↑7.97 | ↑5.94 | ↑6.57 | ↑4.1 | ||
| SEM 1 | ←2.93 | ←1.7 | ←4.86 | |||
| SEM 2 | ↑1.01 | ←1.96 | ||||
| SEM 3 | ←2.77 | |||||
| Model 1 | Model 2 | Model 3 | SEM 1 | SEM 2 | SEM 3 | SEM 4 | |
|---|---|---|---|---|---|---|---|
| McNemar’ s Test | |||||||
| KvasirV2 | ++ | +++ | + | ++++ | +++++ | ++++++ | ++ |
| HyperKvasir | ++ | ++++ | + | +++++++ | +++++ | ++++++ | +++ |
| ROC-AUC | |||||||
| KvasirV2 | +++ | ++++ | ++ | ++++++ | +++++ | +++++++ | + |
| HyperKvasir | +++ | +++++ | ++ | +++++++ | ++++ | ++++++ | + |
| ACC, , JSC, and MCC | |||||||
| KvasirV2 | +++ | ++++ | + | +++++ | ++++++ | +++++++ | ++ |
| HyperKvasir | ++ | ++++ | + | +++++++ | +++++ | ++++++ | +++ |
| Author, Year, Reference | Approach | Results (ACC %) |
|---|---|---|
| KvasirV2 | ||
| Öztürk and Özkaya, 2020 [26] | LSTM based CNN | 97.90 |
| Öztürk and Özkaya, 2021 [27] | Residual LSTM layered CNN | 98.05 |
| Hmoud Al-Adhaileh et al., 2021 [24] | Transfer learning | 97.00 |
| Mohapatra et al., 2021 [32] | 2D-DWT and CNN | 97.25 |
| Khan et al., 2022 [30] | Bayesian optimal deep learning feature selection | 98.02 |
| Yogapriya et al., 2021 [25] | Transfer learning | 96.33 |
| Afriyie et al., 2022 [34] | Dn-CapsNet | 94.16 |
| Khan et al., 2022 [31] | Moth-Crow Optimization with DCCA Fusion | 97.20 |
| Dheir and Abu-Naser, 2022 [23] | Transfer learning | 98.30 |
| HyperKvasir | ||
| Ramamurthy et al., 2022 [29] | Multi-feature fusion method | 97.99 |
| Mohapatra et al., 2022 [33] | EWT and CNN | 96.65 |
| Dutta et al., 2021 [28] | Tiny Darknet | 75.80 (MCC) |
| Borgli et al., 2020 [22] | ResNet-152 + DenseNet-161 | 90.20 (MCC) |
| KvasirV2 + HyperKvasir | ||
| Wang et al., 2022 [35] | Convolutional-capsule network | KvasirV2 94.83; HyperKvasir 85.99 |
| This study | Deep learning-based hybrid stacking ensemble models | KvasirV2 98.42; HyperKvasir 98.53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivari, E.; Bostanci, E.; Guzel, M.S.; Acici, K.; Asuroglu, T.; Ercelebi Ayyildiz, T. A New Approach for Gastrointestinal Tract Findings Detection and Classification: Deep Learning-Based Hybrid Stacking Ensemble Models. Diagnostics 2023, 13, 720. https://doi.org/10.3390/diagnostics13040720
Sivari E, Bostanci E, Guzel MS, Acici K, Asuroglu T, Ercelebi Ayyildiz T. A New Approach for Gastrointestinal Tract Findings Detection and Classification: Deep Learning-Based Hybrid Stacking Ensemble Models. Diagnostics. 2023; 13(4):720. https://doi.org/10.3390/diagnostics13040720
Chicago/Turabian StyleSivari, Esra, Erkan Bostanci, Mehmet Serdar Guzel, Koray Acici, Tunc Asuroglu, and Tulin Ercelebi Ayyildiz. 2023. "A New Approach for Gastrointestinal Tract Findings Detection and Classification: Deep Learning-Based Hybrid Stacking Ensemble Models" Diagnostics 13, no. 4: 720. https://doi.org/10.3390/diagnostics13040720
APA StyleSivari, E., Bostanci, E., Guzel, M. S., Acici, K., Asuroglu, T., & Ercelebi Ayyildiz, T. (2023). A New Approach for Gastrointestinal Tract Findings Detection and Classification: Deep Learning-Based Hybrid Stacking Ensemble Models. Diagnostics, 13(4), 720. https://doi.org/10.3390/diagnostics13040720







