One-Stage Detection without Segmentation for Multi-Type Coronary Lesions in Angiography Images Using Deep Learning
Abstract
:1. Introduction
- This study enrolled angiography images from patients who were candidates for coronary artery bypass (CAB) surgery for the first time to evaluate the detection performance of DL techniques with complex lesions.
- A single-stage detection model by the region-free approach was employed for the first time to detect vascular lesions directly, aiming to improve detection efficiency.
- A more detailed classification of vascular stenoses was performed, providing a comprehensive evaluation of the network model’s performance among different types of lesions.
2. Materials and Methods
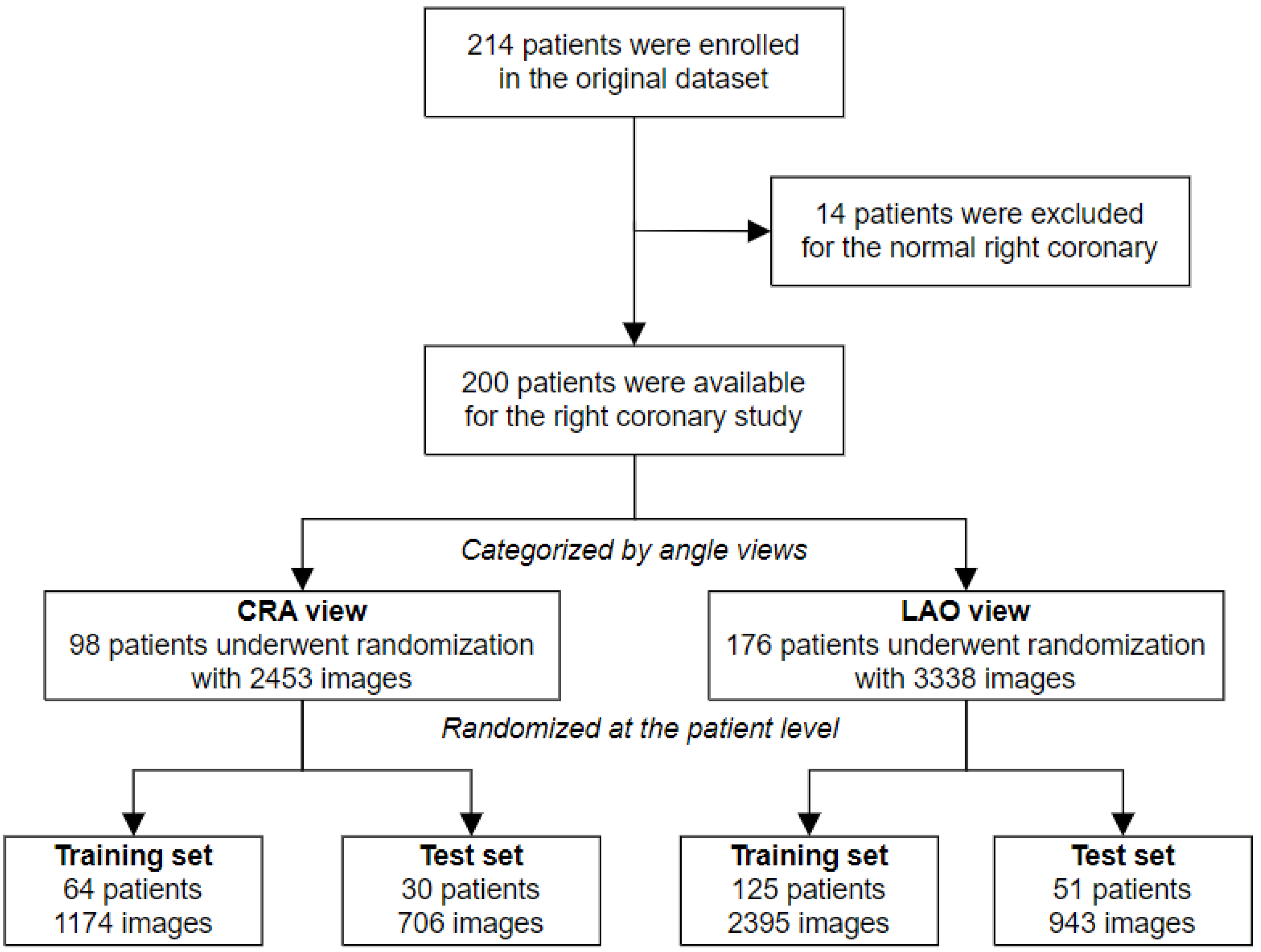
2.1. Dataset Characteristics
2.2. Reference Standard and Annotation Procedures
2.3. Experimental Environment and Methodology
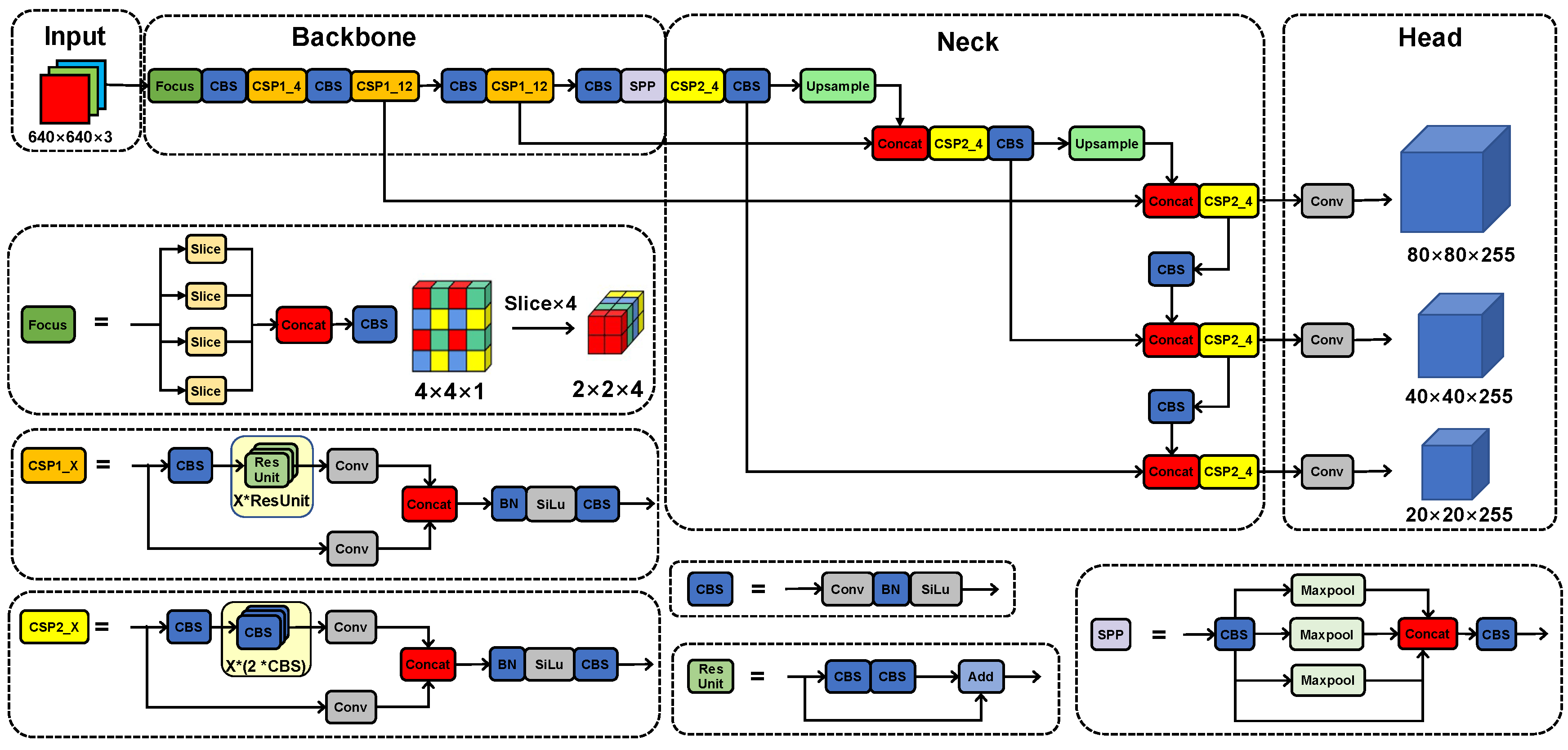
2.4. Architecture of Models
2.4.1. The YOLOv5x Model
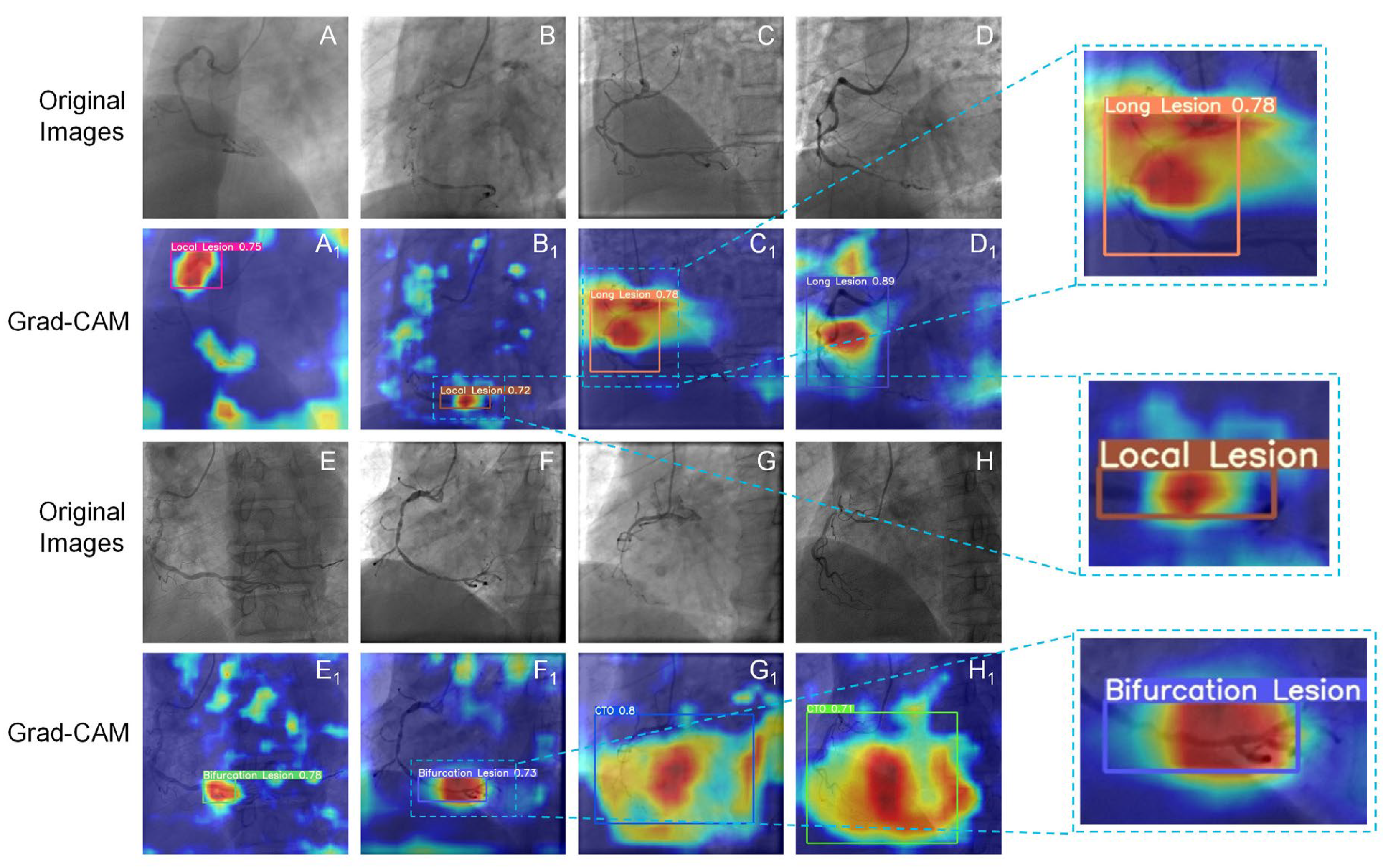
2.4.2. The Grad-CAM Technique
2.5. Performance Evaluation
2.6. Statistics
3. Results
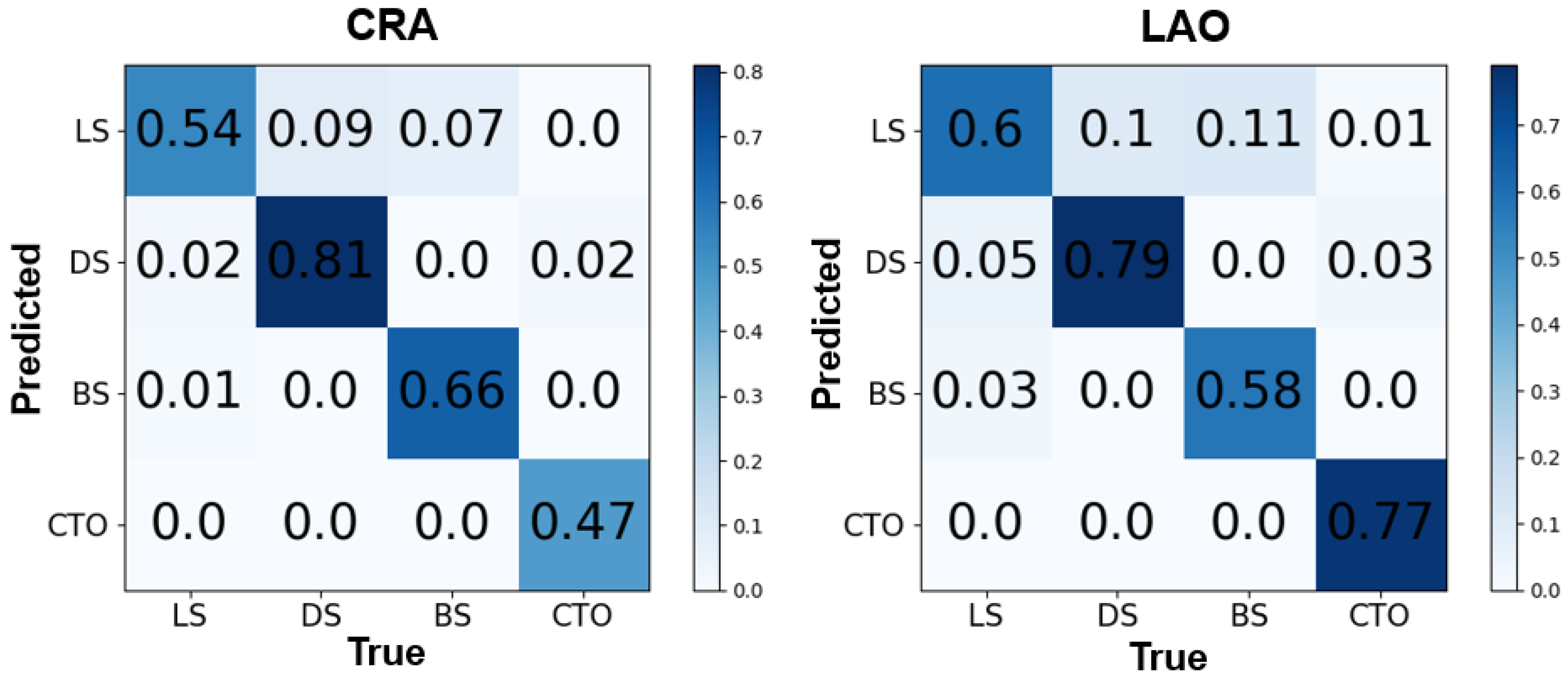
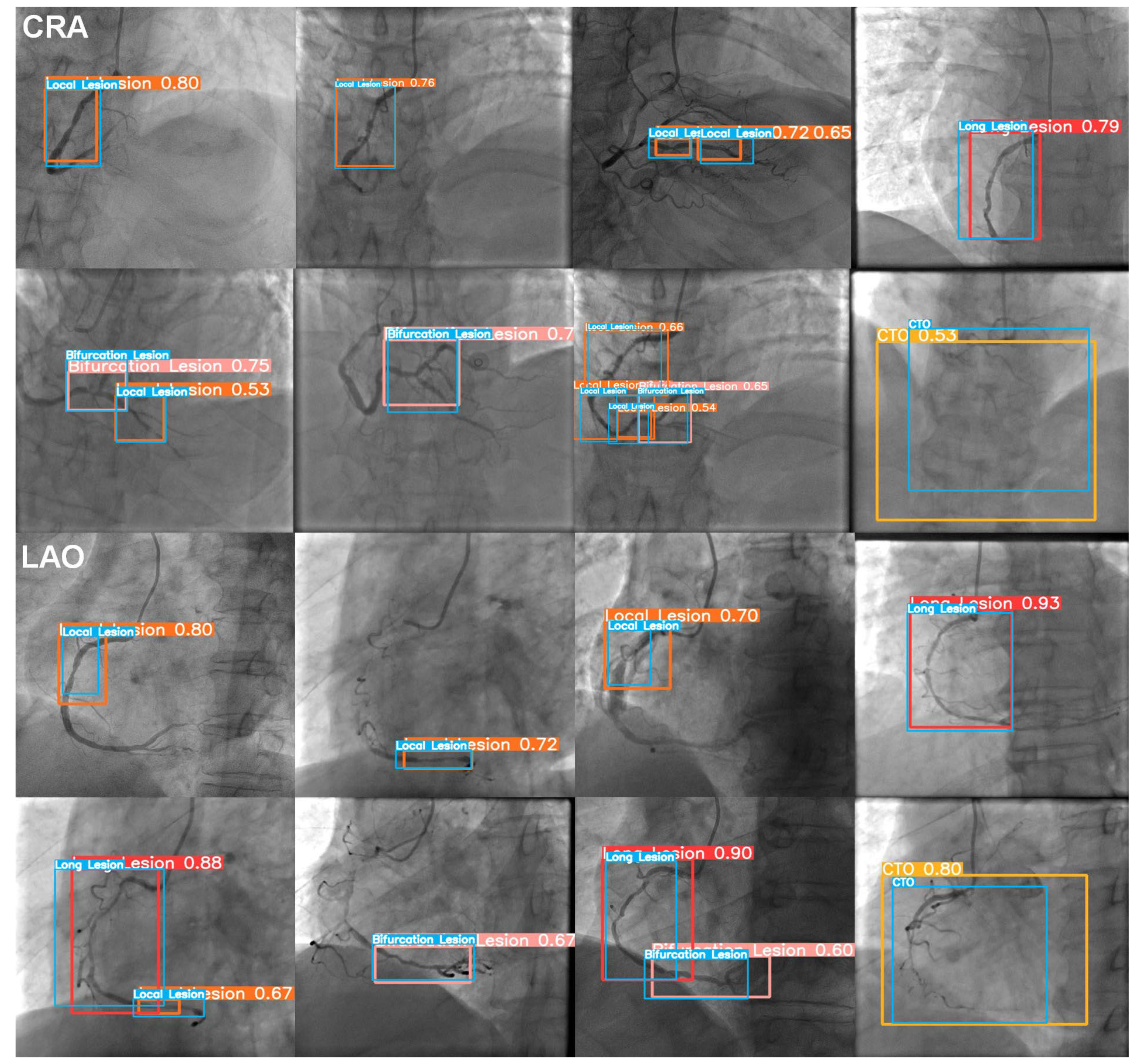
3.1. The Image Level
3.2. The Patient Level
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| BS | Bifurcation Stenosis |
| CAB | Coronary Artery Bypass |
| CAD | Coronary Artery Disease |
| CAG | Coronary AngioGraphy |
| CNN | Convolutional Neural Network |
| CPR | Cardiovascular Pulmonary Resuscitation |
| CRA | CRAnial |
| CTO | Chronic Total Occlusion |
| DICOM | Digital Imaging and COmmunications in Medicine |
| DL | Deep Learning |
| DS | Diffuse Stenosis |
| FN | False Negative |
| FP | False Positive |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| IoU | Intersection over Union |
| LAO | Left Anterior Oblique |
| LS | Local Stenosis |
| mAP | mean Average Precision |
| mFP | mean False Positive |
| PCI | Percutaneous Coronary Intervention |
| PR | Precision-Recall |
| TN | True Negative |
| TP | True Positive |
References
- The Top 10 Causes of Death. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 9 December 2020).
- Collet, J.-P.; Thiele, H.; Barbato, E.; Barthélémy, O.; Bauersachs, J.; Bhatt, D.L.; Dendale, P.; Dorobantu, M.; Edvardsen, T.; Folliguet, T.; et al. 2020 ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation. Eur. Heart J. 2021, 42, 1289–1367. [Google Scholar] [PubMed]
- Lawton, J.S.; Tamis-Holland, J.E.; Bangalore, S.; Bates, E.R.; Beckie, T.M.; Bischoff, J.M.; Bittl, J.A.; Cohen, M.G.; DiMaio, J.M.; Don, C.W.; et al. 2021 ACC/AHA/SCAI Guideline for Coronary Artery Revascularization: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. Circulation 2022, 145, e18–e114. [Google Scholar] [PubMed]
- Knuuti, J.; Wijns, W.; Saraste, A.; Capodanno, D.; Barbato, E.; Funck-Brentano, C.; Prescott, E.; Storey, R.F.; Deaton, C.; Cuisset, T.; et al. 2019 ESC Guidelines for the diagnosis and management of chronic coronary syndromes. Eur. Heart J. 2020, 41, 407–477. [Google Scholar] [PubMed]
- Zhang, D.; Liu, X.; Xia, J.; Gao, Z.; Zhang, H.; de Albuquerque, V.H.C. A Physics-guided Deep Learning Approach for Functional Assessment of Cardiovascular Disease in IoT-based Smart Health. IEEE Internet Things J. 2023, 1. [Google Scholar] [CrossRef]
- Menezes, M.N.; Silva, J.L.; Silva, B.; Rodrigues, T.; Guerreiro, C.; Guedes, J.P.; Santos, M.O.; Oliveira, A.L.; Pinto, F.J. Coronary X-ray angiography segmentation using Artificial Intelligence: A multicentric validation study of a deep learning model. Int. J. Cardiovasc. Imaging 2023, 39, 1385–1396. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, Z.; Zhang, D.; Hau, W.K.; Zhang, H. Progressive Perception Learning for Main Coronary Segmentation in X-Ray Angiography. IEEE Trans. Med. Imaging 2023, 42, 864–879. [Google Scholar]
- Zhao, C.; Vij, A.; Malhotra, S.; Tang, J.; Tang, H.; Pienta, D.; Xu, Z.; Zhou, W. Automatic extraction and stenosis evaluation of coronary arteries in invasive coronary angiograms. Comput. Biol. Med. 2021, 136, 104667. [Google Scholar] [CrossRef]
- Liu, X.; Wang, X.; Chen, D.; Zhang, H. Automatic Quantitative Coronary Analysis Based on Deep Learning. Appl. Sci. 2023, 13, 2975. [Google Scholar] [CrossRef]
- Algarni, M.; Al-Rezqi, A.; Saeed, F.; Alsaeedi, A.; Ghabban, F. Multi-constraints based deep learning model for automated segmentation and diagnosis of coronary artery disease in X-ray angiographic images. PeerJ Comput. Sci. 2022, 8, e933. [Google Scholar] [CrossRef]
- Cong, C.; Kato, Y.; De Vasconcellos, H.D.; Ostovaneh, M.R.; Lima, J.A.C.; Ambale-Venkatesh, B. Deep learning-based end-to-end automated stenosis classification and localization on catheter coronary angiography. Front. Cardiovasc. Med. 2023, 10, 944135. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015. pp. 1–9.
- Moon, J.H.; Lee, D.Y.; Cha, W.C.; Chung, M.J.; Lee, K.-S.; Cho, B.H.; Choi, J.H. Automatic stenosis recognition from coronary angiography using convolutional neural networks. Comput. Methods Programs Biomed. 2020, 198, 105819. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweom, I.S. CBAM: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; Volume 11211, pp. 3–19. [Google Scholar]
- Ling, H.; Chen, B.; Guan, R.; Xiao, Y.; Yan, H.; Chen, Q.; Bi, L.; Chen, J.; Feng, X.; Pang, H.; et al. Deep Learning Model for Coronary Angiography. J. Cardiovasc. Transl. Res. 2023, 16, 896–904. [Google Scholar] [CrossRef] [PubMed]
- Du, T.; Liu, X.; Zhang, H.; Xu, B. Real-time Lesion Detection of Cardiac Coronary Artery Using Deep Neural Networks. In Proceedings of the 2018 International Conference on Network Infrastructure and Digital Content (IC-NIDC), Guiyang, China, 22–24 August 2018; pp. 150–154. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Danilov, V.V.; Klyshnikov, K.Y.; Gerget, O.M.; Kutikhin, A.G.; Ganyukov, V.I.; Frangi, A.F.; Ovcharenko, E.A. Real-time coronary artery stenosis detection based on modern neural networks. Sci. Rep. 2021, 11, 7582. [Google Scholar] [CrossRef]
- Antczak, K.; Liberadzki, A. Stenosis Detection with Deep Convolutional Neural Networks. MATEC Web Conf. 2018, 210, 04001. [Google Scholar] [CrossRef]
- Ovalle-Magallanes, E.; Avina-Cervantes, J.G.; Cruz-Aceves, I.; Ruiz-Pinales, J. Transfer Learning for Stenosis Detection in X-ray Coronary Angiography. Mathematics 2020, 8, 1510. [Google Scholar] [CrossRef]
- Du, T.; Xie, L.; Zhang, H.; Liu, X.; Wang, X.; Chen, D.; Xu, Y.; Sun, Z.; Zhou, W.; Song, L.; et al. Training and validation of a deep learning architecture for the automatic analysis of coronary angiography. EuroIntervention 2021, 17, 32–40. [Google Scholar] [CrossRef]
- Pang, K.; Ai, D.; Fang, H.; Fan, J.; Song, H.; Yang, J. Stenosis-DetNet: Sequence consistency-based stenosis detection for X-ray coronary angiography. Comput. Med. Imaging Graph. 2021, 89, 101900. [Google Scholar] [CrossRef]
- Dingli, P.; Gonzalo, N.; Escaned, J. Intravascular Ultrasound-guided Management of Diffuse Stenosis. Radcl. Cardiol. 2018, 2018, 1–18. [Google Scholar]
- Levine, G.N.; Bates, E.R.; Blankenship, J.C.; Bailey, S.R.; Bittl, J.A.; Cercek, B.; Chambers, C.E.; Ellis, S.G.; Guyton, R.A.; Hollenberg, S.M.; et al. 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation 2011, 124, e574–e651. [Google Scholar]
- Louvard, Y.; Thomas, M.; Dzavik, V.; Hildick-Smith, D.; Galassi, A.R.; Pan, M.; Burzotta, F.; Zelizko, M.; Dudek, D.; Ludman, P.; et al. Classification of coronary artery bifurcation lesions and treatments: Time for a consensus! Catheter. Cardiovasc. Interv. 2007, 71, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Ultralytics. GitHub-Ultralytics/Yolov5: YOLOv5 in PyTorch > ONNX > CoreML > TFLite. 2020. Available online: https://github.com/ultralytics/yolov5 (accessed on 26 June 2020).
- Lin, T.Y.; Dollár, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature Pyramid Networks for Object Detection. arXiv 2016, arXiv:1612.03144. [Google Scholar]
- Liu, S.; Qi, L.; Qin, H.; Shi, J.; Jia, J. Path Aggregation Network for Instance Segmentation. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8759–8768. [Google Scholar]
- Zheng, Z.; Wang, P.; Liu, W.; Li, J.; Ye, R.; Ren, D. Distance-IoU Loss: Faster and Better Learning for Bounding Box Regression. arXiv 2019, arXiv:1911.08287. [Google Scholar] [CrossRef]
- Rezatofighi, H.; Tsoi, N.; Gwak, J.; Sadeghian, A.; Reid, I.; Savarese, S. Generalized Intersection Over Union: A Metric and a Loss for Bounding Box Regression. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 658–666. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-based Localization. arXiv 2016, arXiv:1610.02391. [Google Scholar]
- Dinesh, M.G.; Bacanin, N.; Askar, S.S.; Abouhawwash, M. Diagnostic ability of deep learning in detection of pancreatic tumour. Sci. Rep. 2023, 13, 9725. [Google Scholar] [CrossRef]
- Zahrawi, M.; Shaalan, K. Improving video surveillance systems in banks using deep learning techniques. Sci. Rep. 2023, 13, 7911. [Google Scholar] [CrossRef]
- Chiriboga, M.; Green, C.M.; Hastman, D.A.; Mathur, D.; Wei, Q.; Díaz, S.A.; Medintz, I.L.; Veneziano, R. Rapid DNA origami nanostructure detection and classification using the YOLOv5 deep convolutional neural network. Sci. Rep. 2022, 12, 3871. [Google Scholar] [CrossRef]
- Alyoubi, W.L.; Abulkhair, M.F.; Shalash, W.M. Diabetic Retinopathy Fundus Image Classification and Lesions Localization System Using Deep Learning. Sensors 2021, 21, 3704. [Google Scholar] [CrossRef]
- Santos, C.; Aguiar, M.; Welfer, D.; Belloni, B. A New Approach for Detecting Fundus Lesions Using Image Processing and Deep Neural Network Architecture Based on YOLO Model. Sensors 2022, 22, 6441. [Google Scholar] [CrossRef]
- Li, T.; Bo, W.; Hu, C.; Kang, H.; Liu, H.; Wang, K.; Fu, H. Applications of deep learning in fundus images: A review. Med. Image Anal. 2021, 69, 101971. [Google Scholar] [CrossRef]




| Ref. | Methods | Data | Classes | Results |
|---|---|---|---|---|
| Zhao et al. (2021) [8] | FP-U-Net++, arterial centerline extraction, diameter calculation, arterial stenosis detection | 99 patients, 314 images | 1–24%, 25–49%, 50–69%, 70–100% | Precision = 0.6998, recall = 0.6840, |
| Liu et al. (2023) [9] | AI-QCA | 3275 patients, 13,222 images | 0–100% | Precision = 0.897, recall = 0.879 |
| Algarni et al. (2022) [10] | ASCARIS model | 130 images | normal and abnormal | Accuracy = 97%, recall = 95%, specificity = 93% |
| Cong et al. (2023) [11] | Inception-v3 and LSTM, redundancy training, and Inception-V3, FPN | 230 patients, 14,434 images | <25%, 25–99%, CTO | Accuracy = 0.85, recall = 0.96, AUC = 0.86 |
| Moon et al. (2020) [13] | GoogleNet Inception-v3, CBAM, Grad-CAM | 452 clips | Stenosis ≥ 50% | AUC = 0.971, accuracy = 0.934 |
| Ovalle-Magallanes et al. (2020) [20] | pre-trained CNN via Transfer Learning, CAM | 10,000 artificial images, 250 real images | Stenosis | Accuracy = 0.95, precision = 0.93, sensitivity = 0.98, specificity = 0.92, = 0.95 |
| Antczak et al. (2021) [19] | A patch-based CNN for stenosis detection | 10,000 artificial images, 250 real images | Stenosis | Accuracy = 90% |
| Du et al. (2021) [21] | A DNN for the recognition of lesion morphology | 10,073 patients, 20,612 images | Stenotic lesion, total occlusion, calcification, thrombus, and dissection | = 0.829, 0.810, 0.802, 0.823, 0.854 |
| Ling et al. (2023) [15] | DLCAG diagnose system | 949 patients, 2980 images | Stenosis | mAP = 86.3% |
| Danilov et al. (2021) [18] | Comparison of state-of-the-art CNN (N = 8) | 100 patients, 8325 images | Stenosis ≥ 70% | mAP = 0.94, F1 score = 0.96, prediction speed = 10 fps |
| Pang et al. (2021) [22] | Stenosis-DetNet with SFF and SCA | 166 sequence, 1494 images | Stenosis | Accuracy = 94.87%, sensitivity 82.22% |
| The CRA View | The LAO View | p Value | |
|---|---|---|---|
| Age, years | 63 ± 8 | 64 ± 9 | 0.54 |
| Gender | |||
| Male (%) | 68 (69%) | 118 (67%) | 0.72 |
| Images | 2453 | 3338 | 0.66 |
| Training Set (%) | 1747 | 2395 | |
| Test Set (%) | 706 | 943 | |
| Lesions | |||
| Training Set | 3259 | 1529 | <0.01 |
| LS | 2003 | 1005 | |
| DS | 376 | 96 | |
| BS | 500 | 375 | |
| CTO | 380 | 53 | |
| Test Set | 3874 | 1262 | <0.01 |
| LS | 2187 | 433 | |
| DS | 405 | 273 | |
| BS | 411 | 174 | |
| CTO | 871 | 382 |
| Lesions | Number | Precision | Recall | mAP@0.1 | mAP@0.5 | F1 Score | |
|---|---|---|---|---|---|---|---|
| CRA | LS | 1055 | 0.685 | 0.647 | 0.643 | 0.405 | 0.665 |
| DS | 96 | 0.458 | 0.844 | 0.687 | 0.677 | 0.594 | |
| BS | 374 | 0.656 | 0.658 | 0.675 | 0.625 | 0.657 | |
| CTO | 53 | 0.75 | 0.566 | 0.647 | 0.263 | 0.645 | |
| All | 1578 | 0.637 | 0.679 | 0.663 | 0.493 | 0.657 | |
| LAO | LS | 433 | 0.426 | 0.617 | 0.479 | 0.273 | 0.504 |
| DS | 273 | 0.648 | 0.868 | 0.773 | 0.688 | 0.742 | |
| BS | 174 | 0.699 | 0.655 | 0.694 | 0.521 | 0.676 | |
| CTO | 382 | 0.927 | 0.796 | 0.87 | 0.749 | 0.857 | |
| All | 1262 | 0.675 | 0.734 | 0.704 | 0.558 | 0.703 |
| Lesions | TP + FN | TP | FN | FP | P | R | F1 Score | mFP | |
|---|---|---|---|---|---|---|---|---|---|
| CRA | LS | 59 | 55 | 4 | 44 | 0.556 | 0.932 | 0.696 | 1.467 |
| DS | 6 | 6 | 0 | 8 | 0.429 | 1.000 | 0.600 | 0.267 | |
| BS | 15 | 13 | 2 | 20 | 0.394 | 0.867 | 0.542 | 0.667 | |
| CTO | 6 | 5 | 1 | 2 | 0.714 | 0.833 | 0.769 | 0.067 | |
| All | 86 | 79 | 7 | 74 | 0.523 | 0.908 | 0.652 | 2.467 | |
| LAO | LS | 28 | 24 | 4 | 57 | 0.296 | 0.857 | 0.440 | 1.118 |
| DS | 18 | 18 | 0 | 17 | 0.514 | 1.000 | 0.679 | 0.333 | |
| BS | 11 | 10 | 1 | 16 | 0.385 | 0.909 | 0.541 | 0.314 | |
| CTO | 19 | 19 | 0 | 5 | 0.792 | 1.000 | 0.884 | 0.098 | |
| All | 76 | 71 | 5 | 95 | 0.497 | 0.942 | 0.636 | 1.863 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, H.; Zhao, J.; Li, J.; Zeng, Y.; Wu, W.; Zhou, Z.; Wu, S.; Xu, L.; Song, M.; Yu, Q.; et al. One-Stage Detection without Segmentation for Multi-Type Coronary Lesions in Angiography Images Using Deep Learning. Diagnostics 2023, 13, 3011. https://doi.org/10.3390/diagnostics13183011
Wu H, Zhao J, Li J, Zeng Y, Wu W, Zhou Z, Wu S, Xu L, Song M, Yu Q, et al. One-Stage Detection without Segmentation for Multi-Type Coronary Lesions in Angiography Images Using Deep Learning. Diagnostics. 2023; 13(18):3011. https://doi.org/10.3390/diagnostics13183011
Chicago/Turabian StyleWu, Hui, Jing Zhao, Jiehui Li, Yan Zeng, Weiwei Wu, Zhuhuang Zhou, Shuicai Wu, Liang Xu, Min Song, Qibin Yu, and et al. 2023. "One-Stage Detection without Segmentation for Multi-Type Coronary Lesions in Angiography Images Using Deep Learning" Diagnostics 13, no. 18: 3011. https://doi.org/10.3390/diagnostics13183011
APA StyleWu, H., Zhao, J., Li, J., Zeng, Y., Wu, W., Zhou, Z., Wu, S., Xu, L., Song, M., Yu, Q., Song, Z., & Chen, L. (2023). One-Stage Detection without Segmentation for Multi-Type Coronary Lesions in Angiography Images Using Deep Learning. Diagnostics, 13(18), 3011. https://doi.org/10.3390/diagnostics13183011







