Association between ADAM33 Single-Nucleotide Polymorphisms and Treatment Response to Inhaled Corticosteroids and a Long-Acting Beta-Agonist in Asthma
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Genomic DNA Isolation
2.3. SNP Genotyping
2.4. Analysis of SNPs of the ADAM33 Gene
2.5. Statistical Analysis
3. Results
3.1. Demographic and Clinical Profiles of the Study Population
3.2. Improvement in Lung Function in Various Severity Groups upon ICS and LABA Treatment
3.3. ADAM33 SNP Distribution in the Study Population
3.4. ADAM33 SNP Association with Disease Severity
3.5. ADAM33 SNP Association with Lung Function Improvement in Asthma Patients upon ICS and LABA Treatment
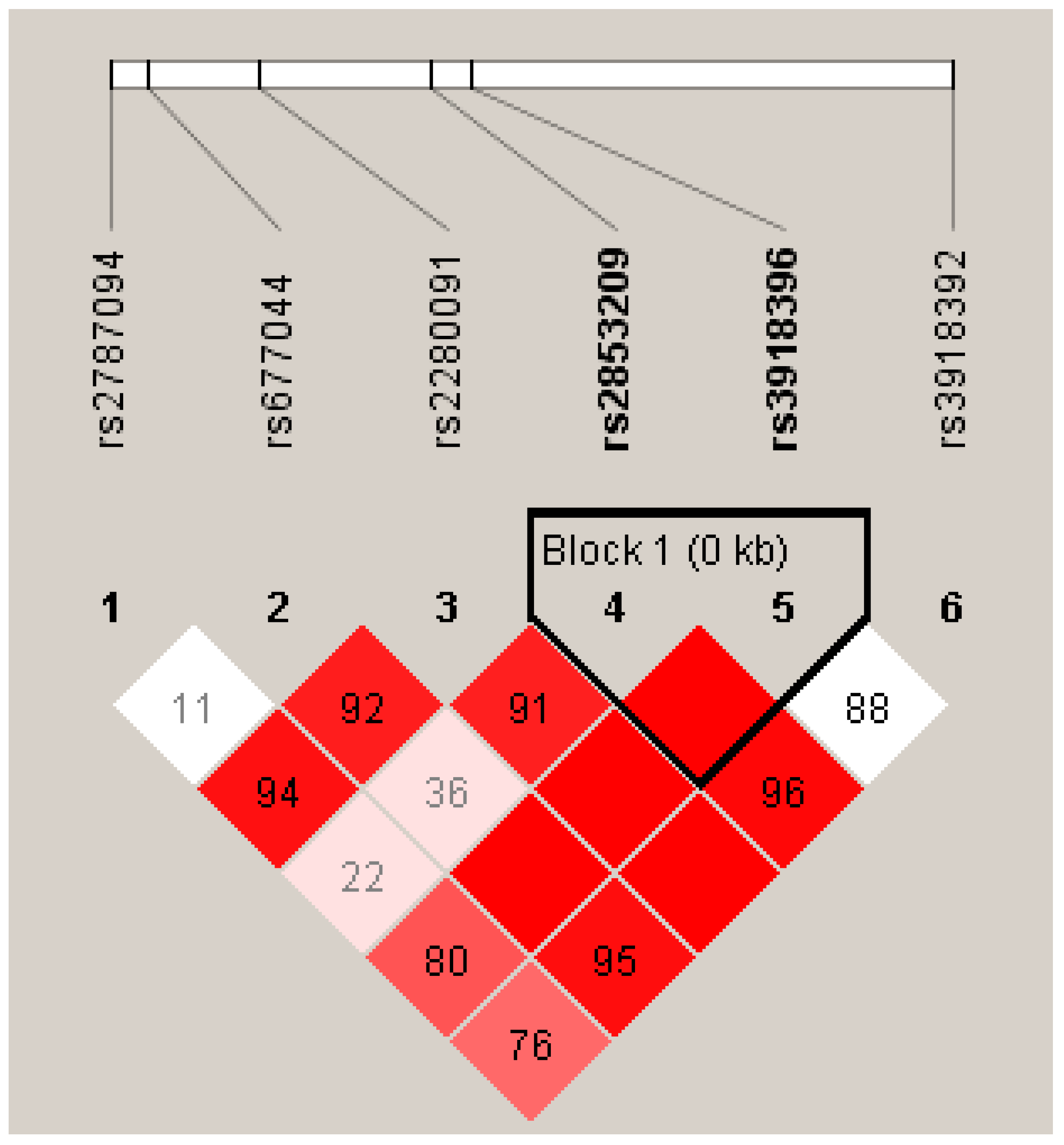
3.6. Haplotype Association of ADAM33 SNPs with Asthma
4. Discussion
Strength and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hargreave, F.E.; Nair, P. The Definition and Diagnosis of Asthma. Clin. Exp. Allergy 2009, 39, 1652–1658. [Google Scholar] [CrossRef]
- Krishna, M.T.; Mahesh, P.A.; Vedanthan, P.K.; Mehta, V.; Moitra, S.; Christopher, D.J. The Burden of Allergic Diseases in the Indian Subcontinent: Barriers and Challenges. Lancet Glob. Health 2020, 8, e478–e479. [Google Scholar] [CrossRef]
- Parthasarathi, A.; Padukudru, S.; Rajgopal, N.; Holla, A.D.; Krishna, M.T.; Mahesh, P.A. Allergic Disease Prevalence in School Children in Bengaluru, India: A Cross-sectional Survey. Clin. Exp. Allergy 2021, 51, 955–958. [Google Scholar] [CrossRef] [PubMed]
- Haughney, J.; Price, D.; Kaplan, A.; Chrystyn, H.; Horne, R.; May, N.; Moffat, M.; Versnel, J.; Shanahan, E.R.; Hillyer, E.V.; et al. Achieving Asthma Control in Practice: Understanding the Reasons for Poor Control. Respir. Med. 2008, 102, 1681–1693. [Google Scholar] [CrossRef] [PubMed]
- Martel, M.-J.; Rey, E.; Malo, J.-L.; Perreault, S.; Beauchesne, M.-F.; Forget, A.; Blais, L. Determinants of the Incidence of Childhood Asthma: A Two-Stage Case-Control Study. Am. J. Epidemiol. 2009, 169, 195–205. [Google Scholar] [CrossRef]
- Parthasarathi, A.; Srinivas, S.; Biligere Siddaiah, J.; Anand Mahesh, P. Local Adverse Drug Reactions in Ambulatory Asthma Patients TreatedWith Inhaled Corticosteroids: An Experience from a South Indian TeachingHospital. Curr. Respir. Med. Rev. 2022, 18, 217–227. [Google Scholar] [CrossRef]
- Vercelli, D. Discovering Susceptibility Genes for Asthma and Allergy. Nat. Rev. Immunol. 2008, 8, 169–182. [Google Scholar] [CrossRef]
- Bijanzadeh, M.; Mahesh, P.A.; Ramachandra, N.B. An Understanding of the Genetic Basis of Asthma. Indian J. Med. Res. 2011, 134, 149–161. [Google Scholar]
- Ober, C. Genome-Wide Search for Asthma Susceptibility Loci in a Founder Population. The Collaborative Study on the Genetics of Asthma. Hum. Mol. Genet. 1998, 7, 1393–1398. [Google Scholar] [CrossRef]
- Teerlink, C.C.; Camp, N.J.; Bansal, A.; Crapo, R.; Hughes, D.; Kort, E.; Rowe, K.; Cannon-Albright, L.A. Significant Evidence for Linkage to Chromosome 5q13 in a Genome-Wide Scan for Asthma in an Extended Pedigree Resource. Eur. J. Hum. Genet. 2009, 17, 636–643. [Google Scholar] [CrossRef]
- Meng, J.-F.; Rosenwasser, L.J. Unraveling the Genetic Basis of Asthma and Allergic Diseases. Allergy Asthma Immunol. Res. 2010, 2, 215–227. [Google Scholar] [CrossRef]
- Bergeron, C.; Al-Ramli, W.; Hamid, Q. Remodeling in Asthma. Proc. Am. Thorac. Soc. 2009, 6, 301–305. [Google Scholar] [CrossRef]
- Davies, D.E. The Role of the Epithelium in Airway Remodeling in Asthma. Proc. Am. Thorac. Soc. 2009, 6, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Hough, K.P.; Curtiss, M.L.; Blain, T.J.; Liu, R.-M.; Trevor, J.; Deshane, J.S.; Thannickal, V.J. Airway Remodeling in Asthma. Front. Med. 2020, 7, 191. [Google Scholar] [CrossRef] [PubMed]
- Chakir, J.; Shannon, J.; Molet, S.; Fukakusa, M.; Elias, J.; Laviolette, M.; Boulet, L.-P.; Hamid, Q. Airway Remodeling-Associated Mediators in Moderate to Severe Asthma: Effect of Steroids on TGF-β, IL-11, IL-17, and Type I and Type III Collagen Expression. J. Allergy Clin. Immunol. 2003, 111, 1293–1298. [Google Scholar] [CrossRef]
- Grainge, C.L.; Lau, L.C.K.; Ward, J.A.; Dulay, V.; Lahiff, G.; Wilson, S.; Holgate, S.; Davies, D.E.; Howarth, P.H. Effect of Bronchoconstriction on Airway Remodeling in Asthma. N. Engl. J. Med. 2011, 364, 2006–2015. [Google Scholar] [CrossRef] [PubMed]
- Van Eerdewegh, P.; Little, R.D.; Dupuis, J.; Del Mastro, R.G.; Falls, K.; Simon, J.; Torrey, D.; Pandit, S.; McKenny, J.; Braunschweiger, K.; et al. Association of the ADAM33 Gene with Asthma and Bronchial Hyperresponsiveness. Nature 2002, 418, 426–430. [Google Scholar] [CrossRef]
- Davies, E.R.; Kelly, J.F.C.; Howarth, P.H.; Wilson, D.I.; Holgate, S.T.; Davies, D.E.; Whitsett, J.A.; Haitchi, H.M. Soluble ADAM33 Initiates Airway Remodeling to Promote Susceptibility for Allergic Asthma in Early Life. JCI Insight 2016, 1. [Google Scholar] [CrossRef]
- Durrani, S.R.; Viswanathan, R.K.; Busse, W.W. What Effect Does Asthma Treatment Have on Airway Remodeling? Current Perspectives. J. Allergy Clin. Immunol. 2011, 128, 439–448. [Google Scholar] [CrossRef]
- Oppenheimer, J. Role of Genetic Polymorphisms in Therapeutic Response to Anti-Asthma Therapy. Allergy Asthma Clin. Immunol. 2007, 3, 50. [Google Scholar] [CrossRef][Green Version]
- Berger, W. New Approaches to Managing Asthma: A US Perspective. Ther. Clin. Risk Manag. 2008, 4, 363–379. [Google Scholar] [CrossRef]
- Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention. 2012. p. 225. Available online: Www.Ginasthma.Org (accessed on 20 September 2022).
- Miller, M.R. Standardisation of Spirometry. Eur. Respir. J. 2005, 26, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Vishweswaraiah, S.; Ramachandra, N.; Jayaraj, B.; Holla, A.; Chakraborty, S.; Agrawal, A.; Mahesh, P. Haplotype Analysis of ADAM33 Polymorphisms in Asthma: A Pilot Study. Indian J. Med. Res. 2019, 150, 272. [Google Scholar] [CrossRef] [PubMed]
- Holgate, S.T. Is Big Beautiful? The Continuing Story of ADAM33 and Asthma. Thorax 2005, 60, 263–264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bijanzadeh, M.; Ramachandra, N.B.; Mahesh, P.A.; Savitha Mysore, R.; Kumar, P.; Manjunath, B.S.; Jayaraj, B.S. Association of IL-4 and ADAM33 Gene Polymorphisms with Asthma in an Indian Population. Lung 2010, 188, 415–422. [Google Scholar] [CrossRef]
- Holgate, S.T.; Davies, D.E.; Rorke, S.; Cakebread, J.; Murphy, G.; Powell, R.M.; Holloway, J.W. ADAM 33 and Its Association with Airway Remodeling and Hyperresponsiveness in Asthma. Clin. Rev. Allergy Immunol. 2004, 27, 023–034. [Google Scholar] [CrossRef] [PubMed]
- Kedda, M.-A.; Duffy, D.L.; Bradley, B.; O’Hehir, R.E.; Thompson, P.J. ADAM33 Haplotypes Are Associated with Asthma in a Large Australian Population. Eur. J. Hum. Genet. 2006, 14, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Vishweswaraiah, S.; Veerappa, A.M.; Mahesh, P.A.; Jayaraju, B.S.; Krishnarao, C.S.; Ramachandra, N.B. Molecular Interaction Network and Pathway Studies of ADAM33 Potentially Relevant to Asthma. Ann. Allergy Asthma. Immunol. 2014, 113, 418–424.e1. [Google Scholar] [CrossRef]
- Mahesh, P.A. Unravelling the Role of ADAM 33 in Asthma. Indian J. Med. Res. 2013, 137, 447–450. [Google Scholar] [PubMed]
- Gauderman, W.J. Sample Size Requirements for Matched Case-Control Studies of Gene-Environment Interaction. Stat. Med. 2002, 21, 35–50. [Google Scholar] [CrossRef]
- Lloyd-Jones, G. Design and Control Issues in Qualitative Case Study Research. Int. J. Qual. Methods 2003, 2, 33–42. [Google Scholar] [CrossRef]
- Iles, M.M. What Can Genome-Wide Association Studies Tell Us about the Genetics of Common Disease? PLoS Genet. 2008, 4, e33. [Google Scholar] [CrossRef] [PubMed]
- Gauderman, W.J.; Murcray, C.; Gilliland, F.; Conti, D.V. Testing Association between Disease and Multiple SNPs in a Candidate Gene. Genet. Epidemiol. 2007, 31, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Tripathi, P.; Ganesh, S.; Husain, N. Association of ADAM33 Gene Polymorphisms with Asthma in Indian Children. J. Hum. Genet. 2011, 56, 188–195. [Google Scholar] [CrossRef]
- Berenguer, A.; Fernandes, A.; Oliveira, S.; Rodrigues, M.; Ornelas, P.; Romeira, D.; Serrão, T.; Rosa, A.; Câmara, R. Genetic Polymorphisms and Asthma: Findings from a Case–Control Study in the Madeira Island Population. Biol. Res. 2014, 47, 40. [Google Scholar] [CrossRef] [PubMed]
- Thongngarm, T.; Jameekornrak, A.; Limwongse, C.; Sangasapaviliya, A.; Jirapongsananuruk, O.; Assawamakin, A.; Chaiyaratana, N.; Luangwedchakarn, V.; Thongnoppakhun, W. Association between ADAM33 Polymorphisms and Asthma in a Thai Population. Asian Pac. J. Allergy Immunol. 2008, 26, 205–211. [Google Scholar]
- Van Den Berge, M.; Ten Hacken, N.; Kerstjens, H.; Postma, D. Management of Asthma with ICS and LABAs: Different Treatment Strategies. Clin. Med. Ther. 2009, 1, CMT.S2283. [Google Scholar] [CrossRef]
- Tamm, M.; Richards, D.H.; Beghé, B.; Fabbri, L. Inhaled Corticosteroid and Long-Acting Β2-Agonist Pharmacological Profiles: Effective Asthma Therapy in Practice. Respir. Med. 2012, 106, S9–S19. [Google Scholar] [CrossRef] [PubMed]
- Lopert, A.; Rijavec, M.; Žavbi, M.; Korošec, P.; Fležar, M. Asthma Treatment Outcome in Adults Is Associated with Rs9910408 in TBX21 Gene. Sci. Rep. 2013, 3, 2915. [Google Scholar] [CrossRef]
- Qiu, Y.; Zhang, D.; Qin, Y.; Yin, K.-S. Effect of Β2 -Adrenergic Receptor Gene Arg16Gly Polymorphisms on Response to Long-Acting Β2-Agonist in Chinese Han Asthmatic Patients. Multidiscip. Respir. Med. 2014, 9, 22. [Google Scholar] [CrossRef]
- Holgate, S.T. ADAM 33: Just Another Asthma Gene or a Breakthrough in Understanding the Origins of Bronchial Hyperresponsiveness? Thorax 2003, 58, 466–469. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, J.H.; Park, H.-S.; Park, S.W.; Jang, A.S.; Uh, S.T.; Rhim, T.; Park, C.-S.; Hong, S.-J.; Holgate, S.T.; Holloway, J.W.; et al. ADAM33 Polymorphism: Association with Bronchial Hyper-Responsiveness in Korean Asthmatics. Clin. Htmlent Glyphamp Asciiamp Exp. Allergy 2004, 34, 860–865. [Google Scholar] [CrossRef] [PubMed]
- Blakey, J.D.; Sayers, I.; Ring, S.M.; Strachan, D.P.; Hall, I.P. Positionally Cloned Asthma Susceptibility Gene Polymorphisms and Disease Risk in the British 1958 Birth Cohort. Thorax 2009, 64, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Jongepier, H.; Boezen, H.M.; Dijkstra, A.; Howard, T.D.; Vonk, J.M.; Koppelman, G.H.; Zheng, S.L.; Meyers, D.A.; Bleecker, E.R.; Postma, D.S. Polymorphisms of the ADAM33 Gene Are Associated with Accelerated Lung Function Decline in Asthma. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2004, 34, 757–760. [Google Scholar] [CrossRef]
- Lee, J.-Y.; Park, S.-W.; Chang, H.K.; Kim, H.Y.; Rhim, T.; Lee, J.-H.; Jang, A.-S.; Koh, E.-S.; Park, C.-S. A Disintegrin and Metalloproteinase 33 Protein in Patients with Asthma: Relevance to Airflow Limitation. Am. J. Respir. Crit. Care Med. 2006, 173, 729–735. [Google Scholar] [CrossRef]
- Drazen, J.M.; Silverman, E.K.; Lee, T.H. Heterogeneity of Therapeutic Responses in Asthma. Br. Med. Bull. 2000, 56, 1054–1070. [Google Scholar] [CrossRef]
- Szostak, E.; Gebauer, F. Translational Control by 3’-UTR-Binding Proteins. Brief. Funct. Genom. 2013, 12, 58–65. [Google Scholar] [CrossRef]
- Abdullah, N.; Borhanuddin, B.; Shah, S.A.; Hassan, T.; Jamal, R. Global Lung Initiative 2012 Spirometry Reference Values in a Large Asian Cohort of Malay, Chinese and Indian Ancestry. Respirology 2018, 23, 1173–1179. [Google Scholar] [CrossRef]
- Balding, D.J. A Tutorial on Statistical Methods for Population Association Studies. Nat. Rev. Genet. 2006, 7, 781–791. [Google Scholar] [CrossRef]
- Stram, D.O.; Seshan, V.E. Multi-SNP Haplotype Analysis Methods for Association Analysis. In Statistical Human Genetics; Elston, R.C., Satagopan, J.M., Sun, S., Eds.; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 850, pp. 423–452. ISBN 978-1-61779-554-1. [Google Scholar]
- Tishkoff, S.A.; Pakstis, A.J.; Ruano, G.; Kidd, K.K. The Accuracy of Statistical Methods for Estimation of Haplotype Frequencies: An Example from the CD4 Locus. Am. J. Hum. Genet. 2000, 67, 518–522. [Google Scholar] [CrossRef]
- Xu, C.-F.; Lewis, K.; Cantone, K.L.; Khan, P.; Donnelly, C.; White, N.; Crocker, N.; Boyd, P.R.; Zaykin, D.V.; Purvis, I.J. Effectiveness of Computational Methods in Haplotype Prediction. Hum. Genet. 2002, 110, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Halldórsson, B.V.; Bafna, V.; Edwards, N.; Lippert, R.; Yooseph, S.; Istrail, S. Combinatorial Problems Arising in SNP and Haplotype Analysis. In Discrete Mathematics and Theoretical Computer Science; Calude, C.S., Dinneen, M.J., Vajnovszki, V., Eds.; Lecture Notes in Computer Science; Springer Berlin Heidelberg: Berlin/Heidelberg, Germany, 2003; Volume 273, pp. 26–47. ISBN 978-3-540-40505-4. [Google Scholar]
- McKeigue, P.M. Efficiency of Estimation of Haplotype Frequencies: Use of Marker Phenotypes of Unrelated Individuals versus Counting of Phase-Known Gametes. Am. J. Hum. Genet. 2000, 67, 1626–1627. [Google Scholar] [CrossRef] [PubMed]
- Vergara, C.I.; Acevedo, N.; Jiménez, S.; Martínez, B.; Mercado, D.; Gusmão, L.; Barnes, K.C.; Caraballo, L. A Six-SNP Haplotype of ADAM33 Is Associated with Asthma in a Population of Cartagena, Colombia. Int. Arch. Allergy Immunol. 2010, 152, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-Wide Association Studies. PLoS Comput. Biol. 2012, 8, e1002822. [Google Scholar] [CrossRef]
- Singh, K.; Singh, V.K.; Agrawal, N.K.; Gupta, S.K.; Singh, K. Association of Toll-Like Receptor 4 Polymorphisms with Diabetic Foot Ulcers and Application of Artificial Neural Network in DFU Risk Assessment in Type 2 Diabetes Patients. BioMed Res. Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Tripathi, P.; Awasthi, S.; Prasad, R.; Husain, N.; Ganesh, S. Association of ADAM33 Gene Polymorphisms with Adult-Onset Asthma and Its Severity in an Indian Adult Population. J. Genet. 2011, 90, 265–273. [Google Scholar] [CrossRef]
- Su, D.; Zhang, X.; Sui, H.; Lü, F.; Jin, L.; Zhang, J. Association of ADAM33 Gene Polymorphisms with Adult Allergic Asthma and Rhinitis in a Chinese Han Population. BMC Med. Genet. 2008, 9, 82. [Google Scholar] [CrossRef]
- Awasthi, S.; Tripathi, P.; Prasad, R.; Ganesh, S. ASSOCIATION OF A DISINTEGRIN AND METALLOPROTEASE 33 GENE POLYMORPHISMS AND THEIR HAPLOTYPES WITH ASTHMA IN THE NORTH-INDIAN POPULATION. Indian J. Med. Sci. 2016, 68, 54. [Google Scholar] [CrossRef]
- Manal, M.K.; Al-damerchi, A.; Kazaal, M.A. Evaluation of A Disintegrin and Metalloprotein33 Gene Polymorphism in Bronchial Asthma. AL-Qadisiya Med. J. 2014, 11, 9. [Google Scholar]
- Sinha, S.; Singh, J.; Kumar Jindal, S. Significant Association of ADAM33 V4C>G Polymorphisms with Asthma in a North Indian Population. Biores. Commun. 2015, 1, 62–68. Available online: https://Www.Bioresearchcommunications.Com/Index.Php/Brc/Article/View/149 (accessed on 15 October 2022).
- Xue, W.; Han, W.; Zhou, Z.-S. ADAM33 Polymorphisms Are Associated with Asthma and a Distinctive Palm Dermatoglyphic Pattern. Mol. Med. Rep. 2013, 8, 1795–1800. [Google Scholar] [CrossRef] [PubMed]
- Bukvic, B.K.; Blekic, M.; Simpson, A.; Marinho, S.; Curtin, J.A.; Hankinson, J.; Aberle, N.; Custovic, A. Asthma Severity, Polymorphisms in 20p13 and Their Interaction with Tobacco Smoke Exposure. Pediatr. Allergy Immunol. 2013, 24, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Miyake, Y.; Tanaka, K.; Arakawa, M. ADAM33 Polymorphisms, Smoking and Asthma in Japanese Women: The Kyushu Okinawa Maternal and Child Health Study. Int. J. Tuberc. Lung Dis. 2012, 16, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Farjadian, S.; Moghtaderi, M.; Hoseini-Pouya, B.-A.; Ebrahimpour, A.; Nasiri, M. ADAM33 Gene Polymorphisms in Southwestern Iranian Patients with Asthma. Iran. J. Basic Med. Sci. 2018, 21, 813–817. [Google Scholar] [CrossRef]
- Tripathi, P.; Awasthi, S.; Prasad, R.; Ganesh, S. Haplotypic Association of ADAM33 (T+1, S+1 and V − 3) Gene Variants in Genetic Susceptibility to Asthma in Indian Population. Ann. Hum. Biol. 2012, 39, 479–483. [Google Scholar] [CrossRef]

| Controls (N = 486) | Asthma Patients (N = 503) | |
|---|---|---|
| Age | ||
| ≤40 years | 291 (59.8) | 305 |
| >40 years | 195 | 198 |
| Gender | ||
| Male | 252 | 253 |
| Female | 234 | 250 |
| Smoking status | ||
| Smoker | - | 57 |
| Non-Smoker | 486 | 446 |
| Family History of Asthma | ||
| Yes | - | 213 |
| No | - | 290 |
| Asthma Duration | ||
| <1 year | - | 61 |
| 1–5 years | - | 196 |
| >5 years | - | 246 |
| Severity according to GINA guidelines | ||
| Mild persistent | - | 125 (24.9%) |
| Moderate persistent | - | 212 (42.3%) |
| Severe persistent | - | 166 (32.8%) |
| Number of Allergens Sensitized | ||
| ≤5 | - | 246 |
| >5 | - | 257 |
| Wheal diameter | ||
| ≤Histamine | - | 307 |
| >Histamine | - | 196 |
| Pulmonary Function Test | Controls | Asthma Patients | |
|---|---|---|---|
| Pre-Bronchodilator | Pre-Bronchodilator | Post-Bronchodilator | |
| FVC (% pred) | 88.44 ± 0.511 | 70.53 ± 0.82 | 76.02 ± 0.79 |
| FEV1 (% pred) | 88.84 ± 0.47 | 66.96 ± 0.89 | 74.23 ± 0.90 |
| FEV1/FVC (% pred) | 104.23 ± 0.35 | 96.69 ± 0.62 | 102.0 ± 0.66 |
| PEF (L/s) | 92.81 ± 0.95 | 71.60 ± 1.0 | 77.96 ± 1.05 |
| Baseline | Follow-Up (Three Months after ICS+LABAs) | |||||
|---|---|---|---|---|---|---|
| Genotype | Asthma Patients | Mean ± SE of Baseline Predicted FVC (%) | 95% CI Lower and Upper Limits | Mean ± SE of FVC (%) | 95% CI Lower and Upper Limits | |
| rs2280091 | AA | 353 | 70.3 ± 0.952 | 68.4–72.2 | 75.8 ± 0.916 | 74.0–77.6 |
| GA | 129 | 70.0 ± 1.70 | 66.7–73.4 | 75.0 ± 1.596 | 71.8–78.2 | |
| GG | 21 | 77.2 ± 4.78 | 67.3–87.2 | 82.0 ± 4.72 | 72.2–91.8 | |
| p-value | 0.293 | 0.141 | ||||
| rs2787094 | GG | 269 | 70.5 ± 1.15 | 68.3–72.8 | 76.2 ± 1.05 | 74.1–78.2 |
| GC | 196 | 71.8 ± 1.28 | 69.3–74.3 | 76.8 ± 1.27 | 74.3–79.3 | |
| CC | 38 | 64.1 ± 2.88 | 58.2–69.9 | 68.4 ± 2.92 | 62.5–74.4 | |
| p-value | 0.043 | 0.048 | ||||
| rs3918396 | GG | 389 | 70.5 ± 0.945 | 68.7–72.4 | 75.8 ± 0.891 | 74.1–77.6 |
| GA | 107 | 71.0 ± 1.72 | 67.6–74.4 | 76.8 ± 1.75 | 73.1–79.9 | |
| AA | 7 | 62.4 ± 7.14 | 45.0–79.9 | 67.4 ± 7.51 | 49.1–85.8 | |
| p-value | 0.442 | 0.222 | ||||
| rs677044 | AA | 327 | 70.4 ± 0.990 | 68.5–72.4 | 76.1 ± 0.945 | 74.1–77.8 |
| AG | 155 | 70.8 ± 1.58 | 67.7–73.9 | 75.7 ± 1.540 | 72.7–78.7 | |
| GG | 21 | 70.0 ± 4.14 | 61.4–78.6 | 75.2 ± 3.349 | 68.2–82.2 | |
| p-value | 0.913 | 0.964 | ||||
| rs2853209 | AA | 247 | 71.1 ± 1.06 | 69.0–73.2 | 77.1 ± 1.07 | 75.0–79.2 |
| AT | 195 | 70.4 ± 1.40 | 67.7–73.2 | 74.9 ± 1.31 | 72.4–77.5 | |
| TT | 61 | 68.5 ± 2.80 | 62.9–74.1 | 74.0 ± 2.50 | 69.0–79.0 | |
| p-value | 0.969 | 0.374 | ||||
| rs3918392 | AA | 435 | 70.3 ± 0.908 | 68.5–72.1 | 75.8 ± 0.853 | 74.2–77.5 |
| AG | 64 | 72.0 ± 1.897 | 68.2–75.8 | 76.2 ± 2.157 | 71.9–80.5 | |
| GG | 4 | 72.8 ± 7.962 | 47.4–98.1 | 72.5 ± 6.035 | 53.3–91.7 | |
| p-value | 0.822 | 0.847 | ||||
| Baseline | Follow-Up (Three Months after ICS+LABAs) | |||||
|---|---|---|---|---|---|---|
| Genotype | Asthma Patients | Mean ± SE of Baseline Predicted FEV1 (%) | 95% CI Lower and Upper Limits | Mean ± SE of FEV1 (%) | 95% CI Lower and Upper Limits | |
| rs2280091 | AA | 353 | 66.3 ± 1.061 | 64.2–68.4 | 74.2 ± 1.10 | 72.0–76.3 |
| GA | 129 | 67.7 ± 1.780 | 64.1–71.2 | 73.6 ± 1.62 | 70.4–76.8 | |
| GG | 21 | 73.6 ± 4.776 | 63.6–83.5 | 78.4 ± 4.54 | 68.9–87.8 | |
| p-value | 0.314 | 0.459 | ||||
| rs2787094 | GG | 269 | 66.9 ± 1.23 | 64.5–69.3 | 74.1 ± 1.16 | 71.8–76.3 |
| GC | 196 | 68.5 ± 1.39 | 65.8–71.3 | 75.9 ± 1.47 | 73.0–78.8 | |
| CC | 38 | 59.1 ± 3.48 | 52.1–66.2 | 66.4 ± 3.78 | 58.8–74.1 | |
| p-value | 0.038 | 0.049 | ||||
| rs3918396 | GG | 389 | 66.8 ± 1.027 | 64.8–68.8 | 73.7 ± 0.990 | 71.7–75.7 |
| GA | 107 | 67.9 ± 1.890 | 64.1–71.6 | 76.0 ± 2.14 | 71.7–80.2 | |
| AA | 7 | 63.4 ± 8.454 | 42.7–84.1 | 73.6 ± 6.37 | 58.0–89.2 | |
| p-value | 0.746 | 0.421 | ||||
| rs677044 | AA | 327 | 66.7 ± 1.063 | 64.6–68.8 | 74.6 ± 1.10 | 72.4–76.7 |
| AG | 155 | 67.4 ± 1.725 | 64.0–70.8 | 73.4 ± 1.64 | 70.2–76.7 | |
| GG | 21 | 67.6 ± 5.187 | 56.8–78.4 | 74.2 ± 4.58 | 64.6–83.7 | |
| p-value | 0.676 | 0.978 | ||||
| rs2853209 | AA | 247 | 66.5 ± 1.19 | 64.2–68.9 | 76.7 ± 1.30 | 74.1–79.3 |
| AT | 195 | 68.0 ± 1.48 | 65.1–70.9 | 72.4 ± 1.36 | 69.7–75.0 | |
| TT | 61 | 65.3 ± 3.02 | 59.3–71.4 | 69.9 ± 2.62 | 64.7–75.1 | |
| p-value | 0.722 | 0.040 | ||||
| rs3918392 | AA | 435 | 66.7 ± 0.975 | 64.7–68.6 | 74.0 ± 0.969 | 72.1–75.9 |
| AG | 64 | 69.2 ± 2.307 | 64.5–73.8 | 75.6 ± 2.37 | 70.8–80.3 | |
| GG | 4 | 64.3 ± 11.842 | 26.6–101.9 | 70.3 ± 11.9 | 32.4–108.1 | |
| p-value | 0.631 | 0.826 | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vishweswaraiah, S.; Ramachandra, N.B.; Joshi, N.; Parthasarathi, A.; Kaleem Ullah, M.; Siddaiah, J.B.; Holla, A.D.; Chakraborty, S.; Agrawal, A.; Mahesh, P.A. Association between ADAM33 Single-Nucleotide Polymorphisms and Treatment Response to Inhaled Corticosteroids and a Long-Acting Beta-Agonist in Asthma. Diagnostics 2023, 13, 405. https://doi.org/10.3390/diagnostics13030405
Vishweswaraiah S, Ramachandra NB, Joshi N, Parthasarathi A, Kaleem Ullah M, Siddaiah JB, Holla AD, Chakraborty S, Agrawal A, Mahesh PA. Association between ADAM33 Single-Nucleotide Polymorphisms and Treatment Response to Inhaled Corticosteroids and a Long-Acting Beta-Agonist in Asthma. Diagnostics. 2023; 13(3):405. https://doi.org/10.3390/diagnostics13030405
Chicago/Turabian StyleVishweswaraiah, Sangeetha, Nallur B. Ramachandra, Neha Joshi, Ashwaghosha Parthasarathi, Mohammed Kaleem Ullah, Jayaraj Biligere Siddaiah, Amrutha D. Holla, Samarpana Chakraborty, Anurag Agrawal, and Padukudru Anand Mahesh. 2023. "Association between ADAM33 Single-Nucleotide Polymorphisms and Treatment Response to Inhaled Corticosteroids and a Long-Acting Beta-Agonist in Asthma" Diagnostics 13, no. 3: 405. https://doi.org/10.3390/diagnostics13030405
APA StyleVishweswaraiah, S., Ramachandra, N. B., Joshi, N., Parthasarathi, A., Kaleem Ullah, M., Siddaiah, J. B., Holla, A. D., Chakraborty, S., Agrawal, A., & Mahesh, P. A. (2023). Association between ADAM33 Single-Nucleotide Polymorphisms and Treatment Response to Inhaled Corticosteroids and a Long-Acting Beta-Agonist in Asthma. Diagnostics, 13(3), 405. https://doi.org/10.3390/diagnostics13030405







