Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Populations and Imaging Parameters
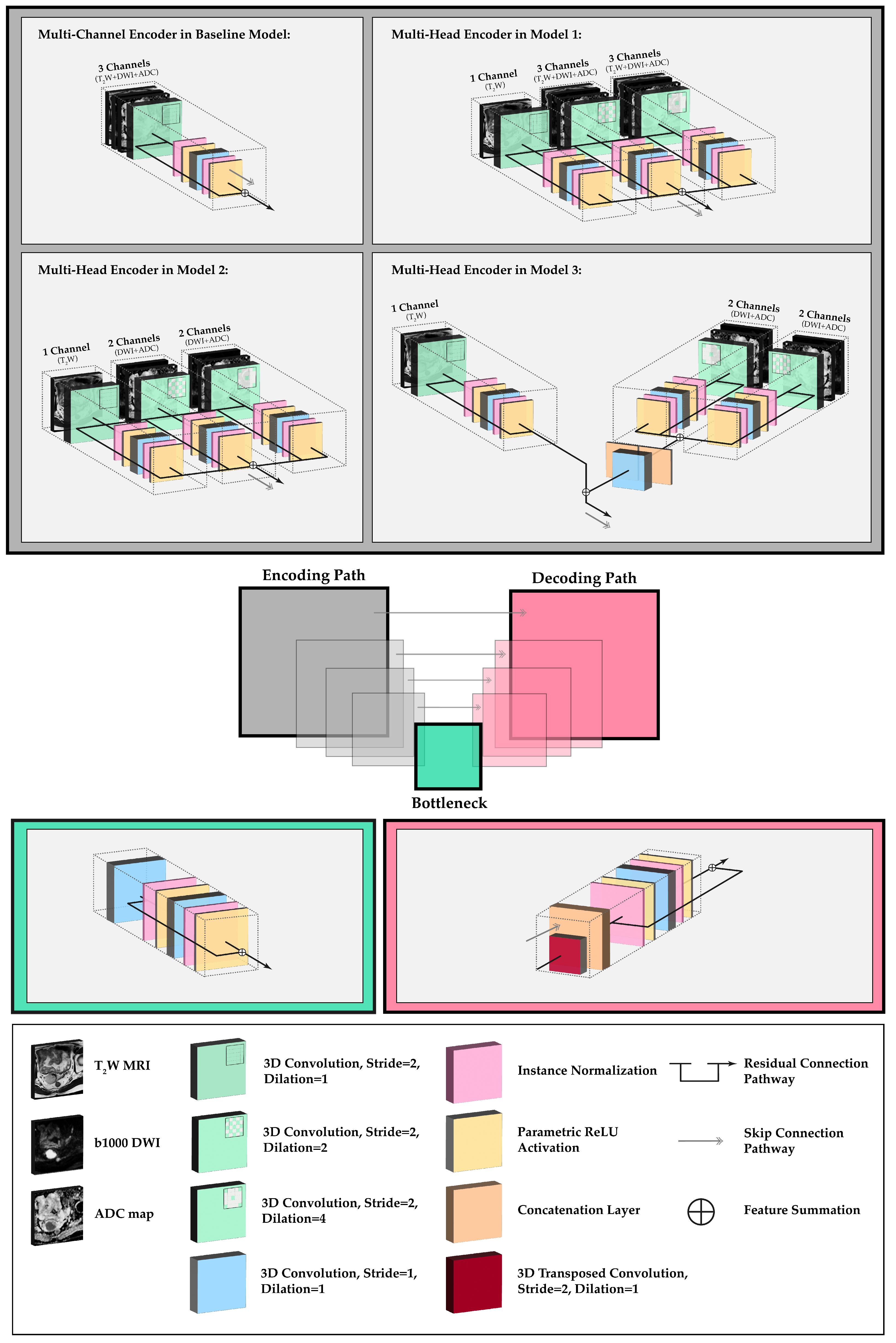
2.2. Network Topology and Architectural Experiments
2.3. Image Pre-Processing and Implementation Details
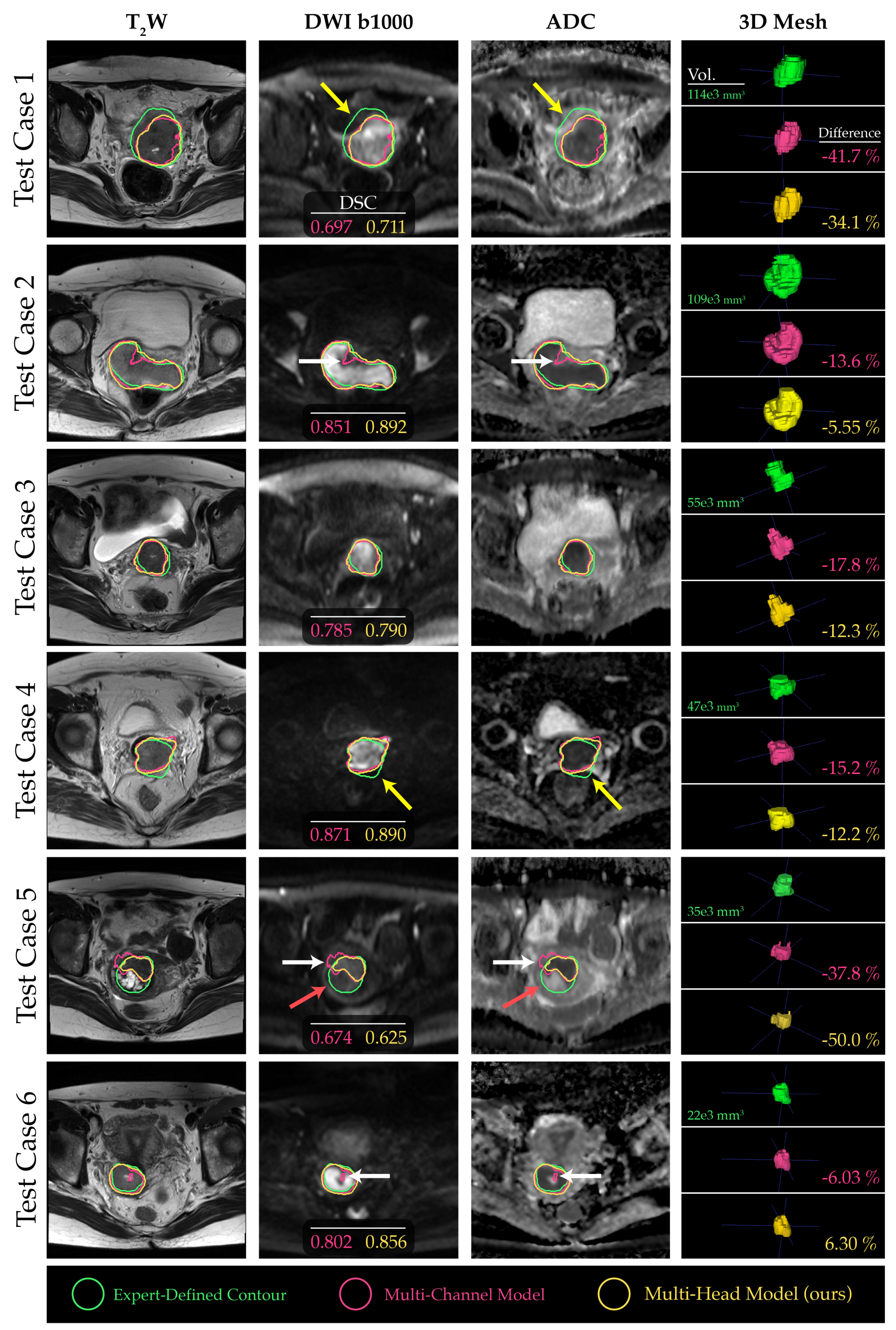
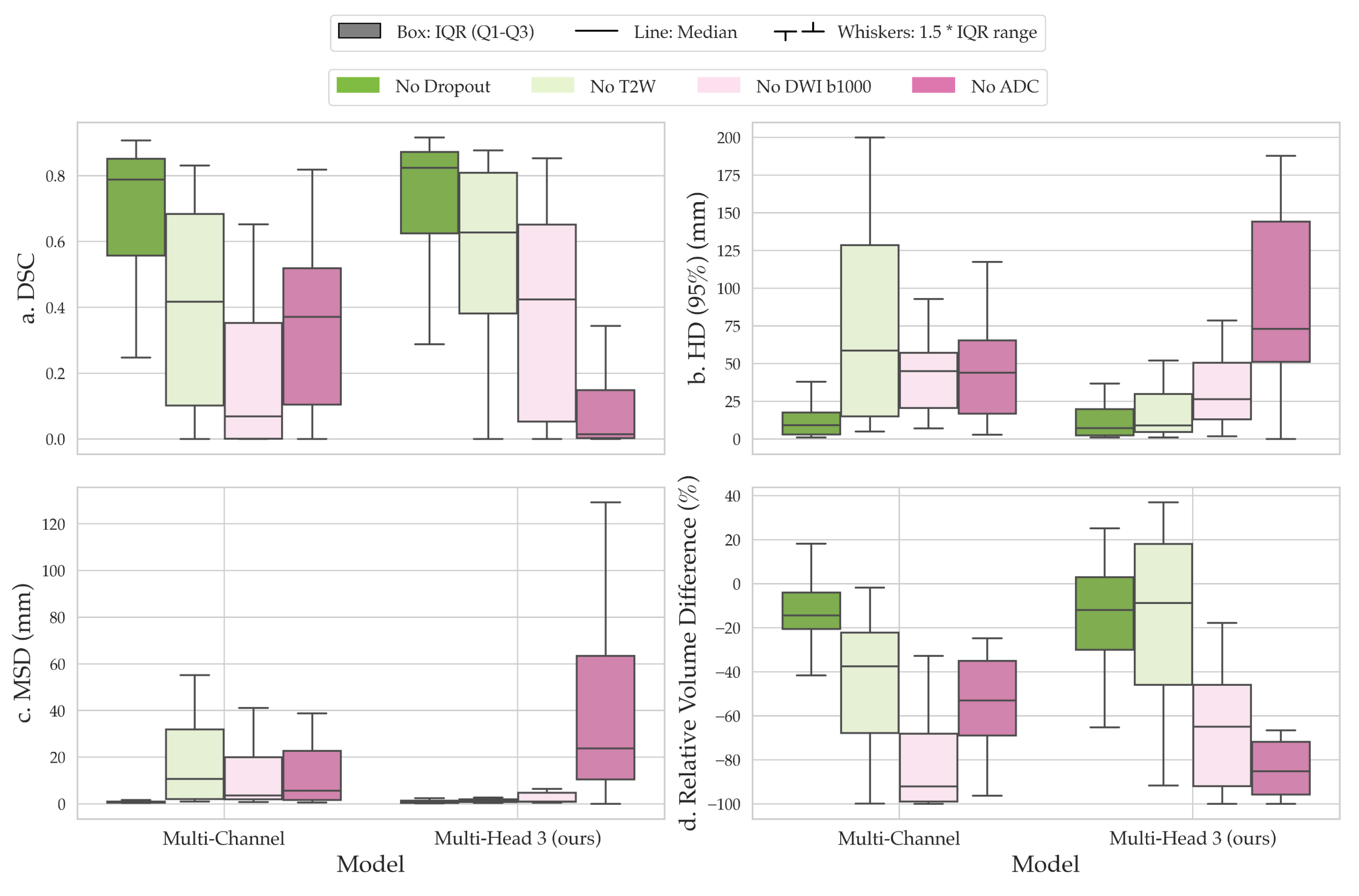
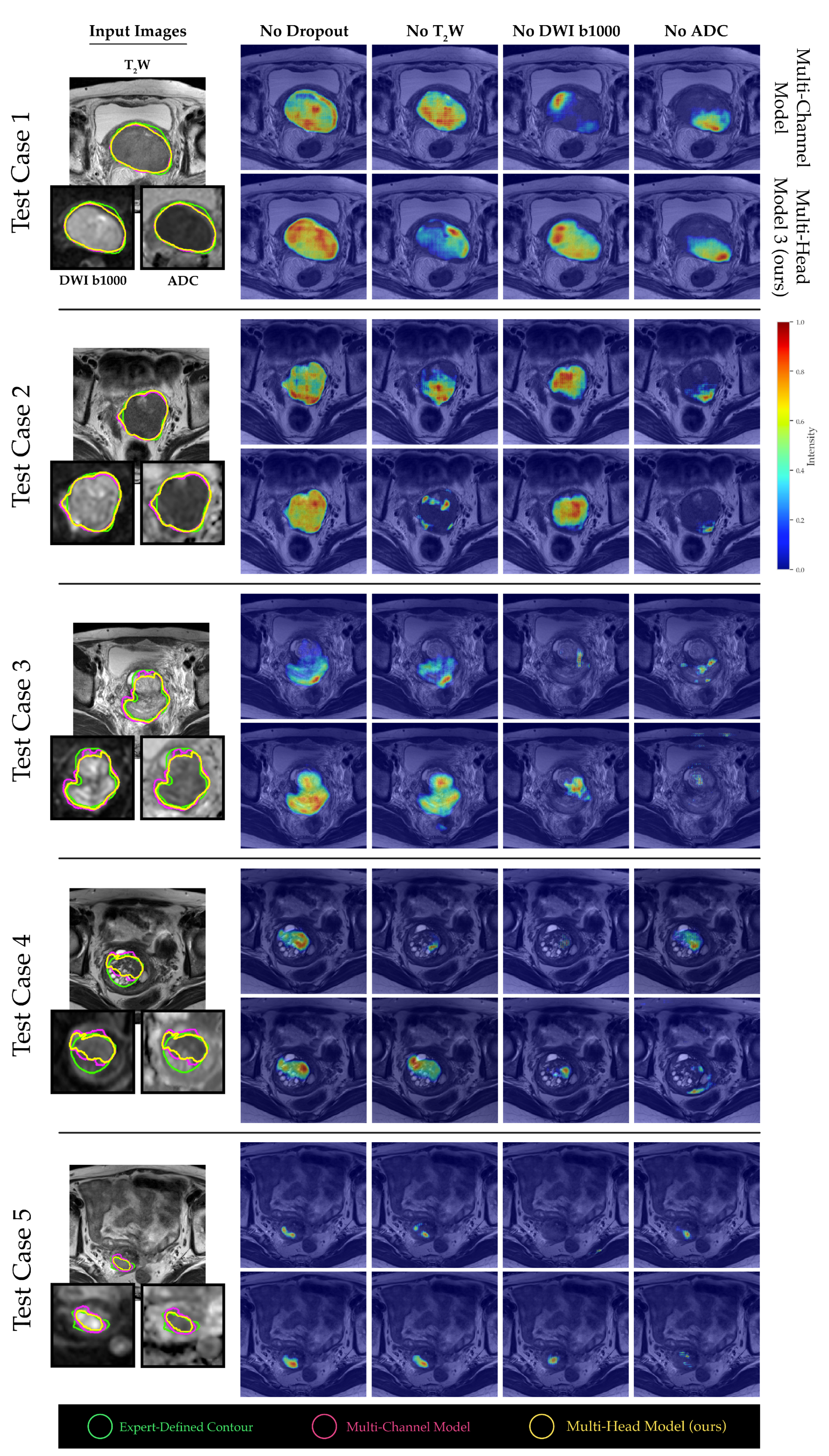
2.4. Channel Sensitivity Analysis and Visualization
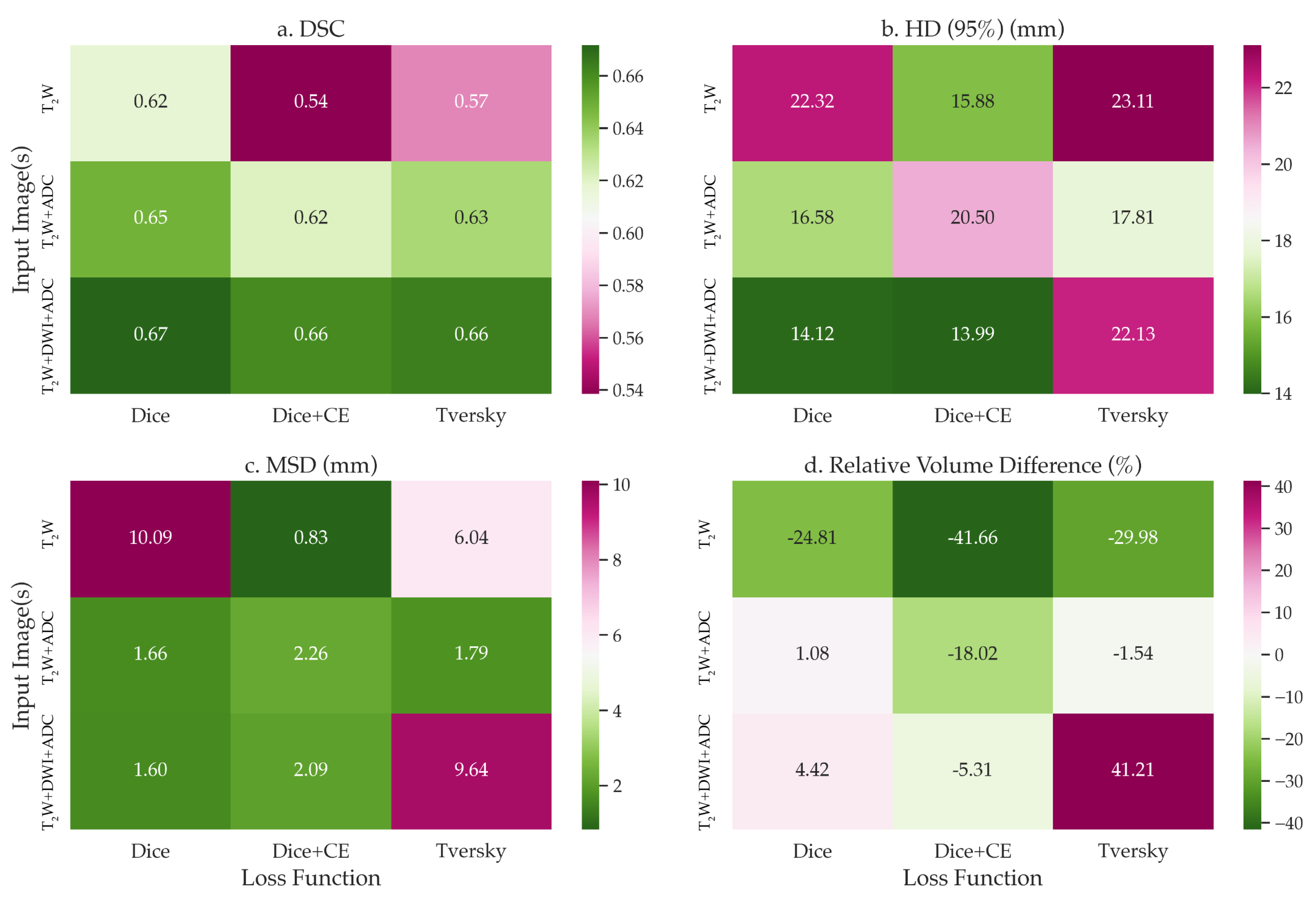
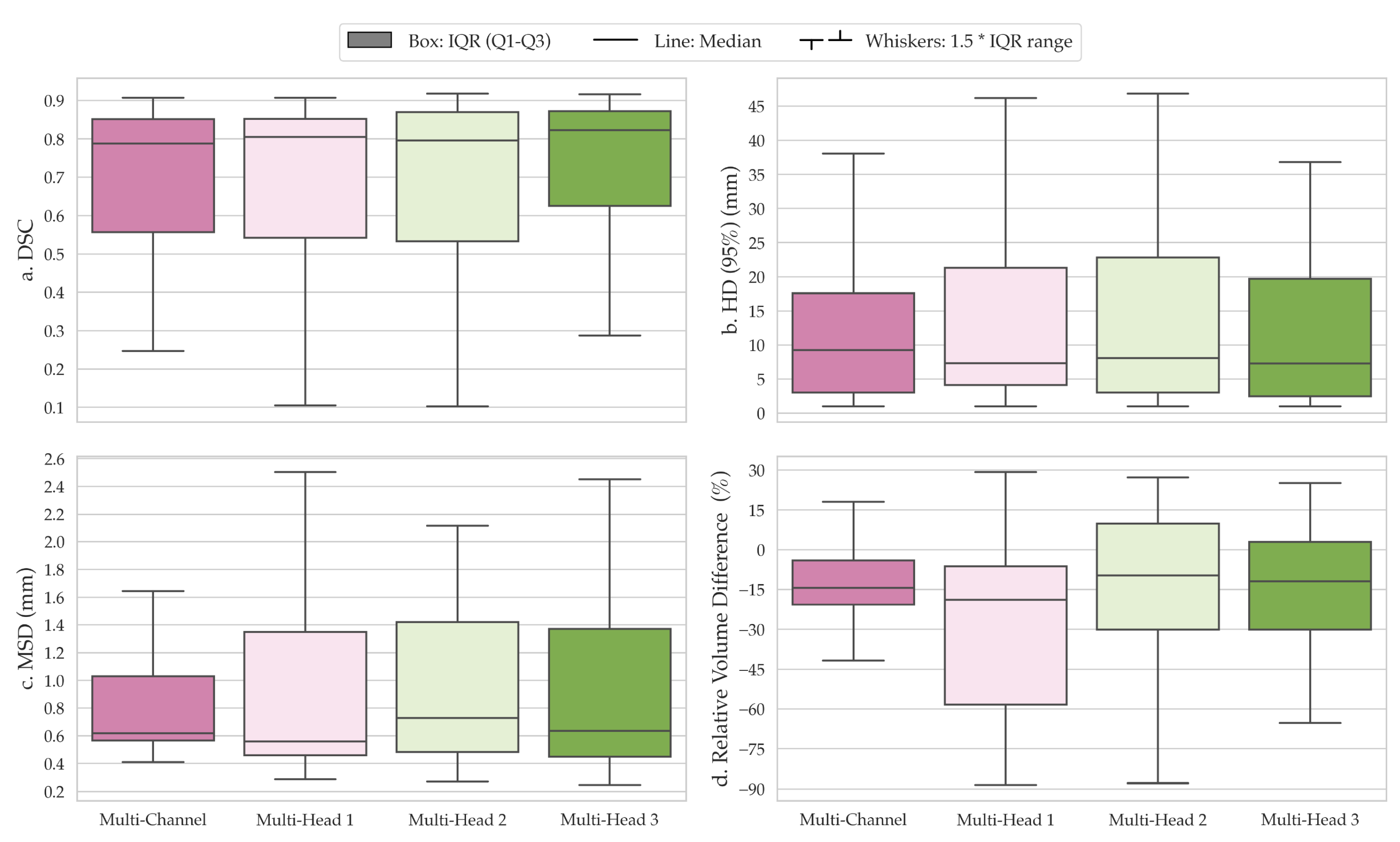
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Vignat, J.; Lorenzoni, V.; Eslahi, M.; Ginsburg, O.; Lauby-Secretan, B.; Arbyn, M.; Basu, P.; Bray, F.; Vaccarella, S. Global estimates of incidence and mortality of cervical cancer in 2020: A baseline analysis of the WHO Global Cervical Cancer Elimination Initiative. Lancet Glob. Health 2023, 11, e197–e206. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Togashi, K.; Nishimura, K.; Sagoh, T.; Minami, S.; Noma, S.; Fujisawa, I.; Nakano, Y.; Konishi, J.; Ozasa, H.; Konishi, I. Carcinoma of the cervix: Staging with MR imaging. Radiology 1989, 171, 245–251. [Google Scholar] [CrossRef] [PubMed]
- Green, J.A.; Kirwan, J.M.; Tierney, J.F.; Symonds, P.; Fresco, L.; Collingwood, M.; Williams, C.J. Survival and recurrence after concomitant chemotherapy and radiotherapy for cancer of the uterine cervix: A systematic review and meta-analysis. Lancet 2001, 358, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Pollard, J.M.; Wen, Z.; Sadagopan, R.; Wang, J.; Ibbott, G.S. The future of image-guided radiotherapy will be MR guided. Br. J. Radiol. 2017, 90, 20160667. [Google Scholar] [CrossRef] [PubMed]
- Koh, D.M.; Thoeny, H.C. Diffusion-Weighted MR Imaging: Applications in the Body; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2010. [Google Scholar]
- Yoshida, S.; Takahara, T.; Kwee, T.C.; Waseda, Y.; Kobayashi, S.; Fujii, Y. DWI as an imaging biomarker for bladder cancer. Am. J. Roentgenol. 2017, 208, 1218–1228. [Google Scholar] [CrossRef]
- Tsuruta, C.; Hirata, K.; Kudo, K.; Masumori, N.; Hatakenaka, M. DWI-related texture analysis for prostate cancer: Differences in correlation with histological aggressiveness and data repeatability between peripheral and transition zones. Eur. Radiol. Exp. 2022, 6, 1–12. [Google Scholar] [CrossRef]
- Higaki, T.; Nakamura, Y.; Tatsugami, F.; Kaichi, Y.; Akagi, M.; Akiyama, Y.; Baba, Y.; Iida, M.; Awai, K. Introduction to the technical aspects of computed diffusion-weighted imaging for radiologists. Radiographics 2018, 38, 1131–1144. [Google Scholar] [CrossRef]
- Abd elsalam, S.M.; Mokhtar, O.; Adel, L.; Hassan, R.; Ibraheim, M.; Kamal, A. Impact of diffusion weighted magnetic resonance imaging in diagnosis of cervical cancer. Egypt. J. Radiol. Nucl. Med. 2020, 51, 23. [Google Scholar] [CrossRef]
- Subak, L.E.; Hricak, H.; Powell, C.B.; Azizi, E.; Stern, J.L. Cervical carcinoma: Computed tomography and magnetic resonance imaging for preoperative staging. Obstet. Gynecol. 1995, 86, 43–50. [Google Scholar] [CrossRef]
- Romesser, P.B.; Tyagi, N.; Crane, C.H. Magnetic resonance imaging-guided adaptive radiotherapy for colorectal liver metastases. Cancers 2021, 13, 1636. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241, Proceedings, Part III 18. [Google Scholar]
- Zhang, J.; Jin, Y.; Xu, J.; Xu, X.; Zhang, Y. Mdu-net: Multi-scale densely connected u-net for biomedical image segmentation. arXiv 2018, arXiv:1812.00352. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Siddiquee, M.M.R.; Tajbakhsh, N.; Liang, J. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans. Med. Imaging 2019, 39, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.U.; Cho, S.; Kim, J.H.; Chong, K.T. Bu-net: Brain tumor segmentation using modified u-net architecture. Electronics 2020, 9, 2203. [Google Scholar] [CrossRef]
- Rehman, M.U.; Cho, S.; Kim, J.; Chong, K.T. Brainseg-net: Brain tumor mr image segmentation via enhanced encoder–decoder network. Diagnostics 2021, 11, 169. [Google Scholar] [CrossRef] [PubMed]
- Mubashar, M.; Ali, H.; Grönlund, C.; Azmat, S. R2U++: A multiscale recurrent residual U-Net with dense skip connections for medical image segmentation. Neural Comput. Appl. 2022, 34, 17723–17739. [Google Scholar] [CrossRef]
- Lu, H.; She, Y.; Tie, J.; Xu, S. Half-UNet: A simplified U-Net architecture for medical image segmentation. Front. Neuroinform. 2022, 16, 911679. [Google Scholar] [CrossRef]
- Ryu, J.; Rehman, M.U.; Nizami, I.F.; Chong, K.T. SegR-Net: A deep learning framework with multi-scale feature fusion for robust retinal vessel segmentation. Comput. Biol. Med. 2023, 163, 107132. [Google Scholar] [CrossRef]
- Rehman, M.U.; Ryu, J.; Nizami, I.F.; Chong, K.T. RAAGR2-Net: A brain tumor segmentation network using parallel processing of multiple spatial frames. Comput. Biol. Med. 2023, 152, 106426. [Google Scholar] [CrossRef]
- Bougourzi, F.; Distante, C.; Dornaika, F.; Taleb-Ahmed, A. PDAtt-Unet: Pyramid dual-decoder attention Unet for Covid-19 infection segmentation from CT-scans. Med. Image Anal. 2023, 86, 102797. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Wang, H.; Xie, S.; Lin, L.; Iwamoto, Y.; Han, X.H.; Chen, Y.W.; Tong, R. Mixed transformer u-net for medical image segmentation. In Proceedings of the ICASSP 2022–2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Virtual, 7–13 May 2022; pp. 2390–2394. [Google Scholar]
- Huang, L.; Zhu, E.; Chen, L.; Wang, Z.; Chai, S.; Zhang, B. A transformer-based generative adversarial network for brain tumor segmentation. Front. Neurosci. 2022, 16, 1054948. [Google Scholar] [CrossRef] [PubMed]
- Rigaud, B.; Anderson, B.M.; Zhiqian, H.Y.; Gobeli, M.; Cazoulat, G.; Söderberg, J.; Samuelsson, E.; Lidberg, D.; Ward, C.; Taku, N.; et al. Automatic segmentation using deep learning to enable online dose optimization during adaptive radiation therapy of cervical cancer. Int. J. Radiat. Oncol. Biol. Phys. 2021, 109, 1096–1110. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.Y.; Zhou, J.Y.; Xu, X.T.; Guo, J.; Han, M.F.; Gao, Y.Z.; Du, H.; Stahl, J.N.; Maltz, J.S. Deep learning-based auto-segmentation of clinical target volumes for radiotherapy treatment of cervical cancer. J. Appl. Clin. Med. Phys. 2022, 23, e13470. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, Y.; Wu, C.; Wang, F. Multimodal MRI analysis of cervical cancer on the basis of artificial intelligence algorithm. Contrast Media Mol. Imaging 2021, 2021, 1673490. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Fang, F.; Zhang, H.; Ling, L.; Hua, K. AugMS-Net: Augmented multiscale network for small cervical tumor segmentation from MRI volumes. Comput. Biol. Med. 2022, 141, 104774. [Google Scholar] [CrossRef]
- Zabihollahy, F.; Viswanathan, A.N.; Schmidt, E.J.; Lee, J. Fully automated segmentation of clinical target volume in cervical cancer from magnetic resonance imaging with convolutional neural network. J. Appl. Clin. Med. Phys. 2022, 23, e13725. [Google Scholar] [CrossRef]
- Lin, Y.C.; Lin, C.H.; Lu, H.Y.; Chiang, H.J.; Wang, H.K.; Huang, Y.T.; Ng, S.H.; Hong, J.H.; Yen, T.C.; Lai, C.H.; et al. Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer. Eur. Radiol. 2020, 30, 1297–1305. [Google Scholar] [CrossRef]
- Kano, Y.; Ikushima, H.; Sasaki, M.; Haga, A. Automatic contour segmentation of cervical cancer using artificial intelligence. J. Radiat. Res. 2021, 62, 934–944. [Google Scholar] [CrossRef]
- Yoganathan, S.; Paul, S.N.; Paloor, S.; Torfeh, T.; Chandramouli, S.H.; Hammoud, R.; Al-Hammadi, N. Automatic segmentation of magnetic resonance images for high-dose-rate cervical cancer brachytherapy using deep learning. Med. Phys. 2022, 49, 1571–1584. [Google Scholar] [CrossRef] [PubMed]
- Hodneland, E.; Kaliyugarasan, S.; Wagner-Larsen, K.S.; Lura, N.; Andersen, E.; Bartsch, H.; Smit, N.; Halle, M.K.; Krakstad, C.; Lundervold, A.S.; et al. Fully Automatic Whole-Volume Tumor Segmentation in Cervical Cancer. Cancers 2022, 14, 2372. [Google Scholar] [CrossRef] [PubMed]
- Yoshizako, T.; Yoshida, R.; Asou, H.; Nakamura, M.; Kitagaki, H. Comparison between turbo spin-echo and echo planar diffusion-weighted imaging of the female pelvis with 3T MRI. Acta Radiol. Open 2021, 10, 2058460121994737. [Google Scholar] [CrossRef] [PubMed]
- Donato, F., Jr.; Costa, D.N.; Yuan, Q.; Rofsky, N.M.; Lenkinski, R.E.; Pedrosa, I. Geometric distortion in diffusion-weighted MR imaging of the prostate—Contributing factors and strategies for improvement. Acad. Radiol. 2014, 21, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Carcangiu, M.L.; Herrington, C.S. World Health Organisation Classification of Tumours of the Female Reproductive Organs; International Agency for Research on Cancer: Lyon, France, 2014. [Google Scholar]
- Saleh, M.; Virarkar, M.; Javadi, S.; Elsherif, S.B.; de Castro Faria, S.; Bhosale, P. Cervical cancer: 2018 revised international federation of gynecology and obstetrics staging system and the role of imaging. Am. J. Roentgenol. 2020, 214, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Tversky, A. Features of similarity. Psychol. Rev. 1977, 84, 327. [Google Scholar] [CrossRef]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Cardoso, M.J.; Li, W.; Brown, R.; Ma, N.; Kerfoot, E.; Wang, Y.; Murrey, B.; Myronenko, A.; Zhao, C.; Yang, D.; et al. MONAI: An open-source framework for deep learning in healthcare. arXiv 2022, arXiv:2211.02701. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- McVeigh, P.Z.; Syed, A.M.; Milosevic, M.; Fyles, A.; Haider, M.A. Diffusion-weighted MRI in cervical cancer. Eur. Radiol. 2008, 18, 1058–1064. [Google Scholar] [CrossRef]
- Kalantar, R.; Lin, G.; Winfield, J.M.; Messiou, C.; Lalondrelle, S.; Blackledge, M.D.; Koh, D.M. Automatic segmentation of pelvic cancers using deep learning: State-of-the-art approaches and challenges. Diagnostics 2021, 11, 1964. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), 8–14 September 2018; pp. 801–818. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Shah, N.A.; Gupta, D.; Lodaya, R.; Baid, U.; Talbar, S. Colorectal cancer segmentation using atrous convolution and residual enhanced unet. In Proceedings of the Computer Vision and Image Processing: 5th International Conference, CVIP 2020, Prayagraj, India, 4–6 December 2020; Springer: Berlin/Heidelberg, Germany, 2021; pp. 451–462, Revised Selected Papers, Part I 5. [Google Scholar]
- Wang, B.; Lei, Y.; Jeong, J.J.; Wang, T.; Liu, Y.; Tian, S.; Patel, P.; Jiang, X.; Jani, A.B.; Mao, H.; et al. Automatic MRI prostate segmentation using 3D deeply supervised FCN with concatenated atrous convolution. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis, San Diego, CA, USA, 16–21 February 2019; SPIE: Bellingham, WA, USA, 2019; Volume 10950, pp. 988–995. [Google Scholar]
- Breto, A.L.; Spieler, B.; Zavala-Romero, O.; Alhusseini, M.; Patel, N.V.; Asher, D.A.; Xu, I.R.; Baikovitz, J.B.; Mellon, E.A.; Ford, J.C.; et al. Deep Learning for Per-Fraction Automatic Segmentation of Gross Tumor Volume (GTV) and Organs at Risk (OARs) in Adaptive Radiotherapy of Cervical Cancer. Front. Oncol. 2022, 12, 854349. [Google Scholar] [CrossRef]
- Lin, Y.C.; Lin, Y.; Huang, Y.L.; Ho, C.Y.; Chiang, H.J.; Lu, H.Y.; Wang, C.C.; Wang, J.J.; Ng, S.H.; Lai, C.H.; et al. Generalizable transfer learning of automated tumor segmentation from cervical cancers toward a universal model for uterine malignancies in diffusion-weighted MRI. Insights Imaging 2023, 14, 14. [Google Scholar] [CrossRef]
| Parameter | T2W MRI | DWI |
|---|---|---|
| Manufacturer Name | Siemens Healthineers | Siemens Healthineers |
| Scanner Model | MAGNETOM Trio | MAGNETOM Trio |
| Magnetic Field Strength (T) | 3 | 3 |
| Sequence | Turbo Spin Echo (TSE) | Echo-Planar Imaging (EPI) |
| Slice Orientation | Axial | Axial |
| Echo Time (ms) | 80–101 | 60–80 |
| Repetition Time (ms) | 3600–8060 | 3300–10,844 |
| Acquired Matrix Size (read) | 224–320 | 128–172 |
| Reconstructed Matrix Size (read) | 256–320 | 240–248 |
| Reconstructed Pixel Size (mm) | 0.5 × 0.5–0.8 × 0.8 | 1.2 × 1.2–1.4 × 1.4 |
| Slice Thickness (mm) | 4.0–5.0 | 4.0–5.0 |
| Flip Angle (°) | 120–160 | 180 |
| Phase Encoding Direction | Anterior–Posterior or Left–Right | Anterior–Posterior |
| Receiver Bandwidth (Hz/pixel) | 190–200 | 1940–2441 |
| b-values (s/mm) | - | [0,1000] or [200,600,1000] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalantar, R.; Curcean, S.; Winfield, J.M.; Lin, G.; Messiou, C.; Blackledge, M.D.; Koh, D.-M. Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging. Diagnostics 2023, 13, 3381. https://doi.org/10.3390/diagnostics13213381
Kalantar R, Curcean S, Winfield JM, Lin G, Messiou C, Blackledge MD, Koh D-M. Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging. Diagnostics. 2023; 13(21):3381. https://doi.org/10.3390/diagnostics13213381
Chicago/Turabian StyleKalantar, Reza, Sebastian Curcean, Jessica M. Winfield, Gigin Lin, Christina Messiou, Matthew D. Blackledge, and Dow-Mu Koh. 2023. "Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging" Diagnostics 13, no. 21: 3381. https://doi.org/10.3390/diagnostics13213381
APA StyleKalantar, R., Curcean, S., Winfield, J. M., Lin, G., Messiou, C., Blackledge, M. D., & Koh, D.-M. (2023). Deep Learning Framework with Multi-Head Dilated Encoders for Enhanced Segmentation of Cervical Cancer on Multiparametric Magnetic Resonance Imaging. Diagnostics, 13(21), 3381. https://doi.org/10.3390/diagnostics13213381









