Deep Learning with Automatic Data Augmentation for Segmenting Schisis Cavities in the Optical Coherence Tomography Images of X-Linked Juvenile Retinoschisis Patients
Abstract
:1. Introduction
2. Methods
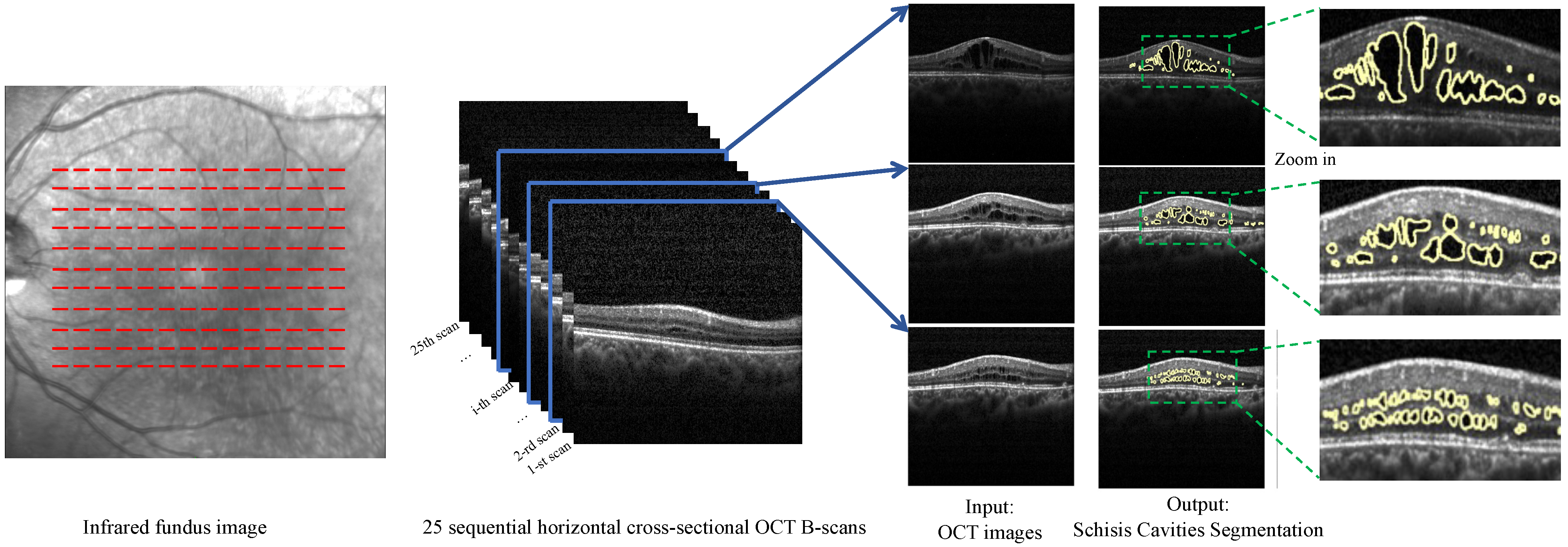
2.1. Data Collection and Annotation
2.1.1. Data Collection
2.1.2. Manual Annotation
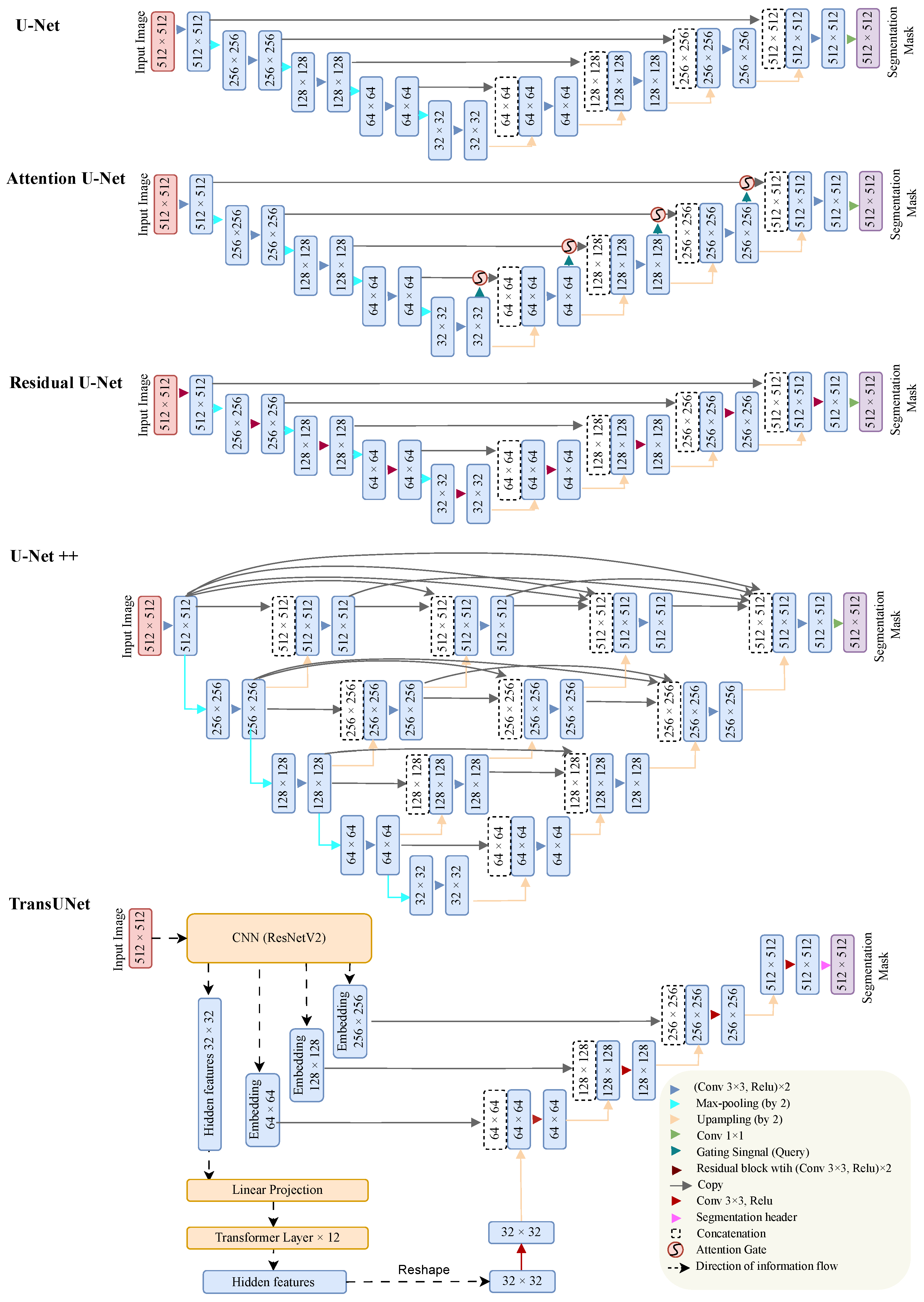
2.2. Deep Learning Model for Schisis Cavity Segmentation in OCT
2.2.1. Neural Network Architecture
2.2.2. Deep Neural Network Training
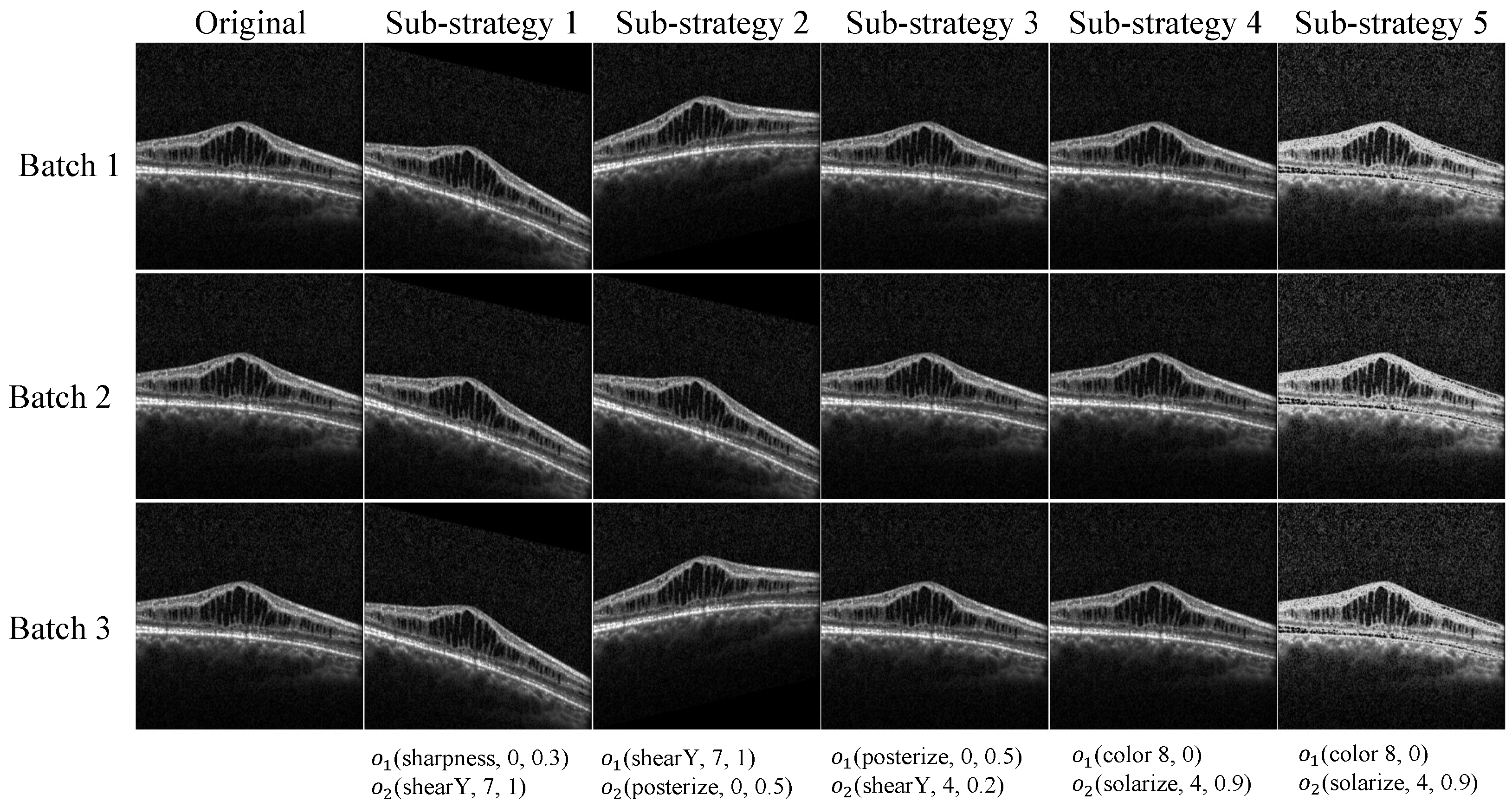
2.3. Data Augmentation
2.3.1. Problem Formulation
2.3.2. Search Space
2.3.3. Optimizing the Augmentation Strategy with Reinforcement Learning
| Algorithm 1 Automated augmentation via DRL for DL segmentation models |
| Initialization: Segmentation DL model , RL augmentation policy controller Input: Training dataset , Validation dataset Output: , 1: for do 2: Sample an augmentation strategy from RL policy , 3: for do 4: for do 5: Choose a sub-strategy f uniformly from F, 6: Augment the mini-batch data by f, ) 7: Update by 8: end for 9: end for 10: Evaluate the loss of the DL model on the validation set, 11: Define the reward for RL agent, 12: Update the policy parameters , by reward R and the gradient 13: end for |
2.4. Evaluation Metrics
2.5. Implementation Details
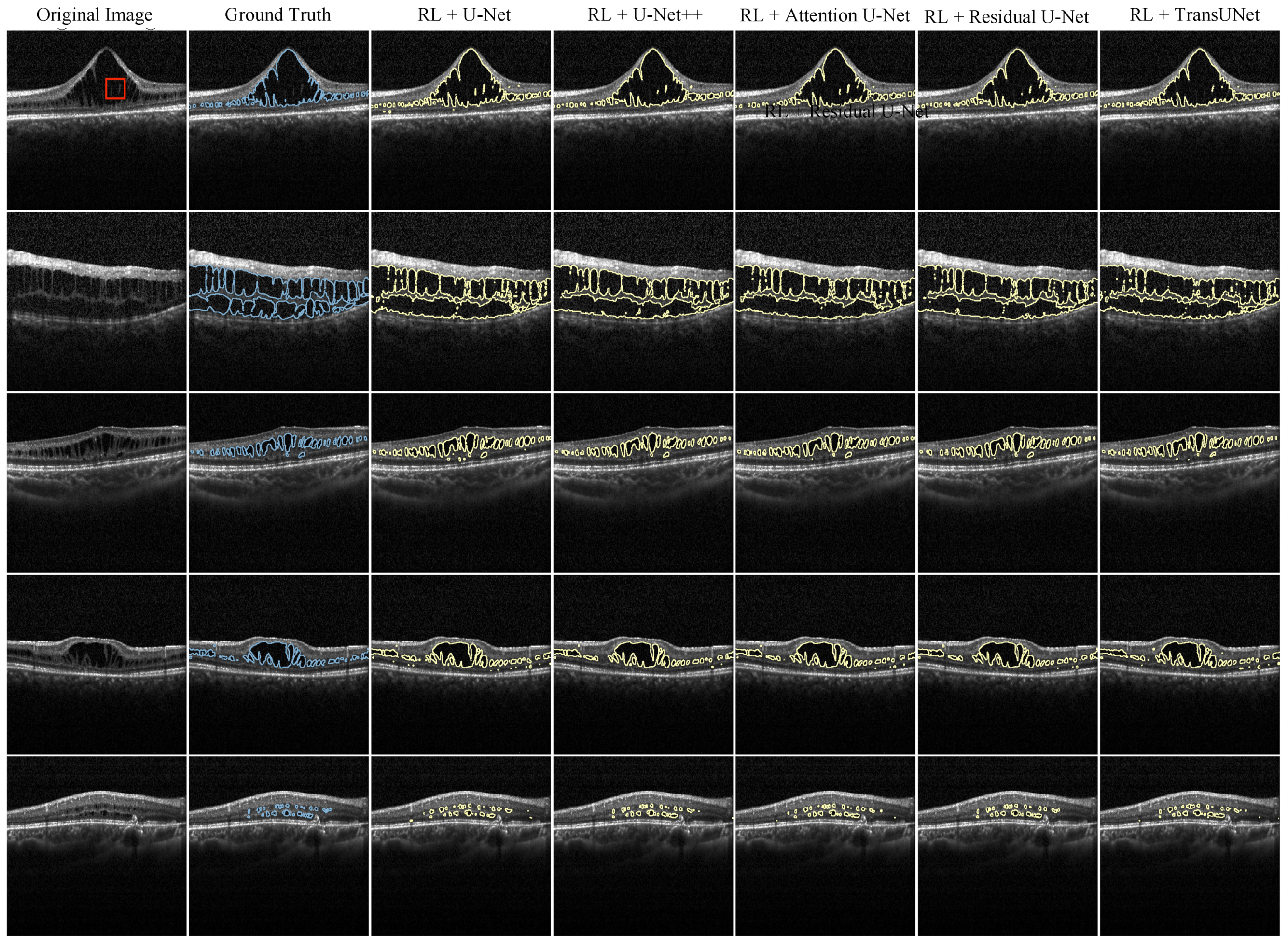
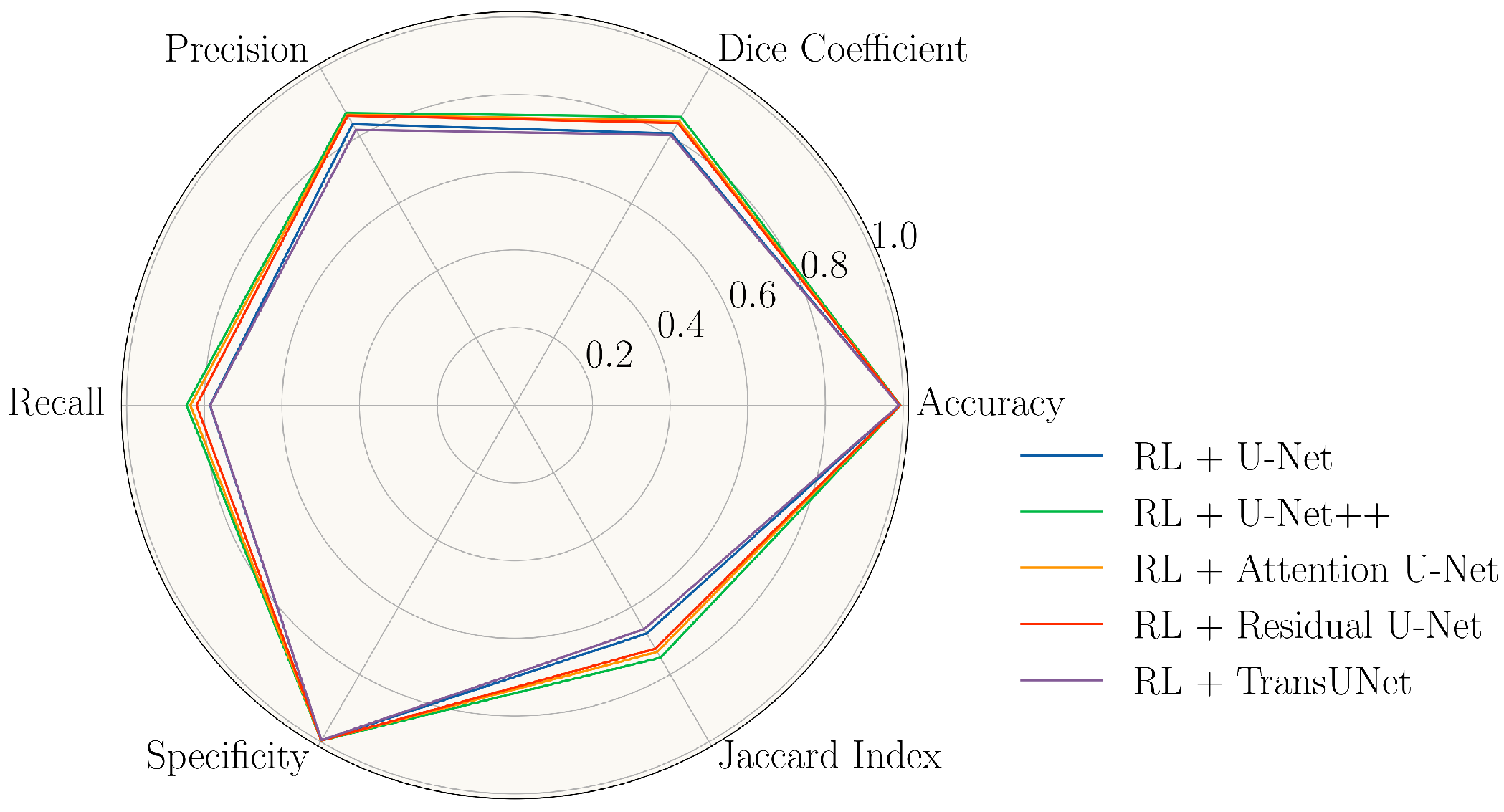
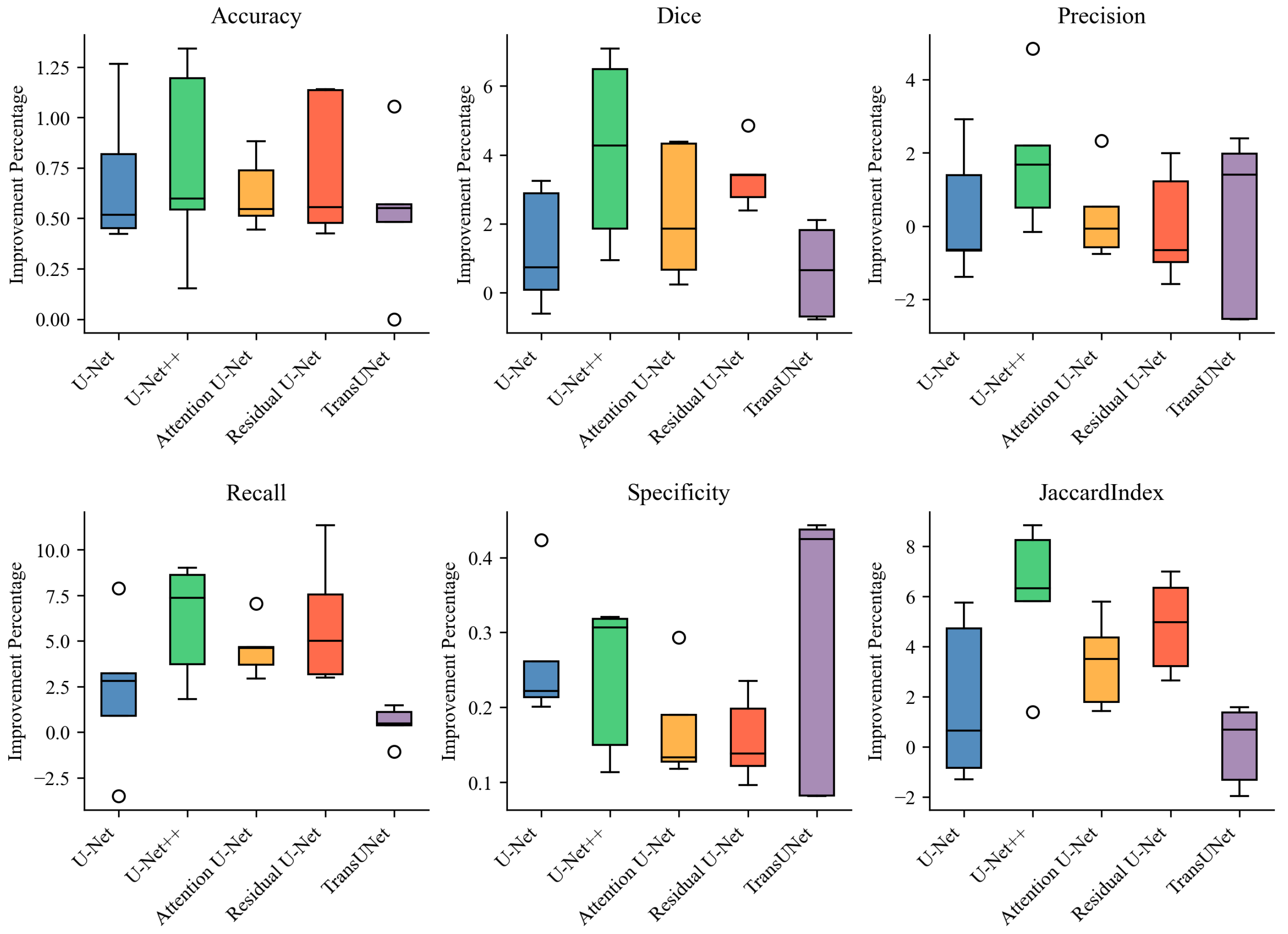
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tantri, A.; Vrabec, T.R.; Cu-Unjieng, A.; Frost, A.; Annesley, W.H., Jr.; Donoso, L.A. X-linked retinoschisis: A clinical and molecular genetic review. Surv. Ophthalmol. 2004, 49, 214–230. [Google Scholar] [CrossRef]
- Tsang, S.H.; Sharma, T. X-linked juvenile retinoschisis. In Atlas of Inherited Retinal Diseases; Springer: Cham, Switzerland, 2018; pp. 43–48. [Google Scholar]
- George, N.; Yates, J.; Moore, A. X linked retinoschisis. Br. J. Ophthalmol. 1995, 79, 697. [Google Scholar] [CrossRef] [PubMed]
- Apushkin, M.A.; Fishman, G.A.; Janowicz, M.J. Correlation of optical coherence tomography findings with visual acuity and macular lesions in patients with X-linked retinoschisis. Ophthalmology 2005, 112, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.M.; Choy, K.W.; Wang, J.; Lam, D.S.; Yip, W.W.; Fu, W.; Pang, C.P. Two cases of X-linked juvenile retinoschisis with different optical coherence tomography findings and RS1 gene mutations. Clin. Exp. Ophthalmol. 2004, 32, 429–432. [Google Scholar] [CrossRef]
- Urrets-Zavalía, J.A.; Venturino, J.P.; Mercado, J.; Urrets-Zavalía, E.A. Macular and extramacular optical coherence tomography findings in X-linked retinoschisis. Ophthalmic Surg. Lasers Imaging Retin. 2007, 38, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Condon, G.P.; Brownstein, S.; Wang, N.S.; Kearns, J.A.F.; Ewing, C.C. Congenital hereditary (juvenile X-linked) retinoschisis: Histopathologic and ultrastructural findings in three eyes. Arch. Ophthalmol. 1986, 104, 576–583. [Google Scholar] [CrossRef]
- Manschot, W.A. Pathology of hereditary juvenile retinoschisis. Arch. Ophthalmol. 1972, 88, 131–138. [Google Scholar] [CrossRef]
- Yanoff, M.; Rahn, E.K.; Zimmerman, L.E. Histopathology of juvenile retinoschisis. Arch. Ophthalmol. 1968, 79, 49–53. [Google Scholar] [CrossRef]
- Apushkin, M.A.; Fishman, G.A.; Rajagopalan, A.S. Fundus findings and longitudinal study of visual acuity loss in patients with X-linked retinoschisis. Retina 2005, 25, 612–618. [Google Scholar] [CrossRef]
- Forsius, H. Visual acuity in 183 cases of X-chromosomal retinoschisis. Can. J. Ophthalmol. 1973, 8, 385–393. [Google Scholar]
- George, N.; Yates, J.; Moore, A. Clinical features in affected males with X-linked retinoschisis. Arch. Ophthalmol. 1996, 114, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Pimenides, D.; George, N.; Yates, J.; Bradshaw, K.; Roberts, S.; Moore, A.; Trump, D. X-linked retinoschisis: Clinical phenotype and RS1 genotype in 86 UK patients. J. Med. Genet. 2005, 42, e35. [Google Scholar] [CrossRef]
- Lin, Z.; Zang, S.; Maman Lawali, D.J.A.; Xiao, Y.; Zeng, X.; Yu, H.; Hu, Y. Investigation of Correlations between Optical Coherence Tomography Biomarkers and Visual Acuity in X-Linked Retinoschisis. Front. Med. 2022, 8, 734888. [Google Scholar] [CrossRef] [PubMed]
- Pennesi, M.E.; Birch, D.G.; Jayasundera, K.T.; Parker, M.; Tan, O.; Gurses-Ozden, R.; Reichley, C.; Beasley, K.N.; Yang, P.; Weleber, R.G.; et al. Prospective evaluation of patients with X-linked retinoschisis during 18 months. Investig. Ophthalmol. Vis. Sci. 2018, 59, 5941–5956. [Google Scholar]
- Wei, X.; Sui, R. A Review of Machine Learning Algorithms for Retinal Cyst Segmentation on Optical Coherence Tomography. Sensors 2023, 23, 3144. [Google Scholar] [CrossRef] [PubMed]
- Bogunović, H.; Venhuizen, F.; Klimscha, S.; Apostolopoulos, S.; Bab-Hadiashar, A.; Bagci, U.; Beg, M.F.; Bekalo, L.; Chen, Q.; Ciller, C.; et al. RETOUCH: The retinal OCT fluid detection and segmentation benchmark and challenge. IEEE Trans. Med. Imaging 2019, 38, 1858–1874. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, X.; Zhu, W.; Cheng, X.; Xiang, D.; Shi, F. Speckle noise reduction in optical coherence tomography images based on edge-sensitive cGAN. Biomed. Opt. Express 2018, 9, 5129–5146. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Wu, J.; Philip, A.M.; Podkowinski, D.; Gerendas, B.S.; Langs, G.; Simader, C.; Waldstein, S.M.; Schmidt-Erfurth, U.M. Multivendor spectral-domain optical coherence tomography dataset, observer annotation performance evaluation, and standardized evaluation framework for intraretinal cystoid fluid segmentation. J. Ophthalmol. 2016, 2016, 3898750. [Google Scholar] [CrossRef]
- Chiu, S.J.; Allingham, M.J.; Mettu, P.S.; Cousins, S.W.; Izatt, J.A.; Farsiu, S. Kernel regression based segmentation of optical coherence tomography images with diabetic macular edema. Biomed. Opt. Express 2015, 6, 1172–1194. [Google Scholar] [CrossRef]
- Rashno, A.; Nazari, B.; Koozekanani, D.D.; Drayna, P.M.; Sadri, S.; Rabbani, H.; Parhi, K.K. Fully-automated segmentation of fluid regions in exudative age-related macular degeneration subjects: Kernel graph cut in neutrosophic domain. PLoS ONE 2017, 12, e0186949. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Bai, W.; Davies, R.H.; Bhuva, A.N.; Manisty, C.H.; Augusto, J.B.; Moon, J.C.; Aung, N.; Lee, A.M.; Sanghvi, M.M.; et al. Improving the generalizability of convolutional neural network-based segmentation on CMR images. Front. Cardiovasc. Med. 2020, 7, 105. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, X.; Yang, D.; Sanford, T.; Harmon, S.; Turkbey, B.; Wood, B.J.; Roth, H.; Myronenko, A.; Xu, D.; et al. Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans. Med. Imaging 2020, 39, 2531–2540. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yu, L.; Li, K.; Yang, X.; Fu, C.W.; Heng, P.A. Dofe: Domain-oriented feature embedding for generalizable fundus image segmentation on unseen datasets. IEEE Trans. Med. Imaging 2020, 39, 4237–4248. [Google Scholar] [CrossRef]
- Liu, Q.; Chen, C.; Qin, J.; Dou, Q.; Heng, P.A. FedDG: Federated domain generalization on medical image segmentation via episodic learning in continuous frequency space. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 1013–1023. [Google Scholar]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le, Q.V. Autoaugment: Learning augmentation strategies from data. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 113–123. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Zhou, Z.; Rahman Siddiquee, M.M.; Tajbakhsh, N.; Liang, J. UNet++: A nested U-Net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer: Cham, Switzerland, 2018; pp. 3–11. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention U-Net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Drozdzal, M.; Vorontsov, E.; Chartrand, G.; Kadoury, S.; Pal, C. The importance of skip connections in biomedical image segmentation. In Proceedings of the Deep Learning and Data Labeling for Medical Applications, Athens, Greece, 21 October 2016; pp. 179–187. [Google Scholar]
- Chen, J.; Lu, Y.; Yu, Q.; Luo, X.; Adeli, E.; Wang, Y.; Lu, L.; Yuille, A.L.; Zhou, Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021, arXiv:2102.04306. [Google Scholar]
- Varnousfaderani, E.S.; Wu, J.; Vogl, W.D.; Philip, A.M.; Montuoro, A.; Leitner, R.; Simader, C.; Waldstein, S.M.; Gerendas, B.S.; Schmidt-Erfurth, U. A novel benchmark model for intelligent annotation of spectral-domain optical coherence tomography scans using the example of cyst annotation. Comput. Methods Programs Biomed. 2016, 130, 93–105. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Colson, B.; Marcotte, P.; Savard, G. An overview of bilevel optimization. Ann. Oper. Res. 2007, 153, 235–256. [Google Scholar] [CrossRef]
- Schulman, J.; Wolski, F.; Dhariwal, P.; Radford, A.; Klimov, O. Proximal policy optimization algorithms. arXiv 2017, arXiv:1707.06347. [Google Scholar]
- Rasti, R.; Biglari, A.; Rezapourian, M.; Yang, Z.; Farsiu, S. RetiFluidNet: A Self-Adaptive and Multi-Attention Deep Convolutional Network for Retinal OCT Fluid Segmentation. IEEE Trans. Med. Imaging 2023, 42, 1413–1423. [Google Scholar] [CrossRef]
- Lim, S.; Kim, I.; Kim, T.; Kim, C.; Kim, S. Fast autoaugment. Adv. Neural Inf. Process. Syst. 2019, 32, 6665–6675. [Google Scholar]
- Yang, D.; Roth, H.; Xu, Z.; Milletari, F.; Zhang, L.; Xu, D. Searching learning strategy with reinforcement learning for 3D medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Shenzhen, China, 13–17 October 2019; Springer: Cham, Switzerland, 2019; pp. 3–11. [Google Scholar]
- Xu, J.; Li, M.; Zhu, Z. Automatic data augmentation for 3D medical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Lima, Peru, 4–8 October 2020; Springer: Cham, Switzerland, 2020; pp. 378–387. [Google Scholar]
- Castro, E.; Cardoso, J.S.; Pereira, J.C. Elastic deformations for data augmentation in breast cancer mass detection. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Las Vegas, NV, USA, 4–7 March 2018; pp. 230–234. [Google Scholar]
- Nalepa, J.; Marcinkiewicz, M.; Kawulok, M. Data augmentation for brain-tumor segmentation: A review. Front. Comput. Neurosci. 2019, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Chen, K.; Pan, M.; Wang, M.; Song, Z. An augmentation strategy for medical image processing based on statistical shape model and 3D thin plate spline for deep learning. IEEE Access 2019, 7, 133111–133121. [Google Scholar] [CrossRef]
- Madumal, P.; Miller, T.; Sonenberg, L.; Vetere, F. Explainable reinforcement learning through a causal lens. Proc. AAAI Conf. Artif. Intell. 2020, 34, 2493–2500. [Google Scholar] [CrossRef]
- Wang, X.; Meng, F.; Liu, X.; Kong, Z.; Chen, X. Causal explanation for reinforcement learning: Quantifying state and temporal importance. Appl. Intell. 2023, 1–19. [Google Scholar] [CrossRef]
- Huang, Q.; Sun, J.; Ding, H.; Wang, X.; Wang, G. Robust liver vessel extraction using 3D U-Net with variant dice loss function. Comput. Biol. Med. 2018, 101, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Fang, B.; Liu, Y.; Gao, M.; Zheng, S.; Wang, Y. Liver vessels segmentation based on 3D residual U-NET. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 250–254. [Google Scholar]
- Kazerouni, A.; Aghdam, E.K.; Heidari, M.; Azad, R.; Fayyaz, M.; Hacihaliloglu, I.; Merhof, D. Diffusion Models for Medical Image Analysis: A Comprehensive Survey. arXiv 2022, arXiv:2211.07804. [Google Scholar]
- Zhang, Z.; Wu, C.; Coleman, S.; Kerr, D. DENSE-INception U-net for medical image segmentation. Comput. Methods Programs Biomed. 2020, 192, 105395. [Google Scholar] [CrossRef] [PubMed]
- Ibtehaz, N.; Rahman, M.S. MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Netw. 2020, 121, 74–87. [Google Scholar] [CrossRef]
- Frid-Adar, M.; Ben-Cohen, A.; Amer, R.; Greenspan, H. Improving the segmentation of anatomical structures in chest radiographs using U-Net with an Imagenet pre-trained encoder. In Image Analysis for Moving Organ, Breast, and Thoracic Images, Proceedings of the 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, 20 September 2018; Springer: Cham, Switzerland, 2018; pp. 159–168. [Google Scholar]
- Waiker, D.; Baghel, P.D.; Varma, K.R.; Sahu, S.P. Effective semantic segmentation of lung X-ray images using U-Net architecture. In Proceedings of the 2020 Fourth International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 11–13 March 2020; pp. 603–607. [Google Scholar]
- Orlando, J.I.; Seeböck, P.; Bogunović, H.; Klimscha, S.; Grechenig, C.; Waldstein, S.; Gerendas, B.S.; Schmidt-Erfurth, U. U2-Net: A bayesian U-Net model with epistemic uncertainty feedback for photoreceptor layer segmentation in pathological OCT scans. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1441–1445. [Google Scholar]
- Asgari, R.; Waldstein, S.; Schlanitz, F.; Baratsits, M.; Schmidt-Erfurth, U.; Bogunović, H. U-Net with spatial pyramid pooling for drusen segmentation in optical coherence tomography. In Proceedings of the International Workshop on Ophthalmic Medical Image Analysis, Shenzhen, China, 17 October 2019; Springer: Cham, Switzerland, 2019; pp. 77–85. [Google Scholar]
- Schlegl, T.; Waldstein, S.M.; Bogunovic, H.; Endstraßer, F.; Sadeghipour, A.; Philip, A.M.; Podkowinski, D.; Gerendas, B.S.; Langs, G.; Schmidt-Erfurth, U. Fully automated detection and quantification of macular fluid in OCT using deep learning. Ophthalmology 2018, 125, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Kim, Y.; Zhou, L.; Plichta, K.; Allen, B.; Buatti, J.; Wu, X. 3D fully convolutional networks for co-segmentation of tumors on PET-CT images. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 228–231. [Google Scholar]
- Wang, H.; Wang, Z.; Wang, J.; Li, K.; Geng, G.; Kang, F.; Cao, X. ICA-Unet: An improved U-net network for brown adipose tissue segmentation. J. Innov. Opt. Health Sci. 2022, 15, 2250018. [Google Scholar] [CrossRef]

| Operation | Magnitude | Operation | Magnitude |
|---|---|---|---|
| ShearX(Y) | [−0.3, 0.3] | Sharpness | [0, 0.9] |
| TranslateX(Y) | [−150, 150] | Brightness | [0, 0.9] |
| Rotate | [−30, 30] | AutoContrast | - |
| Color | [0.1, 0.9] | Equalize | - |
| Posterize | [4, 8] | Invert | - |
| Solarize | [0, 256] | Horizontal Flip | - |
| Contrast | [0, 0.9] | Vertical Flip | - |
| Sub-Strategy Index | Operation 1 | Operation 1 Parameter | Operation 2 | Operation 2 Parameter |
|---|---|---|---|---|
| 1 | Sharpness | 2, 0.5 | AutoContrast | -, 0.4 |
| 2 | Equalize | -, 0.9 | TranslateY | 4, 0.7 |
| 3 | Rotate | 3, 0.5 | Posterize | 9, 0.4 |
| 4 | Sharpness | 8, 0.8 | Invert | -, 0.3 |
| 5 | Contrast | 8, 0.4 | TranslateY | 3, 0.8 |
| 6 | Brightness | 4, 0.7 | Equalize | -, 0.2 |
| 7 | ShearX | 1, 0.2 | Posterize | 3, 0.9 |
| 8 | Solarize | 2, 0.7 | Sharpness | 7, 0.2 |
| 9 | AutoContrast | -, 0.4 | Horizontal Flip | -, 0.4 |
| 10 | TranslateY | 4, 0.7 | Sharpness | 2, 0.8 |
| 11 | Solarize | 7, 0.2 | Brightness | 2, 0.6 |
| 12 | Invert | -, 0.8 | Solarize | 7, 0.2 |
| 13 | Sharpness | 3, 0.6 | Rotate | 8, 0.3 |
| 14 | Equalize | -, 0.4 | Contrast | 8, 0.4 |
| 15 | Solarize | 7, 0.2 | Brightness | 5, 0.2 |
| 16 | Equalize | -, 0.9 | Contrast | 3, 0.6 |
| 17 | Sharpness | 2, 0.2 | Invert | -, 0.7 |
| 18 | Contrast | 3, 0.3 | Invert | -, 0.3 |
| 19 | Sharpness | 2, 0.9 | TranslateY | 7, 0.7 |
| 20 | AutoContrast | -, 0.8 | Posterize | 5, 0.2 |
| 21 | ShearX | 1, 1 | Rotate | 7, 0.7 |
| 22 | Contrast | -, 0.3 | Equalize | -, 0.2 |
| 23 | Equalize | -, 0.3 | Sharpness | 8, 0.8 |
| 24 | TranslateY | 3, 0.1 | Equalize | -, 0.9 |
| 25 | Solarize | 4, 0.1 | Contrast | 2, 0.5 |
| Model | Accuracy | Dice | Precision | Recall | Specificity | Jaccard Index |
|---|---|---|---|---|---|---|
| U-Net | 98.34 ± 0.60 | 79.50 ± 2.50 | 83.26 ± 2.83 | 76.21 ± 4.26 | 99.34 ± 0.19 | 66.04 ± 3.48 |
| RL + U-Net | 99.03 ± 0.20 | 80.77 ± 3.12 | 83.59 ± 5.10 | 78.48 ± 5.36 | 99.60 ± 0.08 | 67.84 ± 4.47 |
| U-Net++ | 98.50 ± 0.52 | 81.55 ± 1.84 | 85.09 ± 3.75 | 78.41 ± 2.55 | 99.42 ± 0.15 | 68.88 ± 2.62 |
| RL + U-Net++ | 99.27 ± 0.14 | 85.68 ± 2.13 | 86.91 ± 1.80 | 84.52 ± 3.02 | 99.66 ± 0.08 | 75.00 ± 3.27 |
| Attention U-Net | 98.59 ± 0.42 | 82.26 ± 2.55 | 86.02 ± 3.62 | 78.94 ± 3.55 | 99.46 ± 0.15 | 69.93 ± 3.69 |
| RL + Attention U-Net | 99.21 ± 0.16 | 84.56 ± 2.42 | 86.31 ± 2.90 | 83.54 ± 4.02 | 99.63 ± 0.08 | 73.31 ± 3.66 |
| Residual U-Net | 98.44 ± 0.61 | 80.56 ± 3.00 | 86.07 ± 4.20 | 75.96 ± 5.14 | 99.49 ± 0.12 | 67.53 ± 4.16 |
| RL + Residual U-Net | 99.19 ± 0.17 | 83.93 ± 2.34 | 86.07 ± 2.80 | 81.97 ± 3.42 | 99.65 ± 0.07 | 72.36 ± 3.51 |
| TransUNet | 98.42 ± 0.24 | 79.69 ± 5.02 | 81.73 ± 5.49 | 77.95 ± 6.49 | 99.26 ± 0.25 | 66.47 ± 7.00 |
| RL + TransUNet | 98.95 ± 0.22 | 80.31 ± 4.22 | 81.87 ± 3.22 | 78.42 ± 6.33 | 99.55 ± 0.12 | 66.54 ± 6.84 |
| RetiFluidNet + Random | 98.42 ± 0.59 | 82.37 ± 3.35 | 83.28 ± 4.80 | 80.38± 4.78 | 98.99 ± 0.27 | 70.03 ± 4.39 |
| Accuracy | Dice | Precision | Recall | Specificity | Jaccard Index | |
|---|---|---|---|---|---|---|
| UNet | 0.040 | 0.497 | 0.904 | 0.480 | 0.023 | 0.496 |
| UNet++ | 0.031 | 0.043 | 0.199 | 0.061 | 0.032 | 0.049 |
| Attention U-Net | 0.006 | 0.837 | 0.660 | 0.909 | 0.045 | 0.587 |
| Residual U-Net | 0.014 | 0.011 | 0.592 | 0.032 | 0.051 | 0.011 |
| TransUNet | 0.013 | 0.182 | 0.357 | 0.009 | 0.014 | 0.184 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, X.; Li, H.; Zhu, T.; Li, W.; Li, Y.; Sui, R. Deep Learning with Automatic Data Augmentation for Segmenting Schisis Cavities in the Optical Coherence Tomography Images of X-Linked Juvenile Retinoschisis Patients. Diagnostics 2023, 13, 3035. https://doi.org/10.3390/diagnostics13193035
Wei X, Li H, Zhu T, Li W, Li Y, Sui R. Deep Learning with Automatic Data Augmentation for Segmenting Schisis Cavities in the Optical Coherence Tomography Images of X-Linked Juvenile Retinoschisis Patients. Diagnostics. 2023; 13(19):3035. https://doi.org/10.3390/diagnostics13193035
Chicago/Turabian StyleWei, Xing, Hui Li, Tian Zhu, Wuyi Li, Yamei Li, and Ruifang Sui. 2023. "Deep Learning with Automatic Data Augmentation for Segmenting Schisis Cavities in the Optical Coherence Tomography Images of X-Linked Juvenile Retinoschisis Patients" Diagnostics 13, no. 19: 3035. https://doi.org/10.3390/diagnostics13193035
APA StyleWei, X., Li, H., Zhu, T., Li, W., Li, Y., & Sui, R. (2023). Deep Learning with Automatic Data Augmentation for Segmenting Schisis Cavities in the Optical Coherence Tomography Images of X-Linked Juvenile Retinoschisis Patients. Diagnostics, 13(19), 3035. https://doi.org/10.3390/diagnostics13193035





