Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review
Abstract
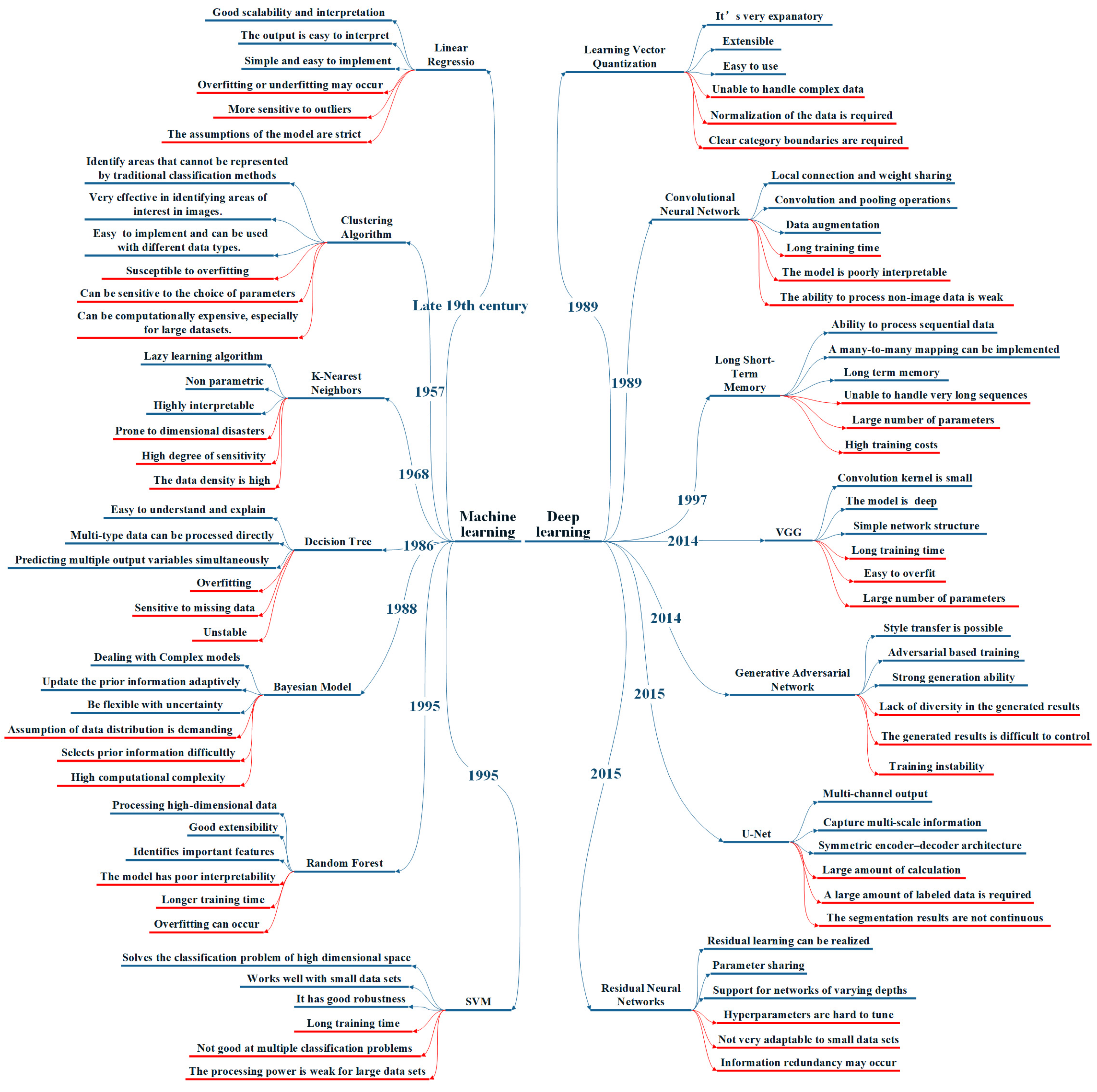
:1. Introduction
2. Materials
2.1. Study Selection
2.2. Datasets
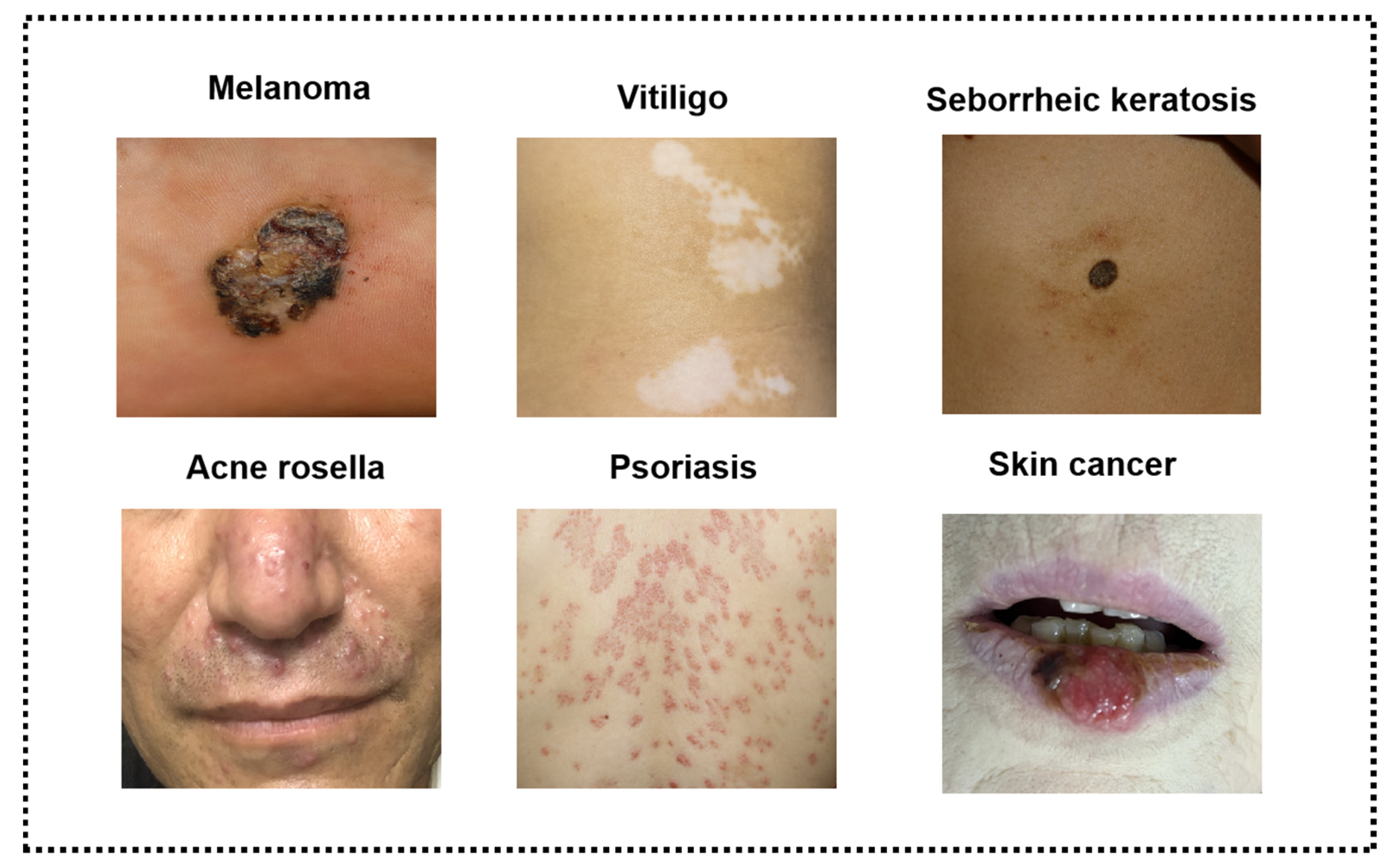
2.3. Selection Criteria of AI Algorithms for Different Types of Skin Images
3. Methods
3.1. Segmentation Methods
3.1.1. Traditional Machine Learning
- (1)
- Enhanced feature learning: DL models possess robust feature learning capabilities, automatically deriving advanced feature representation from data, in contrast to the manually designed features relied upon by ML models.
- (2)
- Improved generalization: DL models exhibit strong generalization capabilities, performing well with small datasets, whereas ML models typically demand extensive manual feature engineering and large datasets.
- (3)
- Scalability: DL models can be efficiently trained on large-scale data and manage such datasets rapidly through distributed computing, whereas ML models struggle due to their need for manual feature design.
- (4)
- Flexibility: DL models offer a more adaptable structure, accommodating a wide range of tasks and data types, unlike ML models, which require manual design for distinct tasks and data types.
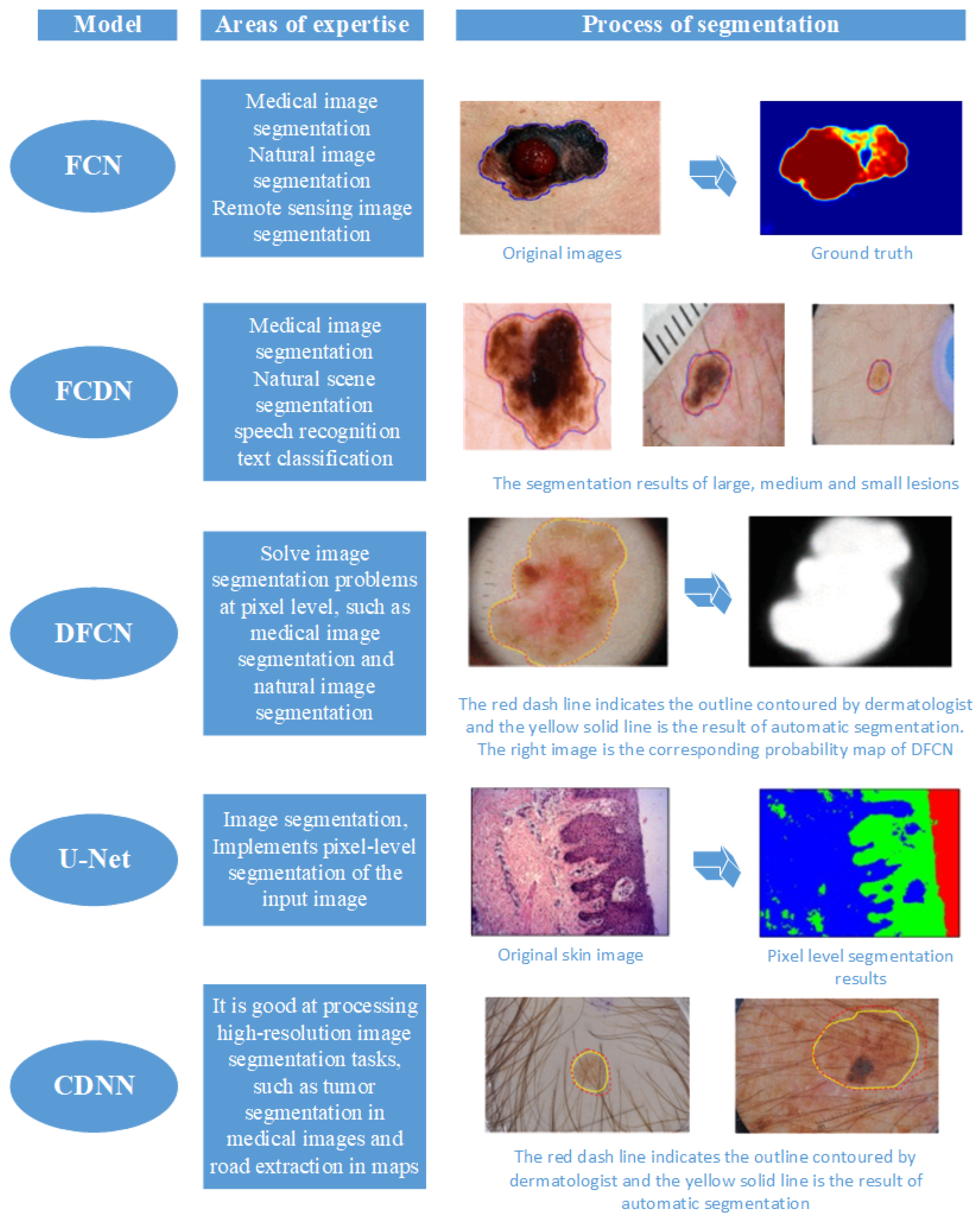
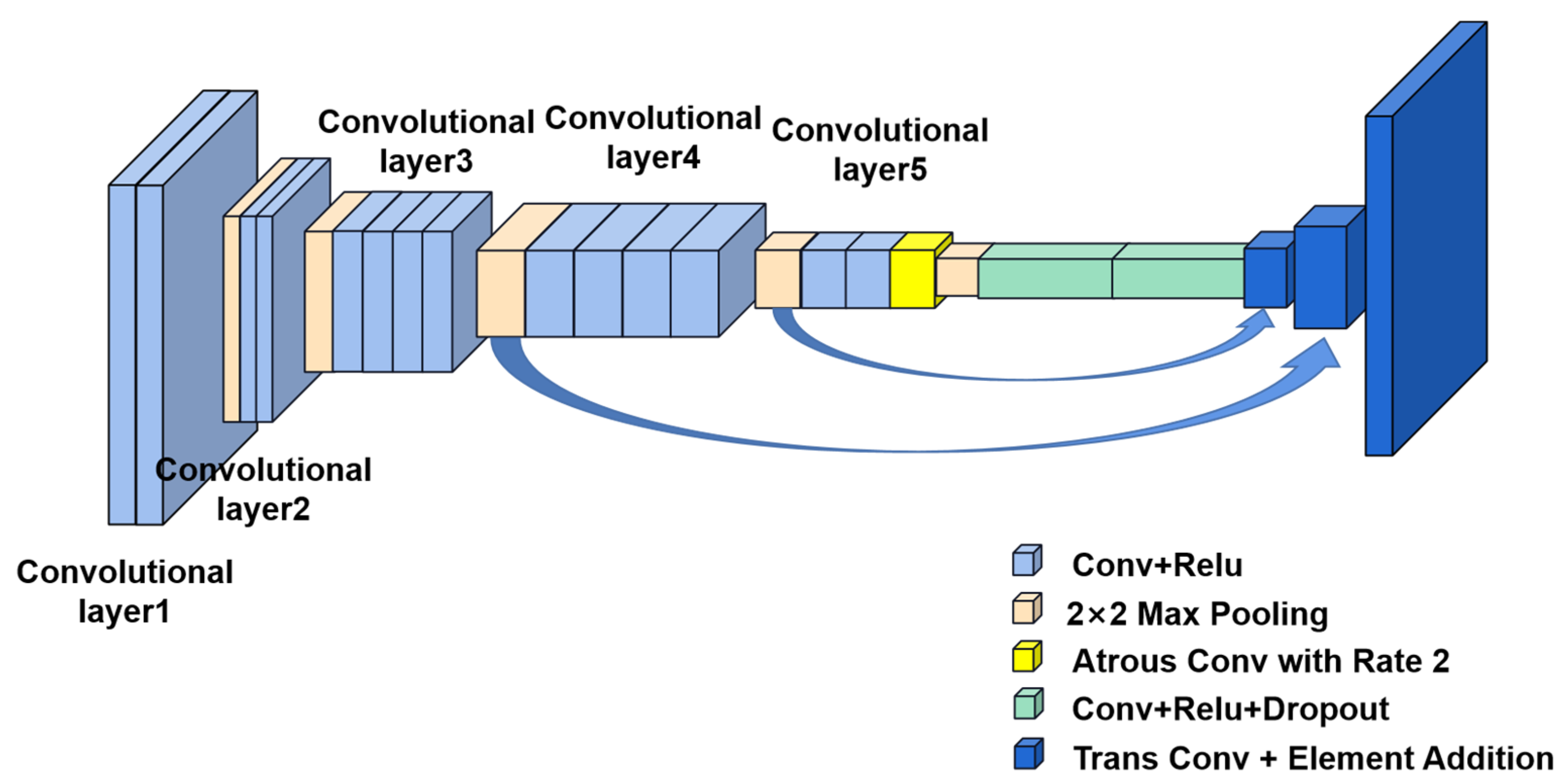
3.1.2. Deep Learning
3.2. Classification Methods
3.2.1. Traditional Machine Learning
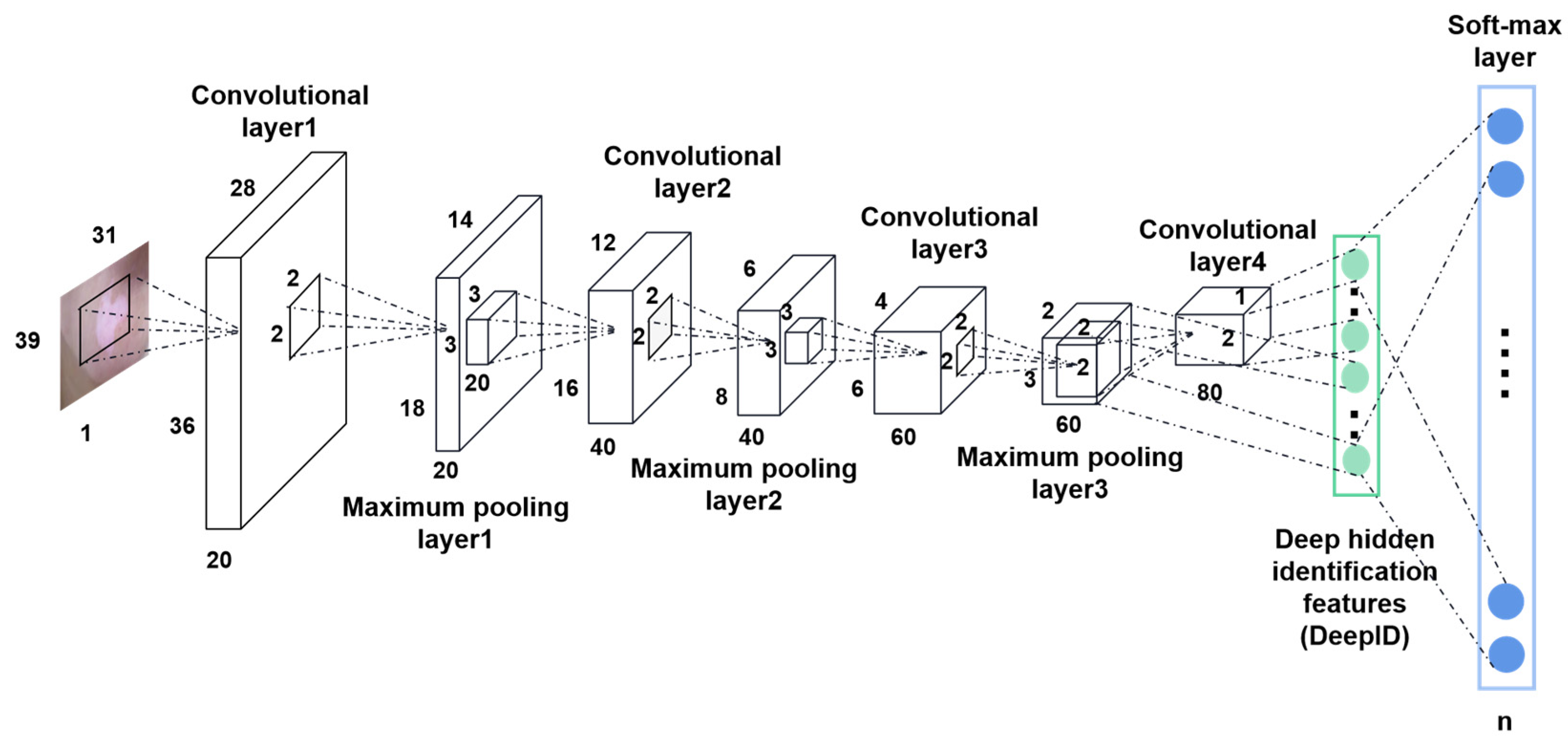
3.2.2. Deep Learning
4. Results
4.1. Indicators of Evaluation
4.2. Analysis of results
5. Discussions
5.1. Current State of Research
5.2. Challenges
5.2.1. Limitations of Datasets
5.2.2. Explainability of Deep Learning Methods
5.2.3. Homogenized Research Directions
5.2.4. More Innovative Algorithms Are Needed
5.3. Future Directions
5.3.1. Establish a Standardized Dermatological Image Dataset
5.3.2. Provide Reasonable Explanations for Predicted Results
5.3.3. Increase the Diversity of the Types of Research
5.3.4. Actively Explore Innovative Models and Methods
- (1)
- Try to use the latest model architecture. The Swin transformer is a new model of computer vision proposed in 2021 with a wide applicability for tasks including image segmentation, image recovery, and image reconstruction [144,145]. Currently, only a limited number of studies have reported the use of this model for medical images [146,147,148]. The Swin transformer, through its hierarchical structure and self-attentive mechanism, has shown powerful feature extraction and modeling capabilities in medical image processing, providing potential opportunities for improvement and innovation in medical image analysis and diagnosis [149]. This suggests that the application of modified models in medicine deserves further research.
- (2)
- Innovate areas of utilization of traditional methods. As mentioned above, there have been many works utilizing transfer learning techniques to improve the performance of DL models in dermatological diagnosis tasks [105,106,107]. At the same time, recent developments regarding transfer learning in other domains can be utilized to facilitate the success of DL in dermatological diagnosis tasks. It is worth mentioning that Reinforcement Learning (RL) also has the potential to be applied to dermatological diagnosis. Recent studies have demonstrated the successful application of RL in different domains [150], partly due to the powerful function approximation capabilities of DL algorithms. RL is very effective in dealing with sequential problems, and many medical decision-making problems fall into this category. Therefore, RL can be used to solve these problems. There have been some works utilizing RL to solve medical image processing tasks with promising results [151]. However, there is no work that applies RL to dermatological diagnostic tasks. Thus, RL may be a potential tool for solving the problems regarding dermatological diagnosis.
- (3)
- Fusion of multimodal data. Integrating multi-source data such as images, text, and biological features can enhance the performance of dermatological diagnostic models. In a clinical setting, accurate dermatological diagnosis is not only dependent on a single skin image, but also requires the consideration of additional clinical information such as medical history, risk factors, and overall skin assessment. Some studies have verified significant improvements in diagnostic performance including these additional data and close-up images [118]. Therefore, this information could be incorporated into the process of model training and testing for skin disease diagnoses. In addition, medical record data can also be processed using techniques such as document analysis [152] and data mining [153], and taken into account in the diagnostic process as well. This multimodal data fusion model has proven its effectiveness in recent studies and can be extended to the field of dermatological diagnosis.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Tschandl, P.; Codella, N.; Akay, B.N.; Argenziano, G.; Braun, R.P.; Cabo, H.; Gutman, D.; Halpern, A.; Helba, B.; Hofmann-Wellenhof, R.; et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: An open, web-based, international, diagnostic study. Lancet Oncol. 2019, 20, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Ezzedine, K.; Hamzavi, I.; Pandya, A.G.; Harris, J.E. New discoveries in the pathogenesis and classification of vitiligo. J. Am. Acad. Dermatol. 2017, 77, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kotnis, A.; Jadeja, S.D.; Mondal, A.; Mansuri, M.S.; Begum, R. Cytokines: The yin and yang of vitiligo pathogenesis. Expert Rev. Clin. Immunol. 2019, 15, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Burlando, M.; Muracchioli, A.; Cozzani, E.; Parodi, A. Psoriasis, Vitiligo, and Biologic Therapy: Case Report and Narrative Review. Case Rep. Dermatol. 2021, 13, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Du-Harpur, X.; Watt, F.M.; Luscombe, N.M.; Lynch, M.D. What is AI? Applications of artificial intelligence to dermatology. Br. J. Dermatol. 2020, 183, 423–430. [Google Scholar] [CrossRef]
- Silverberg, N.B. The epidemiology of vitiligo. Curr. Dermatol. Rep. 2015, 4, 36–43. [Google Scholar] [CrossRef]
- Patel, S.; Wang, J.V.; Motaparthi, K.; Lee, J.B. Artificial intelligence in dermatology for the clinician. Clin. Dermatol. 2021, 39, 667–672. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Int. J. Surg. 2021, 88, 105906. [Google Scholar] [CrossRef] [PubMed]
- Bajwa, M.N.; Muta, K.; Malik, M.I.; Siddiqui, S.A.; Braun, S.E.; Homey, B.; Dengel, A.; Ahmed, S. Computer-aided diagnosis of skin diseases using deep neural networks. Appl. Sci. 2020, 10, 2488. [Google Scholar] [CrossRef]
- Ioannis, G.; Nynke, M.; Sander, L.; Michael, B.; Marcel, F.J.; Nicolai, P. MED-NODE: A computer-assisted melanoma diagnosis system using non-dermoscopic images. Expert Syst. Appl. 2015, 42, 6578–6585. [Google Scholar]
- Filali, I. Contrast Based Lesion Segmentation on DermIS and DermQuest Datasets. Mendeley Data 2019, 2. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci. Data 2018, 5, 180161. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.F.; Gutman, D.; Celebi, M.E.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; et al. Skin lesion analysis toward melanoma detection: A challenge at the 2017 international symposium on biomedical imaging (isbi), hosted by the international skin imaging collaboration (isic). In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; IEEE: New York, NY, USA, 2018; pp. 168–172. [Google Scholar]
- Marc, C.; Noel, C.F.C.; Veronica, R.; Brian, H.; Veronica, V.; Ofer, R.; Cristina, C.; Alicia, B.; Allan, C.H.; Susana, P.; et al. Bcn20000: Dermoscopic lesions in the wild. arXiv 2019, arXiv:1908.02288. [Google Scholar]
- Veronica, R.; Nicholas, K.; Brigid, B.; Liam, C.; Emmanouil, C.; Noel, C.; Marc, C.; Stephen, D.; Pascale, G.; David, G.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Sci. Data 2021, 8, 34. [Google Scholar]
- Iranpoor, R.; Mahboob, A.S.; Shahbandegan, S.; Baniasadi, N. Skin lesion segmentation using convolutional neural networks with improved U-Net architecture. In Proceedings of the 2020 6th Iranian Conference on Signal Processing and Intelligent Systems (ICSPIS), Mashhad, Iran, 23–24 December 2020; IEEE: New York, NY, USA, 2020; pp. 1–5. [Google Scholar]
- Liu, Y.; Jain, A.; Eng, C.; Way, D.H.; Lee, K.; Bui, P.; Kanada, K.; Marinho, G.O.; Gallegos, J.; Gabriele, S.; et al. A deep learning system for differential diagnosis of skin diseases. Nat. Med. 2020, 26, 900–908. [Google Scholar] [CrossRef]
- Tschandl, P.; Rinner, C.; Apalla, Z.; Argenziano, G.; Codella, N.; Halpern, A.; Janda, M.; Lallas, A.; Longo, C.; Malvehy, J.; et al. Human-computer collaboration for skin cancer recognition. Nat. Med. 2020, 26, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef] [PubMed]
- Qaiser, A.; Ramzan, F.; Ghani, M.U. Acral melanoma detection using dermoscopic images and convolutional neural networks. Vis. Comput. Ind. Biomed. Art 2021, 4, 25. [Google Scholar]
- Han, S.S.; Kim, M.S.; Lim, W.; Park, G.H.; Park, I.; Chang, S.E.; Lee, W. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J. Investig. Dermatol. 2018, 138, 1529–1538. [Google Scholar] [CrossRef]
- Brinker, T.J.; Hekler, A.; Enk, A.H.; Klode, J.; Hauschild, A.; Berking, C.; Schilling, B.; Haferkamp, S.; Schadendorf, D.; Holland-Letz, T.; et al. Deep learning outperformed 136 of 157 dermatologists in a head-to-head dermoscopic melanoma image classification task. Eur. J. Cancer 2019, 113, 47–54. [Google Scholar] [CrossRef]
- Shetty, B.; Fernandes, R.; Rodrigues, A.P.; Chengoden, R.; Bhattacharya, S.; Lakshmanna, K. Skin lesion classification of dermoscopic images using machine learning and convolutional neural network. Sci. Rep. 2022, 12, 18134. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Han, H.; Zhou, X.; Liu, L. A novel hybrid deep learning model for skin lesion classification with interpretable feature extraction. Med. Image Anal. 2021, 67, 101831. [Google Scholar]
- Kivanc, K.; Christi, A.; Melissa, G.; Jennifer, G.D.; Dana, H.B.; Milind, R. A machine learning method for identifying morphological patterns in reflectance confocal microscopy mosaics of melanocytic skin lesions in-vivo. In Photonic Therapeutics and Diagnostics XII; SPIE: Bellingham, WA, USA, 2016; Volume 9689. [Google Scholar]
- Alican, B.; Kivanc, K.; Christi, A.; Melissa, G.; Jennifer, G.D.; Dana, H.B.; Milind, R. A multiresolution convolutional neural network with partial label training for annotating reflectance confocal microscopy images of skin. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Proceedings, Part II 11. Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Zorgui, S.; Chaabene, S.; Bouaziz, B.; Batatia, H.; Chaari, L. A convolutional neural network for lentigo diagnosis. In Proceedings of the Impact of Digital Technologies on Public Health in Developed and Developing Countries: 18th International Conference, ICOST 2020, Hammamet, Tunisia,, 24–26 June 2020; Proceedings 18. Springer International Publishing: Cham, Switzerland, 2020; pp. 89–99. [Google Scholar]
- Halimi, A.; Batatia, H.; Le Digabel, J.; Josse, G.; Tourneret, J.Y. An unsupervised Bayesian approach for the joint reconstruction and classification of cutaneous reflectance confocal microscopy images. In Proceedings of the 2017 25th European Signal Processing Conference (EUSIPCO), Kos, Greece, 28 August–2 September 2017; IEEE: New York, NY, USA, 2017; pp. 241–245. [Google Scholar]
- Ribani, R.; Marengoni, M. A survey of transfer learning for convolutional neural networks. In Proceedings of the 2019 32nd SIBGRAPI Conference on Graphics, Patterns and Images Tutorials (SIBGRAPI-T), Rio de Janeiro, Brazil, 28–31 October 2019; IEEE: New York, NY, USA, 2019; pp. 47–57. [Google Scholar]
- Czajkowska, J.; Badura, P.; Korzekwa, S.; Płatkowska-Szczerek, A.; Słowińska, M. Deep learning-based high-frequency ultrasound skin image classification with multicriteria model evaluation. Sensors 2021, 21, 5846. [Google Scholar] [CrossRef] [PubMed]
- Panayides, A.S.; Amini, A.; Filipovic, N.D.; Sharma, A.; Tsaftaris, S.A.; Young, A.; Foran, D.; Do, N.; Golemati, S.; Kurc, T.; et al. AI in medical imaging informatics: Current challenges and future directions. IEEE J. Biomed. Health Inform. 2020, 24, 1837–1857. [Google Scholar] [CrossRef]
- Abdullah, M.N.; Sahari, M.A. Digital image clustering and colour model selection in content-based image retrieval (CBIR) approach for biometric security image. In AIP Conference Proceedings; AIP Publishing: College Park, MD, USA, 2022; Volume 2617. [Google Scholar]
- Silva, J.; Varela, N.; Patiño-Saucedo, J.A.; Lezama, O.B.P. Convolutional neural network with multi-column characteristics extraction for image classification. In Image Processing and Capsule Networks: ICIPCN 2020; Springer International Publishing: Cham, Switzerland, 2021; pp. 20–30. [Google Scholar]
- Wang, P.; Fan, E.; Wang, P. Comparative analysis of image classification algorithms based on traditional machine learning and deep learning. Pattern Recognit. Lett. 2021, 141, 61–67. [Google Scholar] [CrossRef]
- Sreedhar, B.; Be, M.S.; Kumar, M.S. A comparative study of melanoma skin cancer detection in traditional and current image processing techniques. In Proceedings of the 2020 Fourth International Conference on I-SMAC (IoT in Social, Mobile, Analytics and Cloud) (I-SMAC), Palladam, India, 7–9 October 2020; IEEE: New York, NY, USA, 2020; pp. 654–658. [Google Scholar]
- Prinyakupt, J.; Pluempitiwiriyawej, C. Segmentation of white blood cells and comparison of cell morphology by linear and naïve Bayes classifiers. Biomed. Eng. Online 2015, 14, 63. [Google Scholar] [CrossRef]
- Tan, T.Y.; Zhang, L.; Lim, C.P. Adaptive melanoma diagnosis using evolving clustering, ensemble and deep neural networks. Knowl. -Based Syst. 2020, 187, 104807. [Google Scholar] [CrossRef]
- Yoganathan, S.A.; Zhang, R. Segmentation of organs and tumor within brain magnetic resonance images using K-nearest neighbor classification. J. Med. Phys. 2022, 47, 40. [Google Scholar]
- Thamilselvan, P.; Sathiaseelan, J.G.R. Detection and classification of lung cancer MRI images by using enhanced k nearest neighbor algorithm. Indian J. Sci. Technol. 2016, 9, 1–7. [Google Scholar] [CrossRef]
- Song, Y.Y.; Ying, L.U. Decision tree methods: Applications for classification and prediction. Shanghai Arch. Psychiatry 2015, 27, 130. [Google Scholar]
- Gladence, L.M.; Karthi, M.; Anu, V.M. A statistical comparison of logistic regression and different Bayes classification methods for machine learning. ARPN J. Eng. Appl. Sci. 2015, 10, 5947–5953. [Google Scholar]
- Jaiswal, J.K.; Samikannu, R. Application of random forest algorithm on feature subset selection and classification and regression. In Proceedings of the 2017 World Congress on Computing and Communication Technologies (WCCCT), Tiruchirappalli, India, 2–4 February 2017; IEEE: New York, NY, USA, 2017; pp. 65–68. [Google Scholar]
- Seeja, R.D.; Suresh, A. Deep learning based skin lesion segmentation and classification of melanoma using support vector machine (SVM). Asian Pac. J. Cancer Prev. APJCP 2019, 20, 1555. [Google Scholar]
- Sheikh Abdullah, S.N.H.; Bohani, F.A.; Nayef, B.H.; Sahran, S.; Akash, O.A.; Hussain, R.I.; Ismail, F. Round randomized learning vector quantization for brain tumor imaging. Comput. Math. Methods Med. 2016, 2016, 8603609. [Google Scholar] [CrossRef]
- Ji, L.; Mao, R.; Wu, J.; Ge, C.; Xiao, F.; Xu, X.; Xie, L.; Gu, X. Deep Convolutional Neural Network for Nasopharyngeal Carcinoma Discrimination on MRI by Comparison of Hierarchical and Simple Layered Convolutional Neural Networks. Diagnostics 2022, 12, 2478. [Google Scholar] [CrossRef]
- Lv, Q.J.; Chen, H.Y.; Zhong, W.B.; Wang, Y.Y.; Song, J.Y.; Guo, S.D.; Qi, L.X.; Chen, Y.C. A multi-task group Bi-LSTM networks application on electrocardiogram classification. IEEE J. Transl. Eng. Health Med. 2019, 8, 1900111. [Google Scholar] [CrossRef]
- Sengupta, A.; Ye, Y.; Wang, R.; Liu, C.; Roy, K. Going deeper in spiking neural networks: VGG and residual architectures. Front. Neurosci. 2019, 13, 95. [Google Scholar] [CrossRef]
- Fan, C.; Lin, H.; Qiu, Y. U-Patch GAN: A Medical Image Fusion Method Based on GAN. J. Digit. Imaging 2022, 36, 339–355. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.X.; Sun, L.; Fu, Y.; Lu, R.; Zhang, Y. U-Net-Based medical image segmentation. J. Healthc. Eng. 2022, 2022, 4189781. [Google Scholar] [CrossRef] [PubMed]
- Popescu, D.; El-Khatib, M.; El-Khatib, H.; Ichim, L. New trends in melanoma detection using neural networks: A systematic review. Sensors 2022, 22, 496. [Google Scholar] [CrossRef]
- Li, Z.; Chen, J. Superpixel segmentation using linear spectral clustering. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1356–1363. [Google Scholar]
- Alam, M.N.; Munia, T.T.K.; Tavakolian, K.; Vasefi, F.; MacKinnon, N.; Fazel-Rezai, R. Automatic detection and severity measurement of eczema using image processing. In Proceedings of the 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Orlando, FL, USA, 17–20 August 2016; IEEE: New York, NY, USA, 2016; pp. 1365–1368. [Google Scholar]
- Thanh, D.N.H.; Erkan, U.; Prasath, V.B.S.; Kuma, V.; Hien, N.N. A skin lesion segmentation method for dermoscopic images based on adaptive thresholding with normalization of color models. In Proceedings of the 2019 6th International Conference on Electrical and Electronics Engineering (ICEEE), Istanbul, Turkey, 16–17 April 2019; IEEE: New York, NY, USA, 2019; pp. 116–120. [Google Scholar]
- Chica, J.F.; Zaputt, S.; Encalada, J.; Salamea, C.; Montalvo, M. Objective assessment of skin repigmentation using a multilayer perceptron. J. Med. Signals Sens. 2019, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Nurhudatiana, A. A computer-aided diagnosis system for vitiligo assessment: A segmentation algorithm. In Proceedings of the International Conference on Soft Computing, Intelligent Systems, and Information Technology, Bali, Indonesia, 11–14 March 2015; Springer: Berlin, Heidelberg, 2015; pp. 323–331. [Google Scholar]
- Dash, M.; Londhe, N.D.; Ghosh, S.; Shrivastava, V.K.; Sonawane, R.S. Swarm intelligence based clustering technique for automated lesion detection and diagnosis of psoriasis. Comput. Biol. Chem. 2020, 86, 107247. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer—Assisted Intervention, Singapore, 8–12 October 2022; pp. 234–241. [Google Scholar]
- Deng, Z.; Fan, H.; Xie, F.; Cui, Y.; Liu, J. Segmentation of dermoscopy images based on fully convolutional neural network. In Proceedings of the 2017 IEEE International Conference on Image Processing (ICIP), Beijing, China, 17–20 September 2017; IEEE: New York, NY, USA, 2017. [Google Scholar]
- Luo, W.; Meng, Y. Fast skin lesion segmentation via fully convolutional network with residual architecture and CRF. In Proceedings of the 2018 24th International Conference on Pattern Recognition (ICPR), Beijing, China, 20–24 August 2018; IEEE: New York, NY, USA, 2018. [Google Scholar]
- Yuan, Y. Automatic skin lesion segmentation with fully convolutional-deconvolutional networks. arXiv 2017, arXiv:1703.05165. [Google Scholar]
- Yuan, Y.; Chao, M.; Lo, Y.C. Automatic skin lesion segmentation using deep fully convolutional networks with jaccard distance. IEEE Trans. Med. Imaging 2017, 36, 1876–1886. [Google Scholar] [CrossRef]
- Yuan, Y.; Lo, Y.C. Improving dermoscopic image segmentation with enhanced convolutional-deconvolutional networks. IEEE J. Biomed. Health Inform. 2017, 23, 519–526. [Google Scholar] [CrossRef]
- Saood, A.; Hatem, I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med. Imaging 2021, 21, 1–10. [Google Scholar] [CrossRef]
- Pal, A.; Garain, U.; Chandra, A.; Chatterjee, R.; Senapati, S. Psoriasis skin biopsy image segmentation using Deep Convolutional Neural Network. Comput. Methods Programs Biomed. 2018, 159, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Sinz, C.; Kittler, H. Domain-specific classification-pretrained fully convolutional network encoders for skin lesion segmentation. Comput. Biol. Med. 2019, 104, 111–116. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, N.; Wang, Y.; Wang, M. Segmentation of dermoscopy image using adversarial networks. Multimed. Tools Appl. 2019, 78, 10965–10981. [Google Scholar] [CrossRef]
- Adegun, A.; Viriri, S. Deep learning model for skin lesion segmentation: Fully convolutional network. In Proceedings of the International Conference on Image Analysis and Recognition, Taipei, Taiwan, 22–25 September 2019; Springer: Cham, Switzerland, 2019; pp. 232–242. [Google Scholar]
- Thanh, D.N.H.; Thanh, L.T.; Erkan, U.; Khamparia, A.; Prasath, V.B.S. Dermoscopic image segmentation method based on convolutional neural networks. Int. J. Comput. Appl. Technol. 2021, 66, 89–99. [Google Scholar] [CrossRef]
- Zeng, G.; Zheng, G. Multi-scale fully convolutional DenseNets for automated skin lesion segmentation in dermoscopy images. In Proceedings of the International Conference Image Analysis and Recognition, Póvoa de Varzim, Portugal, 27–29 June 2018; Springer: Cham, Switzerland, 2018; pp. 513–521. [Google Scholar]
- Nasr-Esfahani, E.; Rafiei, S.; Jafari, M.H.; Karimi, N.; Wrobel, J.S.; Samavi, S.; Soroushmehr, S.M.R. Dense pooling layers in fully convolutional network for skin lesion segmentation. Comput. Med. Imaging Graph. 2019, 78, 101658. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Shuai, R.; Ma, L.; Liu, W.; Wu, M. Segmentation of dermoscopy images based on deformable 3D convolution and ResU-NeXt++. Med. Biol. Eng. Comput. 2021, 59, 1815–1832. [Google Scholar] [CrossRef] [PubMed]
- Stofa, M.M.; Zulkifley, M.A.; Zainuri, M.A.A.M.; Ibrahim, A.A. U-Net with Atrous Spatial Pyramid Pooling for Skin Lesion Segmentation. In Proceedings of the 6th International Conference on Electrical, Control and Computer Engineering: InECCE2021, Kuantan, Malaysia, 23 August 2022; Springer: Singapore, 2022; pp. 1025–1033. [Google Scholar]
- Yanagisawa, Y.; Shido, K.; Kojima, K.; Yamasaki, K. Convolutional neural network-based skin image segmentation model to improve classification of skin diseases in conventional and non-standardized picture images. J. Dermatol. Sci. 2023, 109, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-López, C.; Ramírez-Cornejo, C.; Marchetti, M.A.; Han, S.S.; Del Barrio-Díaz, P.; Jaque, A.; Uribe, P.; Majerson, D.; Curi, M.; Del Puerto, C.; et al. Performance of a deep neural network in teledermatology: A single-centre prospective diagnostic study. J. Eur. Acad. Dermatol. Venereol. 2021, 35, 546–553. [Google Scholar] [CrossRef]
- Reddy, D.A.; Roy, S.; Kumar, S.; Tripathi, R. Enhanced U-Net segmentation with ensemble convolutional neural network for automated skin disease classification. Knowl. Inf. Syst 2023, 65, 4111–4156. [Google Scholar] [CrossRef]
- Bian, Z.; Xia, S.; Xia, C.; Shao, M. Weakly supervised Vitiligo segmentation in skin image through saliency propagation. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; IEEE: New York, NY, USA, 2019; pp. 931–934. [Google Scholar]
- Low, M.; Huang, V.; Raina, P. Automating Vitiligo skin lesion segmentation using convolutional neural networks. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; IEEE: New York, NY, USA, 2020; pp. 1–4. [Google Scholar]
- Khatibi, T.; Rezaei, N.; Ataei Fashtami, L.; Totonchi, M. Proposing a novel unsupervised stack ensemble of deep and conventional image segmentation (SEDCIS) method for localizing vitiligo lesions in skin images. for localizing vitiligo lesions in skin images. Ski. Res. Technol. 2021, 27, 126–137. [Google Scholar] [CrossRef]
- Yanling, L.I.; Kong, A.W.K.; Thng, S. Segmenting Vitiligo on Clinical Face Images Using CNN Trained on Synthetic and Internet Images. IEEE J. Biomed. Health Inform. 2021, 25, 3082–3093. [Google Scholar]
- Kassem, M.A.; Hosny, K.M.; Damaševičius, R.; Eltoukhy, M.M. Machine learning and deep learning methods for skin lesion classification and diagnosis: A systematic review. Diagnostics 2021, 11, 1390. [Google Scholar] [CrossRef]
- Banditsingha, P.; Thaipisutikul, T.; Shih, T.K.; Lin, C.Y. A Decision Machine Learning Support System for Human Skin Disease Classifier. In Proceedings of the 2022 Joint International Conference on Digital Arts, Media and Technology with ECTI Northern Section Conference on Electrical, Electronics, Computer and Telecommunications Engineering (ECTI DAMT & NCON), Phuket, Thailand, 26–28 January 2022; IEEE: New York, NY, USA, 2022. [Google Scholar]
- Wang, W.; Sun, G. Classification and research of skin lesions based on machine learning. Comput. Mater. Contin. 2020, 62, 1187–1200. [Google Scholar]
- Farzad, S.; Rouhi, A.; Rastegari, R. The Performance of Deep and Conventional Machine Learning Techniques for Skin Lesion Classification. In Proceedings of the 2021 IEEE 18th International Conference on Smart Communities: Improving Quality of Life Using ICT, IoT and AI (HONET), Karachi, Pakistan, 11–13 October 2021; IEEE: New York, NY, USA, 2021. [Google Scholar]
- Parameshwar, H.; Manjunath, R.; Shenoy, M.; Shekar, B.H. Comparison of machine learning algorithms for skin disease classification using color and texture features. In Proceedings of the 2018 International Conference on Advances in Computing, Communications and Informatics (ICACCI), Bangalore, India, 19–22 September 2018; IEEE: New York, NY, USA, 2018. [Google Scholar]
- Nosseir, A.; Shawky, M.A. Automatic classifier for skin disease using k-NN and SVM. In Proceedings of the 2019 8th International Conference on Software and Information Engineering, Cairo, Egypt, 9–12 April 2019; pp. 259–262. [Google Scholar]
- Pennisi, A.; Bloisi, D.D.; Nardi, D.; Giampetruzzi, A.R.; Mondino, C.; Facchiano, A. Melanoma detection using delaunay triangulation. In Proceedings of the 2015 IEEE 27th International Conference on Tools with Artificial Intelligence (ICTAI), Vietri sul Mare, Italy, 9–11 November 2015; IEEE: New York, NY, USA, 2015; pp. 791–798. [Google Scholar]
- Sumithra, R.; Suhil, M.; Guru, D.S. Segmentation and classification of skin lesions for disease diagnosis. Procedia Comput. Sci. 2015, 45, 76–85. [Google Scholar] [CrossRef]
- Suganya, R. An automated computer aided diagnosis of skin lesions detection and classification for dermoscopy images. In Proceedings of the 2016 International Conference on Recent Trends in Information Technology (ICRTIT), Chennai, India, 8–9 April 2016; IEEE: New York, NY, USA, 2016; pp. 1–5. [Google Scholar]
- Rahman, M.A.; Haque, M.T.; Shahnaz, C.; Fattah, S.A.; Zhu, W.P.; Ahmed, M.O. Skin lesions classification based on color plane-histogram-image quality analysis features extracted from digital images. In Proceedings of the 2017 IEEE 60th International Midwest Symposium on Circuits and Systems (MWSCAS), Boston, MA, USA, 6–9 August 2017; IEEE: New York, NY, USA, 2017; pp. 1356–1359. [Google Scholar]
- Hameed, N.; Shabut, A.; Hossain, M.A. A Computer-aided diagnosis system for classifying prominent skin lesions using machine learning. In Proceedings of the 2018 10th Computer Science and Electronic Engineering (CEEC), Colchester, UK, 19–21 September 2018; IEEE: New York, NY, USA, 2018; pp. 186–191. [Google Scholar]
- Murugan, A.; Nair, S.A.H.; Sanal Kumar, K.P. Detection of skin cancer using SVM, random forest and kNN classifiers. J. Med. Syst. 2019, 43, 1–9. [Google Scholar] [CrossRef]
- Balaji, V.R.; Suganthi, S.T.; Rajadevi, R.; Kumar, V.K.; Balaji, B.S.; Pandiyan, S. Skin disease detection and segmentation using dynamic graph cut algorithm and classification through Naive Bayes Classifier. Measurement 2020, 163, 107922. [Google Scholar] [CrossRef]
- Nasiri, S.; Helsper, J.; Jung, M.; Fathi, M. DePicT Melanoma Deep-CLASS: A deep convolutional neural networks approach to classify skin lesion images. BMC Bioinform. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, B.; Usama, M.; Ahmad, T.; Khatoon, S.; Alam, C.M. An ensemble model of convolution and recurrent neural network for skin disease classification. Int. J. Imaging Syst. Technol. 2022, 32, 218–229. [Google Scholar] [CrossRef]
- Karthik, R.; Vaichole, T.S.; Kulkarni, S.K.; Yadav, O.; Khan, F. Eff2Net: An efficient channel attention-based convolutional neural network for skin disease classification. Biomed. Signal Process. Control. 2022, 73, 103406. [Google Scholar] [CrossRef]
- Mohammed, K.K.; Afify, H.M.; Hassanien, A.E. Artificial intelligent system for skin diseases classification. Biomed. Eng. Appl. Basis Commun. 2020, 32, 2050036. [Google Scholar] [CrossRef]
- Zia Ur Rehman, M.; Ahmed, F.; Alsuhibany, S.A.; Jamal, S.S.; Zulfiqar Ali, M.; Ahmad, J. Classification of skin cancer lesions using explainable deep learning. Sensors 2022, 22, 6915. [Google Scholar] [CrossRef]
- Ahmad, B.; Usama, M.; Huang, C.M.; Hwang, K.; Hossain, M.S.; Muhammad, G. Discriminative feature learning for skin disease classification using deep convolutional neural network. IEEE Access 2020, 8, 39025–39033. [Google Scholar] [CrossRef]
- Wu, Z.H.E.; Zhao, S.; Peng, Y.; He, X.; Zhao, X.; Huang, K.; Wu, X.; Fan, W.; Li, F.; Chen, M.; et al. Studies on different CNN algorithms for face skin disease classification based on clinical images. IEEE Access 2019, 7, 66505–66511. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Skin melanoma classification using ROI and data augmentation with deep convolutional neural networks. Multimed. Tools Appl. 2020, 79, 24029–24055. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A.; Foaud, M.M. Classification of skin lesions into seven classes using transfer learning with AlexNet. J. Digit. Imaging 2020, 33, 1325–1334. [Google Scholar] [CrossRef]
- Kassem, M.A.; Hosny, K.M.; Fouad, M.M. Skin lesions classification into eight classes for ISIC 2019 using deep convolutional neural network and transfer learning. IEEE Access 2020, 8, 114822–114832. [Google Scholar] [CrossRef]
- Hosny, K.M.; Kassem, M.A. Refined residual deep convolutional network for skin lesion classification. J. Digit. Imaging 2022, 35, 258–280. [Google Scholar] [CrossRef] [PubMed]
- Reis, H.C.; Turk, V.; Khoshelham, K.; Kaya, S. InSiNet: A deep convolutional approach to skin cancer detection and segmentation. Med. Biol. Eng. Comput. 2022, 60, 643–662. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.G.C.; Renato, A.K. The impact of patient clinical information on automated skin cancer detection. Comput. Biol. Med. 2020, 116, 103545. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Mou, L.; Zhu, X.X.; Manda, M. Automatic skin lesion classification based on mid-level feature learning. Comput. Med. Imaging Graph. 2020, 84, 101765. [Google Scholar] [CrossRef] [PubMed]
- Al-Masni, M.A.; Kim, D.-H.; Kim, T.-S. Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef] [PubMed]
- Jasil, S.P.G.; Ulagamuthalvi, V. A hybrid CNN architecture for skin lesion classification using deep learning. Soft Comput. 2023, 1–10. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Koundal, D.; Singh, K. Fusion of U-Net and CNN model for segmentation and classification of skin lesion from dermoscopy images. Expert Syst. Appl. 2023, 213, 119230. [Google Scholar] [CrossRef]
- Raghavendra, P.V.S.P.; Charitha, C.; Begum, K.G.; Prasath, V.B.S. Deep Learning–Based Skin Lesion Multi-class Classification with Global Average Pooling Improvement. J. Digit. Imaging 2023, 36, 2227–2248. [Google Scholar] [CrossRef]
- Cai, G.; Zhu, Y.; Wu, Y.; Jiang, X.; Ye, J.; Yang, D. A multimodal transformer to fuse images and metadata for skin disease classification. Vis. Comput. 2023, 39, 2781–2793. [Google Scholar] [CrossRef]
- Liu, J.; Yan, J.; Chen, J.; Sun, G.; Luo, W. Classification of vitiligo based on convolutional neural network. In Proceedings of the International Conference on Artificial Intelligence and Security: 5th International Conference, New York, NY, USA, 26–28 July 2019; Springer: Cham, Switzerland, 2019; pp. 214–223. [Google Scholar]
- Luo, W.; Liu, J.; Huang, Y.; Zhao, N. An effective vitiligo intelligent classification system. J. Ambient. Intell. Humaniz. Comput. 2023, 14, 5479–5488. [Google Scholar] [CrossRef]
- Guo, L.; Yang, Y.; Ding, H.; Zheng, H.; Yan, H.; Xie, J.; Li, Y.; Lin, T.; Ge, Y. A deep learning-based hybrid artificial intelligence model for the detection and severity assessment of vitiligo lesions. Ann. Transl. Med. 2022, 10, 590. [Google Scholar] [CrossRef] [PubMed]
- Anthal, J.; Upadhyay, A.; Gupta, A. Detection of vitiligo skin disease using LVQ neural network. In Proceedings of the 2017 International Conference on Current Trends in Computer, Electrical, Electronics and Communication (CTCEEC), Coimbatore, India, 8–9 September 2017; IEEE: New York, NY, USA, 2017; pp. 922–925. [Google Scholar]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Kalloo, A.; Hassen, A.B.; Thomas, L.; Enk, A.; et al. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef]
- Maron, R.C.; Weichenthal, M.; Utikal, J.S.; Hekler, A.; Berking, C.; Hauschild, A.; Enk, A.H.; Haferkamp, S.; Klode, J.; Schadendor, D.; et al. Systematic outperformance of 112 dermatologists in multiclass skin cancer image classification by convolutional neural networks. Eur. J. Cancer 2019, 119, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Hesamian, M.H.; Jia, W.; He, X.; Kennedy, P. Deep learning techniques for medical image segmentation: Achievements and challenges. J. Digit. Imaging 2019, 32, 582–596. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Masaya, T.; Atsushi, S.; Kosuke, S.; Yasuhiro, F.; Kenshi, Y.; Manabu, F.; Kohei, M.; Youichirou, N.; Shin’ichi, S.; Akinobu, S. Classification of large-scale image database of various skin diseases using deep learning. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 1875–1887. [Google Scholar]
- Bozorgtabar, B.; Sedai, S.; Roy, P.K.; Garnavi, R. Skin lesion segmentation using deep convolution networks guided by local unsupervised learning. IBM J. Res. Dev. 2017, 61, 6:1–6:8. [Google Scholar] [CrossRef]
- Thanh, D.N.H.; Hien, N.N.; Surya Prasath, V.B.; Thanh, L.T.; Hai, N.H. Automatic initial boundary generation methods based on edge detectors for the level set function of the Chan-Vese segmentation model and applications in biomedical image processing. In Frontiers in Intelligent Computing: Theory and Applications; Springer: Singapore, 2020; pp. 171–181. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Codella, N.C.F.; Nguyen, Q.B.; Pankanti, S.; Gutman, D.A.; Helba, B.; Halpern, A.C.; Smith, J.R. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J. Res. Dev. 2017, 61, 5:1–5:15. [Google Scholar] [CrossRef]
- Samia, B.; Meftah, B.; Lézoray, O. Multi-features extraction based on deep learning for skin lesion classification. Tissue Cell 2022, 74, 101701. [Google Scholar]
- Lopez, A.R.; Giro-i-Nieto, X.; Burdick, J.; Marques, O. Skin lesion classification from dermoscopic images using deep learning techniques. In Proceedings of the 2017 13th IASTED International Conference on Biomedical Engineering (BioMed), Innsbruck, Austria, 20–21 February 2017; IEEE: New York, NY, USA, 2017; pp. 49–54. [Google Scholar]
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P.C. Techniques and algorithms for computer aided diagnosis of pigmented skin lesions—A review. Biomed. Signal Process. Control 2018, 39, 237–262. [Google Scholar] [CrossRef]
- Codella, N.; Cai, J.; Abedini, M.; Garnavi, R.; Halpern, A.; Smith, J.R. Deep learning, sparse coding, and SVM for melanoma recognition in dermoscopy images. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Munich, Germany, 5 October 2015; Springer International Publishing: Cham, Switzerland, 2015; pp. 118–126. [Google Scholar]
- Celebi, M.E.; Wen, Q.; Iyatomi, H.; Shimizu, K.; Zhou, H.; Schaefer, G. A state-of-the-art survey on lesion border detection in dermoscopy images. Dermoscopy Image Anal. 2015, 10, 97–129. [Google Scholar]
- Salahuddin, Z.; Woodruff, H.C.; Chatterjee, A.; Lambin, P. Transparency of deep neural networks for medical image analysis: A review of interpretability methods. Comput. Biol. Med. 2022, 140, 105111. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Farooq, M.S.; Khelifi, A.; Abid, A. Malignant melanoma classification using deep learning: Datasets, performance measurements, challenges and opportunities. IEEE Access 2020, 8, 110575–110597. [Google Scholar] [CrossRef]
- Yao, K.; Su, Z.; Huang, K.; Yang, X.; Sun, J.; Hussain, A.; Coenen, F. A novel 3D unsupervised domain adaptation framework for cross-modality medical image segmentation. IEEE J. Biomed. Health Inform. 2022, 26, 4976–4986. [Google Scholar] [CrossRef]
- Silberg, J.; Manyika, J. Notes from the AI frontier: Tackling bias in AI (and in humans). McKinsey Glob. Inst. 2019, 1. [Google Scholar]
- Castelvecchi, D. Can we open the black box of AI? Nat. News 2016, 538, 20. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.B.; Papa, J.P.; Pereira, A.S.; Tavares, J.M.R.S. Computational methods for pigmented skin lesion classification in images: Review and future trends. Neural Comput. Appl. 2018, 29, 613–636. [Google Scholar] [CrossRef]
- Muhaba, K.A.; Dese, K.; Aga, T.M.; Zewdu, F.T.; Simegn, G.L. Automatic skin disease diagnosis using deep learning from clinical image and patient information. Ski. Health Dis. 2022, 2, e81. [Google Scholar] [CrossRef]
- Serte, S.; Serener, A.; Al-Turjman, F. Deep learning in medical imaging: A brief review. Trans. Emerg. Telecommun. Technol. 2022, 33, e4080. [Google Scholar] [CrossRef]
- Li, H.; Pan, Y.; Zhao, J.; Zhang, L. Skin disease diagnosis with deep learning: A review. Neurocomputing 2021, 464, 364–393. [Google Scholar] [CrossRef]
- Marcus, G.; Davis, E. Rebooting AI: Building Artificial Intelligence We Can Trust; Vintage: New York, NY, USA, 2019. [Google Scholar]
- Goyal, M.; Knackstedt, T.; Yan, S.; Hassanpour, S. Artificial intelligence-based image classification methods for diagnosis of skin cancer: Challenges and opportunities. Comput. Biol. Med. 2020, 127, 104065. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, X.; Luo, Y.; Zhang, H.; Yang, H.; Ma, J.; Ma, L. Opportunities and challenges: Classification of skin disease based on deep learning. Chin. J. Mech. Eng. 2021, 34, 112. [Google Scholar] [CrossRef]
- Liang, J.; Cao, J.; Sun, G.; Zhang, K.; Van Gool, L.; Timofte, R. Swinir: Image restoration using swin transformer. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, QC, Canada, 10–17 October 2021; pp. 1833–1844. [Google Scholar]
- Cao, H.; Wang, Y.; Chen, J.; Jiang, D.; Zhang, X.; Tian, Q.; Wang, M. Swin-unet: Unet-like pure transformer for medical image segmentation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer Nature: Cham, Switzerland, 2022; pp. 205–218. [Google Scholar]
- Peng, L.; Wang, C.; Tian, G.; Liu, G.; Li, G.; Lu, Y.; Yang, J.; Chen, M.; Li, Z. Analysis of CT scan images for COVID-19 pneumonia based on a deep ensemble framework with DenseNet, Swin transformer, and RegNet. Front. Microbiol. 2022, 13, 995323. [Google Scholar] [CrossRef]
- Chi, J.; Sun, Z.; Wang, H.; Lyu, P.; Yu, X.; Wu, C. CT image super-resolution reconstruction based on global hybrid attention. Comput. Biol. Med. 2022, 150, 106112. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liang, C.; Xue, Y.; Chen, T.; Chen, Y.; Lan, Y.; Wen, J.; Shao, X.; Chen, J. An Intelligent Diagnostic Model for Melasma Based on Deep Learning and Multimode Image Input. Dermatol. Ther. 2023, 13, 569–579. [Google Scholar] [CrossRef] [PubMed]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 102802. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A. Deep reinforcement learning in medicine. Kidney Dis. 2019, 5, 18–22. [Google Scholar] [CrossRef]
- Ghesu, F.C.; Georgescu, B.; Zheng, Y.; Grbic, S.; Maier, A.; Hornegger, J.; Comaniciu, D. Multi-scale deep reinforcement learning for real-time 3D-landmark detection in CT scans. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 41, 176–189. [Google Scholar] [CrossRef]
- Xu, J.; Croft, W.B. Quary expansion using local and global document analysis. In Acm Sigir Forum; ACM: New York, NY, USA, 2017; Volume 51, pp. 168–175. [Google Scholar]
- Wu, W.T.; Li, Y.J.; Feng, A.Z.; Li, L.; Huang, T.; Xu, A.D.; Lyu, J. Data mining in clinical big data: The frequently used databases, steps, and methodological models. Mil. Med. Res. 2021, 8, 44. [Google Scholar] [CrossRef] [PubMed]

| Imaging Equipment | Skin Imaging Standards | Model of Applicability |
|---|---|---|
| Dermoscopy | 1. Overall skin lesion: natural light was used as the light source, and the mode and magnification were noted. 2. Local details: the maximum magnification and clear images of the skin lesions were taken. | AlexNet, VGG, GoogLeNet, ResNet, CNN [20,21,22,23,24]. |
| RCM | 1. Longitudinal scanning: we scanned from the stratum corneum to the superficial dermis; each layer’s thickness was 5 μm. 2. Horizontal scanning: pathological changes in the stratum corneum, stratum granulosum, stratum spinosum, stratum basale, dermo-epidermal junction, and superficial dermis were scanned. 3. Local details: for each layer of pathological changes, photos of local details were taken. | SVM, CNN, InceptionV3, Bayesian model, Nested U-net [25,26,27,28,29]. |
| VHF skin ultrasound | 1. Longitudinal scanning: the lesion area was scanned using high-frequency or ultra-high-frequency ultrasound, and the scanning frequency (20 MHz, 50 MHz, etc.) was marked. 2. Overall and detailed imaging: it was able to clearly display the epidermis, dermis, and subcutaneous tissue, and measure the range, depth, blood flow, and nature of skin lesions and their relationship with surrounding tissues. | DenseNet-201, GoogleNet, Inception-ResNet-v2, ResNet-101, MobileNet [30] |
| Reference | Method | AC | SE | SP | JA | DI |
|---|---|---|---|---|---|---|
| [51] | LSC (ML, 2015) | 96.2% | 92.6% | - | 0.81 | - |
| [52] | K-means (ML, 2016) | 90% | - | - | - | - |
| [53] | RGB threshold (ML, 2019) | - | - | - | 0.789 | 0.876 |
| [53] | XYZ threshold (ML, 2019) | - | - | - | 0.8 | 0.884 |
| [54] | ICA (ML, 2019) | - | 99.49% | 98.46% | 0.7087 | - |
| [56] | FCM (ML, 2020) | 90.89% | 92.84% | 88.27% | - | - |
| [58] | FCN (DL, 2017) | 95.3% | 93.8% | 95.2% | 0.841 | 0.907 |
| [60] | FCDN (DL, 2017) | 99.53% | 87.9% | 97.9% | 0.783 | 0.865 |
| [61] | DFCN (DL, 2017) | 93.4% | 82.5% | 97.5% | 0.765 | 0.849 |
| [62] | FCRN (DL, 2017) | 85.5% | 54.7% | 93.1% | - | - |
| [63] | SegNet (DL, 2021) | - | 95.6% | 95.42% | - | 0.749 |
| [63] | U-Net (DL, 2021) | - | 96.4% | 94.8% | - | 0.733 |
| [64] | FCN (DL, 2018) | - | - | - | 0.884 | - |
| [65] | ResNet34 (DL, 2019) | - | - | - | 0.768 | 0.851 |
| [66] | U-Net (DL, 2019) | 97% | 90% | 99% | 0.88 | 0.94 |
| [67] | FCN U-Net (DL, 2019) | 90% | 96% | - | 0.83 | - |
| [68] | U-Net, VGG-16 (DL, 2021) | 96.7% | 90.4% | 98% | 0.846 | 0.915 |
| [69] | MSFCDN (DL, 2018) | 95.3% | 90.1% | 96.7% | 0.785 | 0.869 |
| [70] | DPFCN (DL, 2019) | 98.9% | 92.4% | 99.6% | 0.852 | 0.916 |
| [71] | ResU-NeXt ++ (DL, 2021) | 96% | - | - | 0.8684 | 0.9235 |
| [72] | U-Net (DL, 2022) | 90.74% | - | - | 0.7572 | - |
| [73] | DeepLabv3 + (DL, 2023) | 95% | 90% | 90% | - | - |
| [75] | W-EFO-E-CNN (DL, 2023) | 98% | 99.54% | 50% | 0.987 | |
| [76] | DCNN (DL, 2019) | - | - | - | 0.714 | - |
| [77] | U-Net (DL, 2020) | - | - | - | 0.887 | - |
| [78] | SEDSIC (DL, 2021) | 97% | 98% | 96% | 0.94 | 0.97 |
| [79] | FCN-UTA (DL, 2021) | - | 86.36% | - | 0.7381 | 0.8493 |
| Reference | Methods | AC | SE | SP | Classes | Data Type | Data Size |
|---|---|---|---|---|---|---|---|
| [86] | Adaboost (ML, 2015) | 89.35% | 93.5% | 85.2% | 2 | Dermoscopic images | - |
| [87] | KNN–SVM (ML, 2015) | 85% | - | - | 5 | Clinical images | 726 |
| [88] | SVM (ML, 2016) | 96.8% | 95.4% | 89.3% | 2 | Dermoscopic images | 320 |
| [89] | KNN–SVM (ML, 2017) | 90% | - | - | 4 | Dermoscopic images | - |
| [90] | SVM (ML, 2018) | 92.3% | - | - | 3 | Dermoscopic images | - |
| [91] | SVM (ML, 2019) | 89.43% | 91.15% | 87.71% | 2 | Dermoscopic images | 1000 |
| [92] | Naive Bayes (ML, 2020) | 72.7% | 91.7% | 70.1% | 6 | Dermoscopic images | 1646 |
| [93] | CNN (DL, 2020) | 75% | 73% | 78% | 2 | Dermoscopic images | 1796 |
| [94] | BLSTM (DL, 2022) | 89.47% | 88.33% | 97.17% | 7 | Dermoscopic images | 10,015 |
| [95] | Eff2Net (DL, 2022) | 84.70% | 84.70% | - | 4 | Clinical images | 17,327 |
| [96] | BPNN (DL, 2020) | 99.7% | 99.4% | 100%% | 2 | Dermoscopic images | 400 |
| [97] | DenseNet201 (DL, 2022) | 95.5% | 93.96% | 97.06% | 2 | Dermoscopic images | 3297 |
| [98] | Inception-ResNet-V2 (DL, 2020) | 87.42% | 97.04% | 96.48% | 4 | Clinical images | 14,000 |
| [99] | Inception-ResNet V2 (DL, 2019) | 89.63% | 77% | - | 6 | Clinical images | 11,445 |
| [100] | GoogleNet (DL, 2020) | 99.29% | 99.22% | 99.38% | 2 | Dermoscopic images | 2376 |
| [101] | AlexNet (DL, 2020) | 98.7% | 95.6% | 99.27% | 7 | Dermoscopic images | 10,015 |
| [102] | GoogleNet (DL, 2020) | 94.92% | 79.8% | 97% | 8 | Dermoscopic images | 29,439 |
| [103] | RDCNN (DL, 2022) | 97% | 94% | 98% | 2 | Dermoscopic images | 2206 |
| [104] | InSiNet (DL, 2022) | 94.59% | 97.5% | 91.18% | 2 | Dermoscopic images | 1471 |
| [105] | GoogleNet(Inception-V3) (DL, 2020) | 83.78% | 87.5% | 79.41% | 2 | Dermoscopic images | 1471 |
| [106] | DenseNet-201 (DL, 2020) | 87.84% | 95% | 79.41% | 2 | Dermoscopic images | 1471 |
| [107] | ResNet152V2 (DL, 2020) | 86.49% | 92.5% | 79.41% | 2 | Dermoscopic images | 1471 |
| [108] | DenseNet, ResNet (DL, 2023) | 95.1% | 92% | 98.8% | 7 | Dermoscopic images | 10,015 |
| [109] | U-Net, CNN (DL, 2023) | 97.96% | 84.86% | 97.93% | 7 | Dermoscopic images | 10,015 |
| [110] | DCNN (DL, 2023) | 97.204% | 97% | - | 7 | Dermoscopic images | 10,015 |
| [111] | Visual Transformer (DL, 2023) | 93.81% | 90.14% | 98.36% | 7 | Dermoscopic images | 10,015 |
| [112] | Resnet50, VGG16, Inception v2 (DL, 2019) | 87.8% | 90.9% | 91.9% | 2 | Clinical images | 38,677 |
| [113] | Cycle GAN, ADRD, Resnet50 (DL, 2020) | 85.69% | - | 90.92% | 2 | Wood lamp images | 10,000 |
| [114] | YOLO v3, PSPNet, UNet ++ (DL, 2022) | 85.02% | 92.91% | - | 3 | Clinical images | 2720 |
| [115] | LVQ Neural Network (DL, 2017) | 92.22% | - | - | 3 | Clinical images | 1002 |
| Year | 2018 | 2019 | 2020 | 2021 | 2022 | 2023 |
|---|---|---|---|---|---|---|
| Publication No. | 104 | 127 | 161 | 190 | 245 | 233 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhong, F.; He, K.; Ji, M.; Li, S.; Li, C. Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review. Diagnostics 2023, 13, 3506. https://doi.org/10.3390/diagnostics13233506
Zhang J, Zhong F, He K, Ji M, Li S, Li C. Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review. Diagnostics. 2023; 13(23):3506. https://doi.org/10.3390/diagnostics13233506
Chicago/Turabian StyleZhang, Junpeng, Fan Zhong, Kaiqiao He, Mengqi Ji, Shuli Li, and Chunying Li. 2023. "Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review" Diagnostics 13, no. 23: 3506. https://doi.org/10.3390/diagnostics13233506
APA StyleZhang, J., Zhong, F., He, K., Ji, M., Li, S., & Li, C. (2023). Recent Advancements and Perspectives in the Diagnosis of Skin Diseases Using Machine Learning and Deep Learning: A Review. Diagnostics, 13(23), 3506. https://doi.org/10.3390/diagnostics13233506