Multilevel Threshold Segmentation of Skin Lesions in Color Images Using Coronavirus Optimization Algorithm
Abstract
:1. Introduction
- COVIDOA is shown to deal with MLT in image segmentation.
- The hybridization of Otsu, Kapur, and Tsallis as a fitness function was used to present a skin cancer segmentation technique.
- Various segmentation levels are employed to assess the proposed technique’s performance.
- The proposed technique is compared to numerous popular meta-heuristics techniques.
- The effectiveness of the segmentation technique is validated by utilizing the MSE, PSNR, FSIM, and NCC matrices.
- The proposed technique may be expanded to accommodate various medical imaging diagnoses and used for additional benchmark images.
2. Literature Review
3. Materials and Methods
3.1. Multilevel Thresholding
3.1.1. Otsu’s (Between-Class Variance) Method
3.1.2. Kapur’s Entropy (Maximum Entropy Method)
3.1.3. T’sallis Entropy Method
3.1.4. Proposed Fitness Function
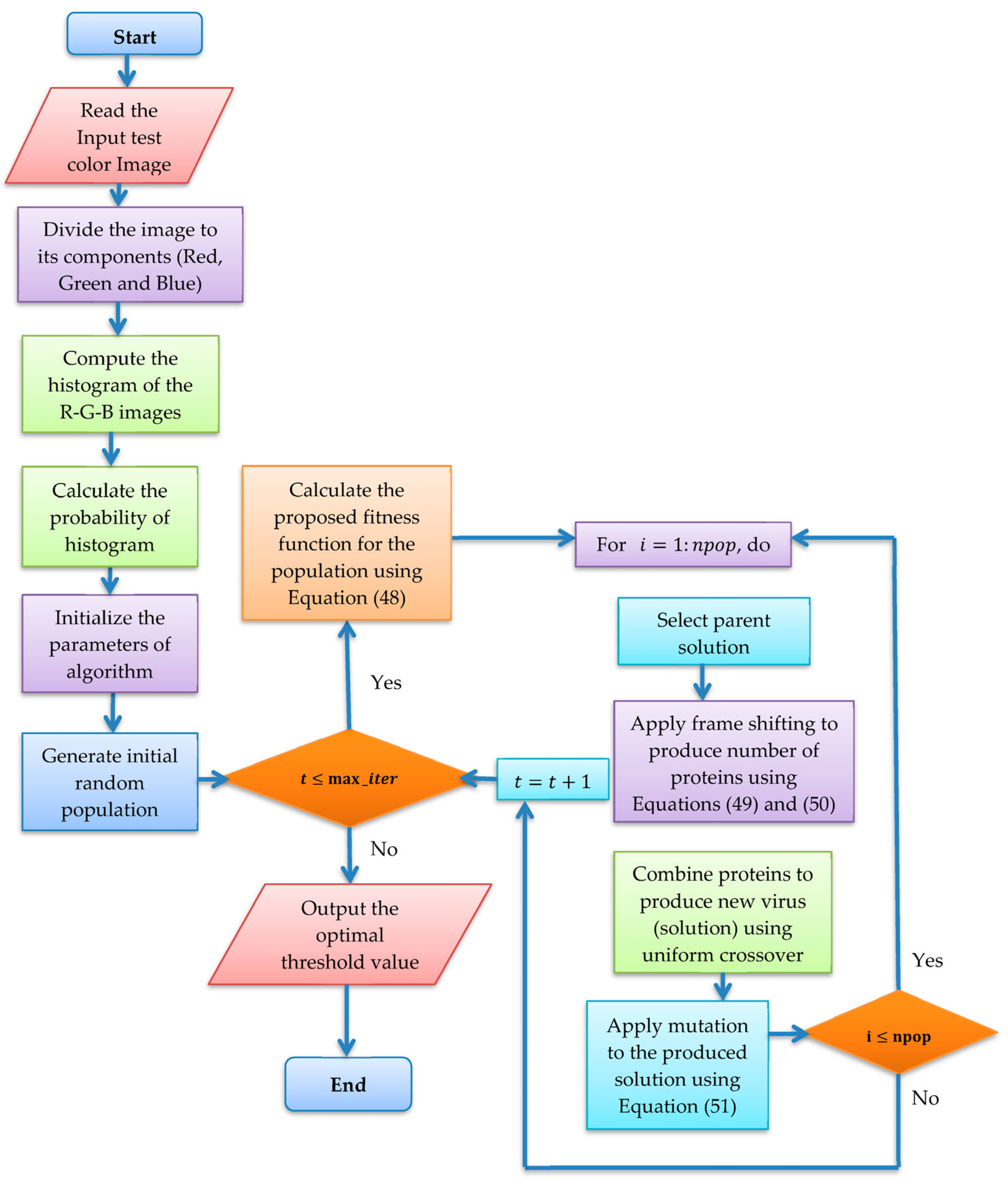
4. COVID Optimization Algorithm with the Proposed Fitness Function
- Virus entry and uncoating
- 2.
- Virus replication
- ▪ +1 frameshifting technique
- 3.
- Virus mutation
- 4
- New virion release
Computational Complexity Analysis
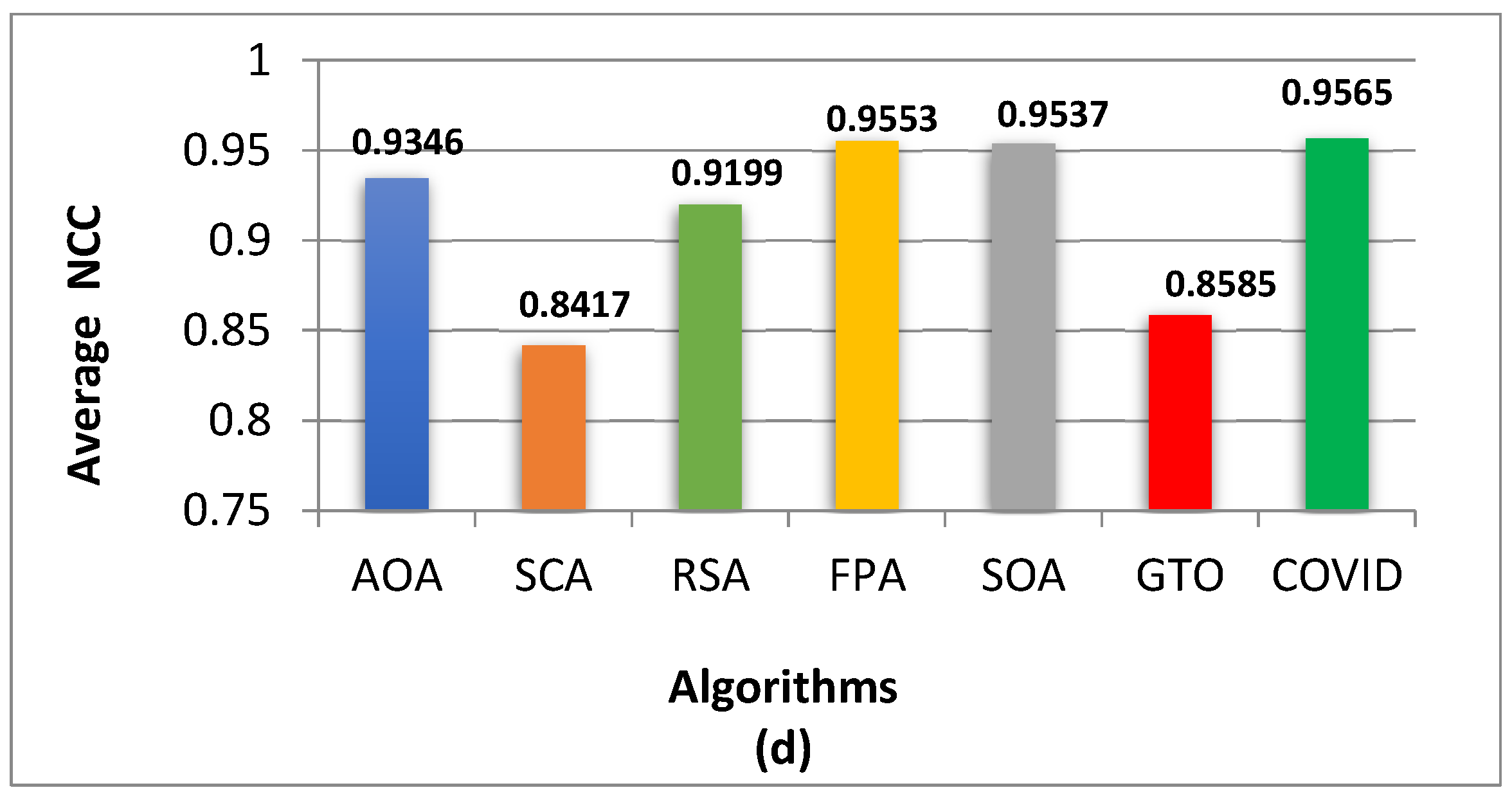
5. Experimental Results and Discussion
5.1. Dataset
5.2. Parameter Setting
- They have demonstrated their superior capacity to solve several optimization challenges, particularly image segmentation.
- The majority of them are current and have been published in reliable sources.
- Their MATLAB implementations are freely accessible on the MATLAB website (https://matlab.mathworks.com/ accessed on 18 August 2023).
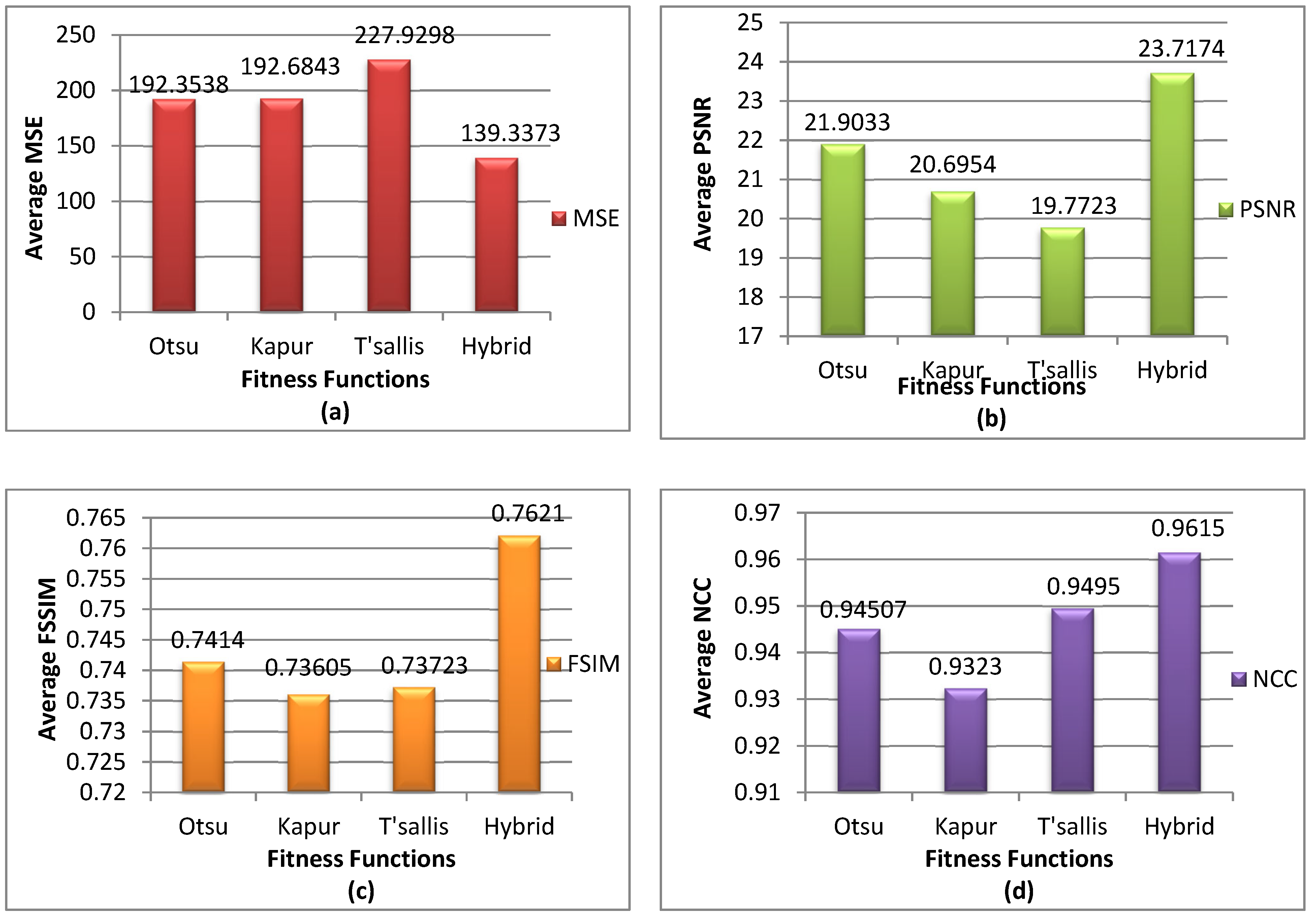
5.3. Performance Evaluation Criteria
5.3.1. Mean Square Error (MSE)
5.3.2. Peak Signal-to-Noise Ratio (PSNR)
5.3.3. Feature Similarity Index Metric (FSIM)
5.3.4. Normalized Correlation Coefficient (NCC)
5.4. Experimental Results
6. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SC | Skin Cancer |
| UVR | Ultraviolet Radiation |
| MLT | Multilevel Thresholding |
| COVIDOA | Coronavirus Disease Optimization Algorithm |
| AOA | Arithmetic Optimization Algorithm |
| SCA | Sine Cosine Algorithm |
| RSA | Reptile Search Algorithm |
| FPA | Flower Pollination Algorithm |
| SOA | Seagull Optimization Algorithm |
| GTO | Gorilla Troops Optimizer |
| MSE | Mean Square Error |
| PSNR | Peak Signal-to-Noise Ratio |
| FSIM | Feature Similarity Index Metric |
| NCC | Normalized Correlation Coefficient |
| CAD | Computer-Aided Diagnosis |
| AI | Artificial Intelligence |
| PSO | Particle Swarm Optimization |
| WOA | Whale Optimization Algorithm |
| CSA | Cuckoo Search Algorithm |
| HHOA | Harris Hawks Optimization Algorithm |
| GWOA | Gray Wolf Optimization Algorithm |
| EOA | Equilibrium Optimization Algorithm |
| COA | Chimp Optimization Algorithm |
| MRFOA | Manta Ray Foraging Optimization Algorithm |
| SMA | Slime Mould Algorithm |
| MPA | Marine Predators Algorithm |
| BWOA | Black Widow Optimization Algorithm |
| MGWO | Multistage Grey Wolf Optimizer |
| VCS | Virus Colony Search |
| SSA | Salp Swarm Algorithm |
| FA | Firefly Algorithm |
| OBL | Opposition-Based Learning |
| ABC | Artificial Bee Colony |
| KHO | Krill Herd Optimization |
| DBN | Deep Belief Network |
| MEFOA | Modified Electromagnetic Field Optimization Algorithm |
| MAFBUZO | Multi-Agent Fuzzy Buzzard Algorithm |
| ISIC | International Skin Imaging Collaboration |
References
- Pathan, S.; Prabhu, K.G.; Siddalingaswamy, P.C. Techniques and algorithms for computer-aided diagnosis of pigmented skin lesions—A review. Biomed. Signal Process. Control 2018, 39, 237–262. [Google Scholar] [CrossRef]
- Dey, N.; Rajinikanth, V.; Ashour, A.S.; Tavares, J.M. Social group optimization supported segmentation and evaluation of skin melanoma images. Symmetry 2018, 10, 51. [Google Scholar] [CrossRef]
- Saba, T. Computer vision for microscopic skin cancer diagnosis using handcrafted and non-handcrafted features. Microsc. Res. Tech. 2021, 84, 1272–1283. [Google Scholar] [CrossRef] [PubMed]
- Pham, D.L.; Xu, C.; Prince, J.L. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng. 2000, 2, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Tung, Y.S.; Chen, J.C.; Wang, S.W.; Wu, J.L. An adaptive edge detection-based colorization algorithm and its applications. In Proceedings of the 13th Annual ACM International Conference on Multimedia, Singapore, 6–11 November 2011; pp. 351–354. [Google Scholar]
- Abonyi, J.; Feil, B.; Nemeth, S.; Arva, P. Fuzzy clustering based segmentation of time-series. In Advances in Intelligent Data Analysis V; Proceedings of the 5th International Symposium on Intelligent Data Analysis, IDA 2003; Berlin, Germany, 28–30 August 2003, Springer: Berlin/Heidelberg, Germany, 2003; pp. 275–285. [Google Scholar]
- Kohler, R. A segmentation system based on thresholding. Comput. Graph. Image Process. 1981, 15, 319–338. [Google Scholar] [CrossRef]
- Kuruvilla, J.; Sukumaran, D.; Sankar, A.; Joy, S.P. A review on image processing and image segmentation. In Proceedings of the 2016 International IEEE Conference on Data Mining and Advanced Computing (SAPIENCE), Ernakulam, India, 16–18 March 2016; pp. 198–203. [Google Scholar]
- Toma, M.; Lu, Y.; Zhou, H.; Garcia, J.D. Thresholding segmentation errors and uncertainty with patient-specific geometries. J. Biomed. Phys. Eng. 2021, 11, 115–122. [Google Scholar] [CrossRef]
- Li, W.; Raj, A.N.; Tjahjadi, T.; Zhuang, Z. Digital hair removal by deep learning for skin lesion segmentation. Pattern Recognit. 2021, 117, 107994. [Google Scholar] [CrossRef]
- Chai, Y.; Lempitsky, V.; Zisserman, A. Bicos: A bi-level co-segmentation method for image classification. In Proceedings of the 2011 International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; pp. 2579–2586. [Google Scholar]
- Horng, M.-H. Multilevel thresholding selection based on the artificial bee colony algorithm for image segmentation. Expert Syst. Appl. 2011, 38, 13785–13791. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Oliva, D.; Elaziz, M.A.; Hinojosa, S. Fuzzy entropy approaches for image segmentation. In Metaheuristic Algorithms for Image Segmentation: Theory and Applications; Springer: Cham, Switzerland, 2019; pp. 141–147. [Google Scholar]
- Kapur, J.N.; Sahoo, P.K.; Wong, A.K. A new method for gray-level picture thresholding using the entropy of the histogram. Comput. Vis. Graph. Image Process. 1985, 29, 273–285. [Google Scholar] [CrossRef]
- Abdollahzadeh, B.; Soleimanian Gharehchopogh, F.; Mirjalili, S. Artificial gorilla troops optimizer: A new nature-inspired metaheuristic algorithm for global optimization problems. Int. J. Intell. Syst. 2021, 36, 5887–5958. [Google Scholar] [CrossRef]
- Gharehchopogh, F.S.; Maleki, I.; Dizaji, Z.A. Chaotic vortex search algorithm: Meta-heuristic algorithm for feature selection. Evol. Intell. 2022, 15, 1777–1808. [Google Scholar] [CrossRef]
- Halim, Z. Optimizing the DNA fragment assembly using meta-heuristic-based overlap layout consensus approach. Appl. Soft Comput. 2020, 92, 106256. [Google Scholar]
- Meera, S.; Sundar, C. A hybrid meta-heuristic approach for efficient feature selection methods in big data. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 3743–3751. [Google Scholar] [CrossRef]
- SaiSindhuTheja, R.; Shyam, G.K. An efficient meta-heuristic algorithm-based feature selection and recurrent neural network for DoS attack detection in cloud computing environment. Appl. Soft Comput. 2021, 100, 106997. [Google Scholar] [CrossRef]
- Ramadas, M.; Abraham, A. Meta-Heuristics for Data Clustering and Image Segmentation; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Borjigin, S.; Sahoo, P.K. Color image segmentation based on multi-level Tsallis–Havrda–Charvát entropy and 2D histogram using PSO algorithms. Pattern Recognit. 2019, 92, 107–118. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Mohamed, R.; AbdelAziz, N.M.; Abouhawwash, M. HWOA: A hybrid whale optimization algorithm with a novel local minima avoidance method for multi-level thresholding color image segmentation. Expert Syst. Appl. 2022, 190, 116145. [Google Scholar] [CrossRef]
- Agrawal, S.; Panda, R.; Choudhury, P.; Abraham, A. Dominant color component and adaptive whale optimization algorithm for multilevel thresholding of color images. Knowl.-Based Syst. 2022, 240, 108172. [Google Scholar] [CrossRef]
- Eberhart, R.; Kennedy, J. Particle swarm optimization. In Proceedings of the 1995 IEEE International Conference on Neural Networks, Perth, Australia, 27 November–1 December 1995; Volume 4, pp. 1942–1948. [Google Scholar]
- Mirjalili, S.; Lewis, A. The whale optimization algorithm. Adv. Eng. Softw. 2016, 95, 51–67. [Google Scholar] [CrossRef]
- Yang, X.S.; Deb, S. Cuckoo search via Lévy flights. In Proceedings of the 2009 World Congress on Nature & Biologically Inspired Computing (NaBIC), Coimbatore, India, 9–11 December 2009; pp. 210–214. [Google Scholar]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris Hawks optimization: Algorithm and applications. Future Gener. Comput. Syst. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Mirjalili, S.; Mirjalili, S.M.; Lewis, A. Grey wolf optimizer. Adv. Eng. Softw. 2014, 69, 46–61. [Google Scholar] [CrossRef]
- Abdel-Basset, M.; Chang, V.; Mohamed, R. A novel equilibrium optimization algorithm for multi-thresholding image segmentation problems. Neural Comput. Appl. 2021, 33, 10685–10718. [Google Scholar] [CrossRef]
- Khishe, M.; Mosavi, M.R. Chimp optimization algorithm. Expert Syst. Appl. 2020, 149, 113338. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, Z.; Wang, L. Manta ray foraging optimization: An effective bio-inspired optimizer for engineering applications. Eng. Appl. Artif. Intell. 2020, 87, 103300. [Google Scholar] [CrossRef]
- Li, S.; Chen, H.; Wang, M.; Heidari, A.A.; Mirjalili, S. Slime mould algorithm: A new method for stochastic optimization. Future Gener. Comput. Syst. 2020, 111, 300–323. [Google Scholar] [CrossRef]
- Faramarzi, A.; Heidarinejad, M.; Mirjalili, S.; Gandomi, A.H. Marine Predators Algorithm: A nature-inspired meta-heuristic. Expert Syst. Appl. 2020, 152, 113377. [Google Scholar] [CrossRef]
- Hayyolalam, V.; Kazem, A.A.P. Black widow optimization algorithm: A novel meta-heuristic approach for solving engineering optimization problems. Eng. Appl. Artif. Intell. 2020, 87, 103249. [Google Scholar] [CrossRef]
- Khalid, A.M.; Hosny, K.M.; Mirjalili, S. COVIDOA: A novel evolutionary optimization algorithm based on coronavirus disease replication lifecycle. Neural Comput. Appl. 2022, 34, 22465–22492. [Google Scholar] [CrossRef]
- Rai, R.; Das, A.; Dhal, K.G. Nature-inspired optimization algorithms and their significance in multi-thresholding image segmentation: An inclusive review. Evol. Syst. 2022, 13, 889–945. [Google Scholar] [CrossRef]
- Sharma, A.; Chaturvedi, R.; Bhargava, A. A novel opposition-based improved firefly algorithm for multilevel image segmentation. Multimed. Tools Appl. 2022, 81, 15521–15544. [Google Scholar] [CrossRef]
- Yu, H.; Song, J.; Chen, C.; Heidari, A.A.; Liu, J.; Chen, H.; Zaguia, A.; Mafarja, M. Image segmentation of Leaf Spot Diseases on Maize using multi-stage Cauchy-enabled grey wolf algorithm. Eng. Appl. Artif. Intell. 2022, 109, 104653. [Google Scholar] [CrossRef]
- Hussien, A.G.; Heidari, A.A.; Ye, X.; Liang, G.; Chen, H.; Pan, Z. Boosting whale optimization with evolution strategy and Gaussian random walks: An image segmentation method. Eng. Comput. 2023, 39, 1935–1979. [Google Scholar] [CrossRef]
- Yu, M.; Han, M.; Li, X.; Wei, X.; Jiang, H.; Chen, H.; Yu, R. Adaptive soft erasure with edge self-attention for weakly supervised semantic segmentation: Thyroid ultrasound image case study. Comput. Biol. Med. 2022, 144, 105347. [Google Scholar] [CrossRef]
- Qi, A.; Zhao, D.; Yu, F.; Heidari, A.A.; Wu, Z.; Cai, Z.; Alenezi, F.; Mansour, R.F.; Chen, H.; Chen, M. Directional mutation and crossover boosted ant colony optimization with application to COVID-19 X-ray image segmentation. Comput. Biol. Med. 2022, 148, 105810. [Google Scholar] [CrossRef]
- Vijh, S.; Saraswat, M.; Kumar, S. Automatic multilevel image thresholding segmentation using hybrid bio-inspired algorithm and artificial neural network for histopathology images. Multimed. Tools Appl. 2023, 82, 4979–5010. [Google Scholar] [CrossRef]
- Bhavani, H.R.; Champa, H.N. A multilevel thresholding method based on HPSO for the segmentation of various objective functions. In Proceedings of the 2022 International Conference on Communication, Computing, and Internet of Things (IC3IoT), Chennai, India, 10–11 March 2022; pp. 1–5. [Google Scholar]
- Choudhury, A.; Samanta, S.; Pratihar, S.; Bandyopadhyay, O. Multilevel segmentation of Hippocampus images using global steered quantum inspired firefly algorithm. Appl. Intell. 2022, 52, 7339–7372. [Google Scholar] [CrossRef]
- Dinkar, S.K.; Deep, K.; Mirjalili, S.; Thapliyal, S. Opposition-based Laplacian equilibrium optimizer with application in image segmentation using multilevel thresholding. Expert Syst. Appl. 2021, 174, 114766. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, X.; Ling, Y.; Zhang, J. Meta-heuristic moth swarm algorithm for multilevel thresholding image segmentation. Multimed. Tools Appl. 2018, 77, 23699–23727. [Google Scholar] [CrossRef]
- Zhang, S.; Jiang, W.; Satoh, S.I. Multilevel thresholding color image segmentation using a modified artificial bee colony algorithm. IEICE Trans. Inf. Syst. 2018, 101, 2064–2071. [Google Scholar] [CrossRef]
- Chakraborty, F.; Roy, P.K.; Nandi, D. Oppositional elephant herding optimization with dynamic Cauchy mutation for multilevel image thresholding. Evol. Intell. 2019, 12, 445–467. [Google Scholar] [CrossRef]
- Yan, Z.; Zhang, J.; Yang, Z.; Tang, J. Kapur’s entropy for underwater multilevel thresholding image segmentation based on whale optimization algorithm. IEEE Access 2020, 9, 41294–41319. [Google Scholar] [CrossRef]
- Resma, K.B.; Nair, M.S. Multilevel thresholding for image segmentation using Krill Herd Optimization algorithm. J. King Saud Univ.-Comput. Inf. Sci. 2021, 33, 528–541. [Google Scholar]
- Chakraborty, S.; Mali, K.; Banerjee, A.; Bhattacharjee, M.; Chatterjee, S. Image segmentation based on galactic swarm optimization. In Advances in Smart Communication Technology and Information Processing: OPTRONIX 2020; Springer: Singapore, 2021; pp. 251–258. [Google Scholar]
- Anitha, J.; Pandian, S.I.A.; Agnes, S.A. An efficient multilevel color image thresholding based on modified whale optimization algorithm. Expert Syst. Appl. 2021, 178, 115003. [Google Scholar] [CrossRef]
- Abualigah, L.; Habash, M.; Hanandeh, E.S.; Hussein, A.M.; Al Shinwan, M.; Abu Zitar, R.; Jia, H. Improved Reptile Search algorithm by Salp Swarm algorithm for medical image segmentation. J. Bionic Eng. 2023, 20, 1766–1790. [Google Scholar] [CrossRef] [PubMed]
- Hao, S.; Huang, C.; Heidari, A.A.; Chen, H.; Li, L.; Algarni, A.D.; Elmannai, H.; Xu, S. Salp swarm algorithm with iterative mapping and local escaping for multi-level threshold image segmentation: A skin cancer dermoscopic case study. J. Comput. Des. Eng. 2023, 10, 655–693. [Google Scholar] [CrossRef]
- Huang, Q.; Ding, H.; Sheykhahmad, F.R. A skin cancer diagnosis system for dermoscopy images according to deep training and metaheuristics. Biomed. Signal Process. Control 2023, 83, 104705. [Google Scholar] [CrossRef]
- Emam, M.M.; Houssein, E.H.; Ghoniem, R.M. A modified reptile search algorithm for global optimization and image segmentation: Case study brain MRI images. Comput. Biol. Med. 2023, 152, 106404. [Google Scholar] [CrossRef] [PubMed]
- Razmjooy, N.; Arshaghi, A. Application of Multilevel Thresholding and CNN for the Diagnosis of Skin Cancer Utilizing a Multi-Agent Fuzzy Buzzard Algorithm. Biomed. Signal Process. Control 2023, 84, 104984. [Google Scholar] [CrossRef]
- Hosny, K.M.; Khalid, A.M.; Hamza, H.M.; Mirjalili, S. Multilevel segmentation of 2D and volumetric medical images using hybrid Coronavirus Optimization Algorithm. Comput. Biol. Med. 2022, 150, 106003. [Google Scholar] [CrossRef] [PubMed]
- Hosny, K.M.; Khalid, A.M.; Hamza, H.M.; Mirjalili, S. Multilevel thresholding satellite image segmentation using chaotic coronavirus optimization algorithm with hybrid fitness function. Neural Comput. Appl. 2022, 35, 855–886. [Google Scholar] [CrossRef] [PubMed]
- Sezgin, M.; Sankur, B.L. Survey over image thresholding techniques and quantitative performance evaluation. J. Electron. Imaging 2004, 13, 146–168. [Google Scholar]
- Pare, S.; Kumar, A.; Bajaj, V.; Singh, G.K. A multilevel color image segmentation technique based on cuckoo search algorithm and energy curve. Appl. Soft Comput. 2016, 47, 76–102. [Google Scholar] [CrossRef]
- Pare, S.; Kumar, A.; Bajaj, V.; Singh, G.K. An efficient method for multilevel color image thresholding using cuckoo search algorithm based on minimum cross entropy. Appl. Soft Comput. 2017, 61, 570–592. [Google Scholar] [CrossRef]
- Agrawal, S.; Panda, R.; Bhuyan, S.; Panigrahi, B.K. Tsallis entropy-based optimal multilevel thresholding using cuckoo search algorithm. Swarm Evol. Comput. 2013, 11, 16–30. [Google Scholar] [CrossRef]
- Khalid, A.M.; Hamza, H.M.; Mirjalili, S.; Hosny, K.M. BCOVIDOA: A novel binary coronavirus disease optimization algorithm for feature selection. Knowl.-Based Syst. 2022, 248, 108789. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.-G.; Lee, W.; Choi, J.-K.; Kim, S.-J.; Plant, E.P.; Almazán, F.; Taylor, D.R.; Enjuanes, L.; Oh, J.-W. Interference of ribosomal frameshifting by antisense peptide nucleic acids suppresses SARS coronavirus replication. Antivir. Res. 2011, 91, 1–10. [Google Scholar] [CrossRef]
- Kelly, J.A.; Olson, A.N.; Neupane, K.; Munshi, S.; San Emeterio, J.; Pollack, L.; Woodside, M.T.; Dinman, J.D. Structural and functional conservation of the programmed− 1 ribosomal frameshift signal of SARS coronavirus 2 (SARS-CoV-2). J. Biol. Chem. 2020, 295, 10741–10748. [Google Scholar] [CrossRef] [PubMed]
- Brian, D.A.; Baric, R.S. Coronavirus genome structure and replication. In Coronavirus Replication and Reverse Genetics; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–30. [Google Scholar]
- Khan, M.I.; Khan, Z.A.; Baig, M.H.; Ahmad, I.; Farouk, A.E.; Song, Y.G.; Dong, J.-J. Comparative genome analysis of novel coronavirus (SARS-CoV-2) from different geographical locations and the effect of mutations on major target proteins: An in silico insight. PLoS ONE 2020, 15, e0238344. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.; Rotemberg, V.; Tschandl, P.; Celebi, M.E.; Dusza, S.; Gutman, D.; Helba, B.; Kalloo, A.; Liopyris, K.; Marchetti, M.; et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (ISIC). arXiv 2019, arXiv:1902.03368. [Google Scholar]
- Abualigah, L.; Diabat, A.; Mirjalili, S.; Abd Elaziz, M.; Gandomi, A.H. The arithmetic optimization algorithm. Comput. Methods Appl. Mech. Eng. 2021, 376, 113609. [Google Scholar] [CrossRef]
- Mirjalili, S. SCA: A sine cosine algorithm for solving optimization problems. Knowl.-Based Syst. 2016, 96, 120–133. [Google Scholar] [CrossRef]
- Abualigah, L.; Abd Elaziz, M.; Sumari, P.; Geem, Z.W.; Gandomi, A.H. Reptile Search Algorithm (RSA): A nature-inspired meta-heuristic optimizer. Expert Syst. Appl. 2022, 191, 116158. [Google Scholar] [CrossRef]
- Shen, L.; Fan, C.; Huang, X. Multi-level image thresholding using modified flower pollination algorithm. IEEE Access 2018, 6, 30508–30519. [Google Scholar] [CrossRef]
- Wang, Y. Otsu image threshold segmentation method based on seagull optimization algorithm. J. Phys. Conf. Ser. 2020, 1650, 032181. [Google Scholar] [CrossRef]
- Sara, U.; Akter, M.; Uddin, M.S. Image quality assessment through FSIM, SSIM, MSE, and PSNR—A comparative study. J. Comput. Commun. 2019, 7, 8–18. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Mou, X.; Zhang, D. FSIM: A feature similarity index for image quality assessment. IEEE Trans. Image Process. 2011, 20, 2378–2386. [Google Scholar] [CrossRef] [PubMed]

 | 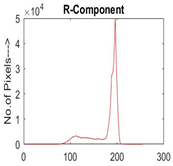 | 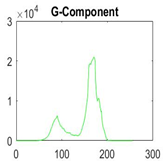 | 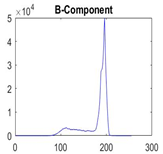 |
| Img1 | --- Pixel Intensity (0–255) ---> | ||
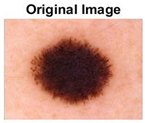 | 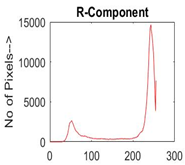 | 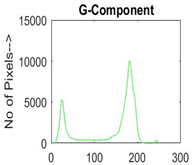 | 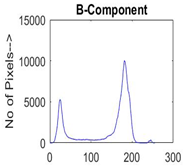 |
| Img2 | --- Pixel Intensity (0–255) ---> | ||
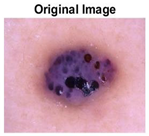
 | 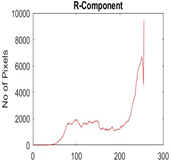 | 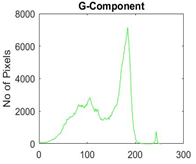 | 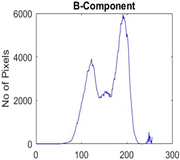 |
| Img3 | --- Pixel Intensity (0–255) ---> | ||
 | 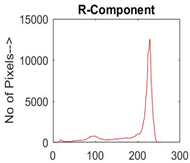 | 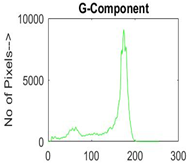 | 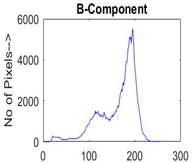 |
| Img4 | --- Pixel Intensity (0–255) ---> | ||
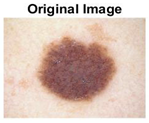 | 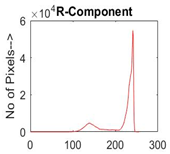 | 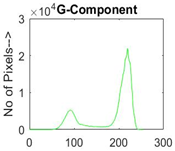 | 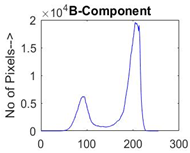 |
| Img5 | --- Pixel Intensity (0–255) ---> | ||
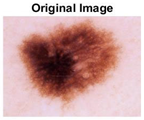 |  |  |  |
| Img6 | --- Pixel Intensity (0–255) ---> | ||
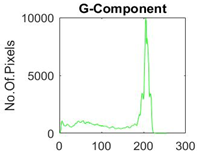
 | 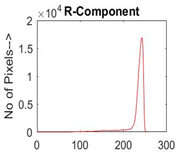 | 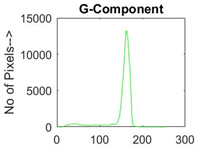 |  |
| Img7 | --- Pixel Intensity (0–255) ---> | ||
 | 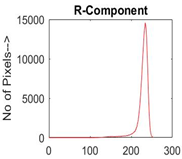 | 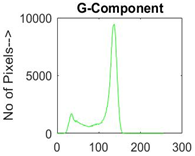 | 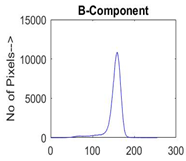 |
| Img8 | --- Pixel Intensity (0–255) ---> | ||
 | 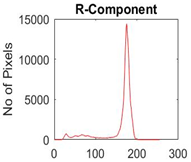 | 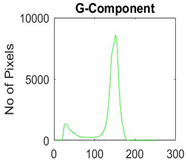 | 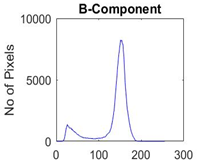 |
| Img9 | --- Pixel Intensity (0–255) ---> | ||
 |  | 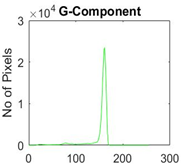 |  |
| Img10 | --- Pixel Intensity (0–255) ---> | ||
| Image | Th | MSE | PSNR | FSIM | NCC | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Otsu | Kapur | T’sallis | Hybrid | Otsu | Kapur | T’sallis | Hybrid | Otsu | Kapur | T’sallis | Hybrid | Otsu | Kapur | T’sallis | Hybrid | ||
| Img1 | 2 | 176.899 | 229.458 | 231.769 | 244.547 | 13.6755 | 13.8419 | 12.5877 | 12.8173 | 0.7278 | 0.7259 | 0.7268 | 0.7387 | 0.9463 | 0.9030 | 0.8988 | 0.9152 |
| 3 | 201.123 | 191.807 | 198.215 | 196.075 | 15.1048 | 15.4739 | 17.6411 | 16.2881 | 0.7543 | 0.7320 | 0.7387 | 0.7654 | 0.9365 | 0.9083 | 0.9055 | 0.9238 | |
| 4 | 153.886 | 193.375 | 167.093 | 196.123 | 16.5593 | 17.5392 | 20.7069 | 20.9091 | 0.7577 | 0.7528 | 0.7866 | 0.7999 | 0.9304 | 0.9173 | 0.9303 | 0.9372 | |
| 5 | 136.530 | 177.843 | 197.954 | 162.939 | 19.1578 | 18.9927 | 21.7588 | 22.6851 | 0.7868 | 0.7584 | 0.8109 | 0.8179 | 0.9371 | 0.9244 | 0.9396 | 0.9530 | |
| Img2 | 2 | 229.326 | 235.223 | 245.039 | 241.141 | 14.5593 | 14.9518 | 15.273 | 15.3446 | 0.6429 | 0.6352 | 0.6436 | 0.6503 | 0.9584 | 0.9304 | 0.9534 | 0.9627 |
| 3 | 230.973 | 234.952 | 231.555 | 222.253 | 18.0646 | 16.2142 | 17.7582 | 18.1292 | 0.6773 | 0.6528 | 0.6646 | 0.6688 | 0.9732 | 0.9596 | 0.9799 | 0.9808 | |
| 4 | 213.243 | 211.400 | 215.130 | 206.814 | 19.4249 | 18.6225 | 19.6770 | 19.4283 | 0.7082 | 0.6696 | 0.7021 | 0.7018 | 0.9836 | 0.9772 | 0.9849 | 0.9852 | |
| 5 | 225.660 | 195.168 | 198.232 | 186.758 | 20.7957 | 19.8970 | 20.2702 | 20.6270 | 0.7367 | 0.6966 | 0.7180 | 0.7244 | 0.9872 | 0.9832 | 0.9862 | 0.9879 | |
| Img3 | 2 | 229.326 | 235.223 | 245.039 | 241.141 | 14.5593 | 14.9518 | 15.273 | 15.3446 | 0.6429 | 0.6352 | 0.6436 | 0.6503 | 0.9584 | 0.9304 | 0.9534 | 0.9627 |
| 3 | 230.973 | 234.952 | 231.555 | 222.253 | 18.0646 | 16.2412 | 17.7582 | 18.1292 | 0.6673 | 0.6528 | 0.6646 | 0.6688 | 0.9732 | 0.9596 | 0.9799 | 0.9808 | |
| 4 | 213.243 | 211.400 | 215.130 | 206814 | 19.4249 | 18.6225 | 19.6770 | 19.4283 | 0.7082 | 0.6696 | 0.7021 | 0.7018 | 0.9836 | 0.9772 | 0.9849 | 0.9852 | |
| 5 | 172.591 | 187.591 | 117.673 | 167.079 | 17.7406 | 19.7514 | 21.058 | 21.3038 | 0.7268 | 0.7090 | 0.7370 | 0.7308 | 0.9444 | 0.9598 | 0.9667 | 0.96804 | |
| Img4 | 2 | 225.398 | 232.498 | 234.029 | 202.004 | 13.8852 | 12.5221 | 15.8050 | 14.4077 | 0.6709 | 0.6771 | 0.6831 | 0.6717 | 0.9167 | 0.7978 | 0.9205 | 0.9292 |
| 3 | 202.052 | 224.445 | 211.753 | 196.021 | 17.4387 | 17.7139 | 18.9657 | 19.5479 | 0.6943 | 0.6893 | 0.7111 | 0.7169 | 0.9535 | 0.9485 | 0.9594 | 0.9621 | |
| 4 | 193.891 | 222.139 | 212.506 | 187.264 | 20.2741 | 18.8596 | 19.9225 | 20.7163 | 0.7443 | 0.7117 | 0.7178 | 0.7359 | 0.9719 | 0.9624 | 0.9676 | 0.9693 | |
| 5 | 158.853 | 183.872 | 171.182 | 21.6327 | 21.5364 | 21.5364 | 21.8322 | 21.1087 | 0.7657 | 0.7505 | 0.7461 | 0.7643 | 0.9748 | 0.9760 | 0.9739 | 0.9763 | |
| Img5 | 2 | 175.242 | 225.645 | 231.323 | 223.039 | 12.4456 | 11.9108 | 12.6701 | 12.8356 | 0.6563 | 0.6351 | 0.6576 | 0.6830 | 0.9646 | 0.9099 | 0.9613 | 0.9658 |
| 3 | 205.294 | 223.118 | 220.808 | 189.994 | 13.6245 | 14.7228 | 15.4734 | 15.5796 | 0.7088 | 0.6913 | 0.7326 | 0.7141 | 0.9578 | 0.9657 | 0.9474 | 0.9506 | |
| 4 | 175.611 | 189.348 | 208.988 | 167.488 | 16.5684 | 16.4125 | 20.0959 | 21.1263 | 0.7286 | 0.6869 | 0.7745 | 0.7572 | 0.9641 | 0.9648 | 0.9620 | 0.9692 | |
| 5 | 163.711 | 198.021 | 150.913 | 172.097 | 17.750 | 18.2216 | 22.6659 | 22.8863 | 0.7464 | 0.7092 | 0.7886 | 0.8277 | 0.9706 | 0.9741 | 0.9772 | 0.9802 | |
| Img6 | 2 | 247.319 | 233.448 | 239.073 | 232.398 | 13.7352 | 14.2525 | 14.4127 | 14.2946 | 0.6704 | 0.6500 | 0.6861 | 0.6681 | 0.9436 | 0.9367 | 0.9621 | 0.94803 |
| 3 | 224.428 | 235.524 | 227.418 | 221.359 | 16.2308 | 16.2704 | 17.2171 | 18.3288 | 0.7005 | 0.6772 | 0.7212 | 0.7161 | 0.9701 | 0.9656 | 0.9790 | 0.98003 | |
| 4 | 228.110 | 222.288 | 213.250 | 197.527 | 19.9904 | 18.8871 | 19.7375 | 20.3376 | 0.7274 | 0.7152 | 0.7426 | 0.7464 | 0.9825 | 0.9797 | 0.9847 | 0.98443 | |
| 5 | 166.596 | 168.018 | 173.173 | 188.119 | 20.9364 | 21.0873 | 21.3780 | 21.7540 | 0.7445 | 0.7456 | 0.7496 | 0.7801 | 0.9858 | 0.9853 | 0.9887 | 0.9884 | |
| Img7 | 2 | 219.250 | 220.827 | 234.351 | 197.279 | 15.1137 | 18.7515 | 18.0671 | 18.4292 | 0.6839 | 0.7035 | 0.6965 | 0.7096 | 0.9146 | 0.9237 | 0.9379 | 0.9415 |
| 3 | 210.507 | 213.080 | 191.698 | 183.035 | 20.2030 | 17.8985 | 21.1190 | 21.2213 | 0.7755 | 0.6989 | 0.7290 | 0.7200 | 0.9591 | 0.8945 | 0.9526 | 0.9589 | |
| 4 | 112.010 | 227143 | 163.328 | 148.629 | 20.4950 | 20.3347 | 22.7456 | 23.1965 | 0.7654 | 0.7041 | 0.7469 | 0.7731 | 0.9627 | 0.9602 | 0.9563 | 0.9680 | |
| 5 | 131.061 | 201.400 | 123.087 | 126.241 | 23.0062 | 21.9584 | 23.0524 | 23.8382 | 0.8116 | 0.7197 | 0.8050 | 0.8041 | 0.9617 | 0.9651 | 0.9369 | 0.9688 | |
| Img8 | 2 | 145.128 | 244.11 | 244.005 | 224.643 | 17.3964 | 16.5393 | 16.4024 | 16.5517 | 0.7213 | 0.6558 | 0.6601 | 0.6806 | 0.7792 | 0.8071 | 0.8040 | 0.8245 |
| 3 | 152.507 | 226.214 | 206.732 | 179.34 | 19.1499 | 18.3084 | 20.3699 | 21.2332 | 0.7624 | 0.6933 | 0.7557 | 0.7739 | 0.8436 | 0.8263 | 0.8623 | 0.8704 | |
| 4 | 218.206 | 223.677 | 181.118 | 157.058 | 19.7812 | 19.1445 | 22.6370 | 23.0862 | 0.7774 | 0.7077 | 0.8201 | 0.8464 | 0.9114 | 0.8387 | 0.8902 | 0.9064 | |
| 5 | 116.378 | 203.504 | 172.312 | 117.823 | 23.5620 | 21.2489 | 23.1626 | 25.2347 | 0.8702 | 0.7796 | 0.8343 | 0.8712 | 0.9384 | 0.8771 | 0.9005 | 0.9176 | |
| Img9 | 2 | 216.044 | 232.309 | 234.950 | 215.202 | 15.9663 | 17.1397 | 18.3113 | 18.8499 | 0.6649 | 0.6607 | 0.6869 | 0.6962 | 0.9278 | 0.9222 | 0.9501 | 0.9583 |
| 3 | 198.979 | 238.836 | 229.886 | 184.955 | 17.9418 | 16.6143 | 18.9293 | 21.2396 | 0.6867 | 0.6671 | 0.7076 | 0.7444 | 0.9495 | 0.9232 | 0.9634 | 0.9729 | |
| 4 | 158.772 | 234.294 | 221.921 | 157.763 | 21.4965 | 18.4320 | 19.7220 | 22.6152 | 0.7671 | 0.6854 | 0.7271 | 0.7872 | 0.9739 | 0.9602 | 0.9671 | 0.9785 | |
| 5 | 121.525 | 204.242 | 163.736 | 134.367 | 22.886 | 20.7396 | 21.4092 | 23.8510 | 0.808 | 0.7397 | 0.7386 | 0.8225 | 0.9789 | 0.9712 | 0.9550 | 0.9829 | |
| Img10 | 2 | 249.483 | 165.215 | 243.583 | 170.210 | 15.3763 | 20.3859 | 19.4787 | 20.8147 | 0.7201 | 0.7388 | 0.7290 | 0.7381 | 0.8960 | 0.9308 | 0.9489 | 0.9470 |
| 3 | 165.470 | 218.479 | 243.311 | 136.404 | 21.7684 | 18.7126 | 19.4453 | 23.9158 | 0.7348 | 07296 | 0.7346 | 0.7617 | 0.9500 | 0.9110 | 0.9509 | 0.9581 | |
| 4 | 172.672 | 198.025 | 217.935 | 133.846 | 24.6356 | 21.2853 | 19.3489 | 24.9056 | 0.7969 | 0.7360 | 0.7369 | 0.7672 | 0.9614 | 0.9417 | 0.9532 | 0.9665 | |
| 5 | 181.790 | 189.018 | 206.890 | 116.888 | 25.8329 | 22.3978 | 20.8161 | 25.2335 | 0.7139 | 0.7393 | 0.7484 | 0.7814 | 0.9729 | 0.9458 | 0.9451 | 0.9743 | |
| Original Image | Th2 | Th3 | Th4 | Th5 |
|---|---|---|---|---|
 Img1 |  |  |  |  |
 Img2 |  |  | 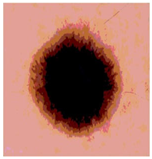 | 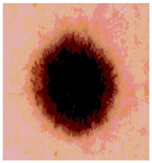 |
 Img3 |  | 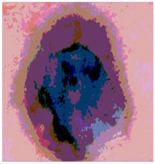 | 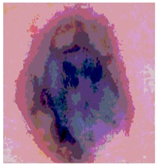 | 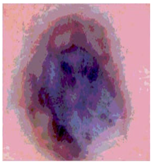 |
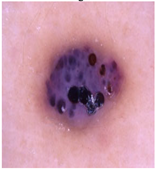 Img4 | 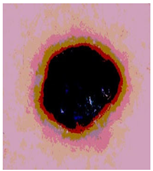 | 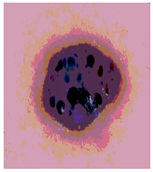 | 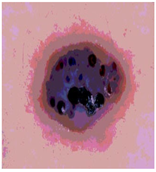 |  |
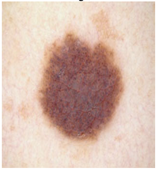 Img5 | 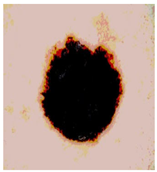 | 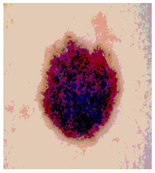 |  |  |
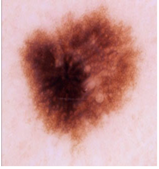 Img6 |  | 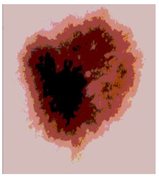 |  | 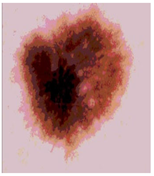 |
 Img7 | 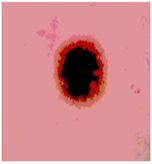 | 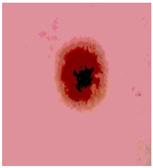 | 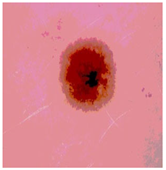 | 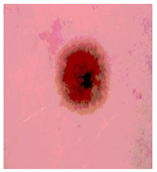 |
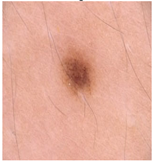 Img8 | 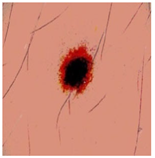 | 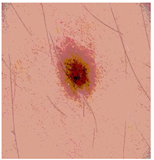 | 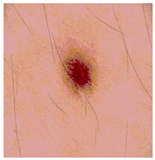 | 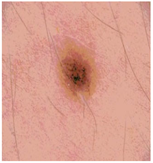 |
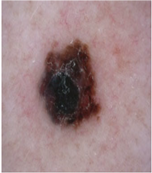 Img9 | 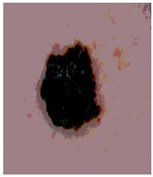 | 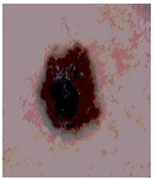 | 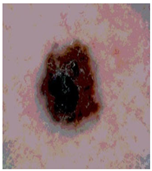 |  |
 Img10 |  |  |  |  |
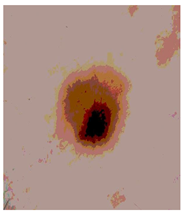
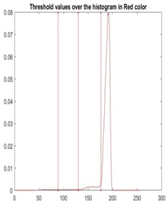
| Segmented Image | R | G | B |
|---|---|---|---|
 | 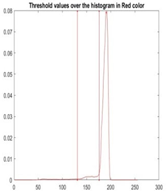 | 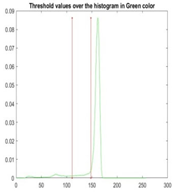 | 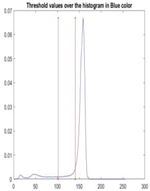 |
 |  | 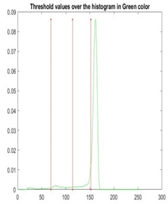 | 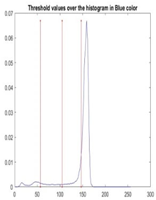 |
 | 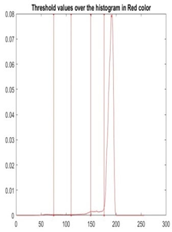 |  | 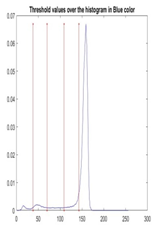 |
 | 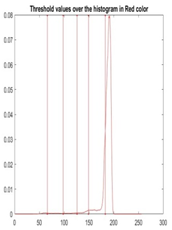 | 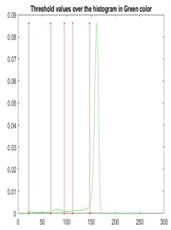 | 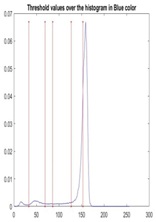 |
| Image | Th | AOA | SCA | RSA | FPA | SOA | GTO | COVIDOA |
|---|---|---|---|---|---|---|---|---|
| Img1 | 2 | 247.6006 | 246.1469 | 247.8406 | 245.1597 | 246.5855 | 245.7669 | 244.5473 |
| 3 | 225.2764 | 214.8453 | 205.2064 | 212.5436 | 223.9644 | 208.1518 | 196.0748 | |
| 4 | 221.2930 | 180.7156 | 203.4535 | 141.8333 | 205.8136 | 196.2350 | 196.1226 | |
| 5 | 241.1227 | 144.8201 | 214.7789 | 186.0322 | 198.2510 | 176.3300 | 162.9383 | |
| Img2 | 2 | 241.9144 | 241.8842 | 246.9748 | 240.1664 | 245.1630 | 242.1664 | 241.1411 |
| 3 | 238.1451 | 237.3881 | 237.8582 | 239.1858 | 240.1110 | 239.1467 | 222.2529 | |
| 4 | 223.7759 | 233.3980 | 226.9811 | 228.1738 | 231.0375 | 220.5672 | 206.8136 | |
| 5 | 245.6072 | 216.9570 | 201.0338 | 205.5512 | 218.0571 | 209.0686 | 186.7584 | |
| Img3 | 2 | 236.8195 | 236.9835 | 222.2556 | 237.1442 | 237.0373 | 237.1505 | 228.0216 |
| 3 | 216.8246 | 230.1775 | 209.0135 | 217.5782 | 217.9692 | 218.2340 | 201.2815 | |
| 4 | 207.5125 | 207.5971 | 205.1720 | 206.8667 | 207.8523 | 199.8962 | 195.8324 | |
| 5 | 223.7134 | 188.6496 | 190.1130 | 192.8761 | 193.0601 | 188.1727 | 167.0796 | |
| Img4 | 2 | 227.0835 | 230.8520 | 227.5441 | 230.5200 | 231.4647 | 230.5200 | 202.0041 |
| 3 | 211.8023 | 220.8783 | 216.0401 | 224.9724 | 195.7951 | 221.5360 | 196.0211 | |
| 4 | 200.8875 | 223.9088 | 219.8616 | 206.1327 | 223.6879 | 216.2579 | 187.2641 | |
| 5 | 247.7828 | 201.5145 | 203.8915 | 206.1327 | 205.0691 | 207.6387 | 171.1820 | |
| Img5 | 2 | 222.5750 | 229.2426 | 238.2147 | 226.4986 | 227.2117 | 227.2265 | 223.0393 |
| 3 | 209.4685 | 230.7308 | 177.2353 | 218.2365 | 226.2237 | 221.8623 | 189.9940 | |
| 4 | 215.9736 | 231.2013 | 222.5808 | 212.2887 | 218.8509 | 208.3670 | 167.4879 | |
| 5 | 250.5794 | 198.7918 | 182.3691 | 199.6067 | 197.7102 | 198.5641 | 172.0972 | |
| Img6 | 2 | 232.2780 | 233.2146 | 237.3292 | 235.6143 | 236.0539 | 235.6143 | 232.2780 |
| 3 | 225.3670 | 219.0977 | 227.9831 | 226.8610 | 228.2006 | 228.8451 | 221.3586 | |
| 4 | 215.5507 | 226.2049 | 212.0640 | 217.9596 | 219.7537 | 210.2340 | 197.5274 | |
| 5 | 236.4972 | 226.2049 | 214.2859 | 202.7830 | 204.4251 | 199.7237 | 188.1148 | |
| Img7 | 2 | 222.5029 | 226.2049 | 248.0666 | 213.2920 | 219.6254 | 215.7291 | 197.2790 |
| 3 | 212.5238 | 243.0814 | 239.0463 | 217.4714 | 222.2559 | 211.3120 | 183.0348 | |
| 4 | 189.3138 | 185.1541 | 204.6421 | 202.6493 | 206.4996 | 208.6859 | 148.6285 | |
| 5 | 205.7216 | 186.6668 | 231.7075 | 174.5481 | 219.3802 | 202.3541 | 126.2406 | |
| Img8 | 2 | 227.6083 | 251.3774 | 219.0853 | 232.5211 | 229.8166 | 229.8120 | 224.6433 |
| 3 | 204.9689 | 221.0381 | 247.8927 | 217.7156 | 224.2208 | 216.4100 | 179.3438 | |
| 4 | 153.7039 | 229.0897 | 214.5952 | 212.1802 | 202.8934 | 208.5816 | 157.0576 | |
| 5 | 163.6398 | 198.8157 | 154.5615 | 207.6764 | 187.3008 | 197.6310 | 117.8226 | |
| Img9 | 2 | 222.6554 | 224.6326 | 247.6065 | 223.4470 | 224.5672 | 223.4491 | 215.2015 |
| 3 | 204.5241 | 228.3561 | 237.1668 | 224.3819 | 214.2435 | 211.5460 | 184.9549 | |
| 4 | 189.5535 | 219.7749 | 180.9008 | 199.3643 | 219.6227 | 202.3142 | 157.7634 | |
| 5 | 246.8424 | 203.0891 | 187.9011 | 171.0936 | 201.3815 | 198.2314 | 134.3671 | |
| Img10 | 2 | 193.9262 | 219.9844 | 241.2282 | 167.2653 | 201.5983 | 191.9097 | 170.2101 |
| 3 | 164.1696 | 251.9099 | 207.3865 | 183.4998 | 195.1959 | 193.4991 | 136.4044 | |
| 4 | 180.4451 | 249.6355 | 241.0450 | 177.0260 | 193.8249 | 177.1338 | 133.8458 | |
| 5 | 249.0814 | 219.0693 | 239.9401 | 218.1823 | 196.4945 | 152.3470 | 116.8888 |
| Image | Th | AOA | SCA | RSA | FPA | SOA | GTO | COVIDOA |
|---|---|---|---|---|---|---|---|---|
| Img1 | 2 | 12.6916 | 12.7582 | 12.7703 | 12.8928 | 12.5227 | 12.7635 | 12.8157 |
| 3 | 15.479 | 15.6818 | 15.5330 | 15.3459 | 14.6225 | 15.5920 | 16.2881 | |
| 4 | 11.9246 | 21.1715 | 18.3366 | 20.4388 | 20.7199 | 19.6530 | 20.9091 | |
| 5 | 14.1807 | 20.0059 | 17.0006 | 21.8298 | 21.3421 | 22.9359 | 22.6851 | |
| Img2 | 2 | 15.2921 | 15.1074 | 13.4111 | 15.1709 | 14.5427 | 15.1708 | 15.3446 |
| 3 | 16.6603 | 17.1524 | 17.1305 | 16.8841 | 16.7627 | 16.9005 | 18.1292 | |
| 4 | 18.1235 | 17.7695 | 19.6477 | 18.5581 | 18.3501 | 18.5623 | 19.4283 | |
| 5 | 13.5758 | 19.5727 | 18.8905 | 20.1187 | 19.6857 | 20.2480 | 20.6270 | |
| Img3 | 2 | 10.2965 | 10.3048 | 09.9944 | 10.3140 | 10.9370 | 10.3359 | 10.8181 |
| 3 | 14.3920 | 13.1320 | 13.1290 | 14.6335 | 14.7912 | 14.9568 | 16.6610 | |
| 4 | 14.4746 | 16.8630 | 17.2929 | 19.0142 | 18.8696 | 19.0124 | 19.3445 | |
| 5 | 13.8634 | 17.3687 | 18.5697 | 20.7029 | 20.0024 | 21.3329 | 21.3038 | |
| Img4 | 2 | 14.0519 | 14.0302 | 14.5456 | 14.0325 | 14.0121 | 14.0325 | 14.4077 |
| 3 | 18.1824 | 18.4383 | 17.1256 | 18.3327 | 18.1885 | 18.9654 | 19.5479 | |
| 4 | 18.8780 | 17.8194 | 17.3093 | 20.0309 | 18.4881 | 19.6989 | 20.7163 | |
| 5 | 10.9544 | 20.4764 | 18.0280 | 21.0277 | 20.6490 | 21.3457 | 22.1088 | |
| Img5 | 2 | 12.8383 | 12.7648 | 12.2583 | 12.8072 | 12.7768 | 12.7933 | 12.8356 |
| 3 | 14.4969 | 14.1322 | 14.8782 | 15.6127 | 16.3258 | 16.4677 | 15.5796 | |
| 4 | 16.8143 | 14.6929 | 17.1395 | 20.4125 | 20.2356 | 21.0231 | 21.1263 | |
| 5 | 12.4279 | 20.8377 | 20.0745 | 21.7622 | 21.7227 | 21.9568 | 22.8863 | |
| Img6 | 2 | 14.1149 | 14.2493 | 12.4859 | 14.1924 | 14.2035 | 14.1924 | 14.2946 |
| 3 | 17.6128 | 17.5604 | 15.9501 | 17.9081 | 17.8064 | 18.0124 | 18.3288 | |
| 4 | 19.2894 | 16.3362 | 18.0414 | 20.4329 | 19.2129 | 19.9823 | 20.3376 | |
| 5 | 13.6432 | 15.9573 | 17.5619 | 21.1660 | 21.3138 | 21.5138 | 21.7540 | |
| Img7 | 2 | 17.8573 | 9.0647 | 15.1141 | 18.1887 | 18.1174 | 18.2218 | 18.4292 |
| 3 | 20.9240 | 15.7042 | 18.4234 | 20.9894 | 20.7422 | 20.4210 | 21.2213 | |
| 4 | 23.5151 | 20.3822 | 18.7204 | 21.9646 | 22.1361 | 22.0069 | 23.1965 | |
| 5 | 18.4159 | 21.7143 | 19.3929 | 22.9927 | 21.0572 | 22.8410 | 23.8382 | |
| Img8 | 2 | 16.3356 | 13.4726 | 14.8376 | 16.4363 | 16.6193 | 15.0121 | 16.5517 |
| 3 | 20.7668 | 16.5391 | 16.4506 | 20.4295 | 20.1469 | 19.2310 | 21.2332 | |
| 4 | 22.6643 | 16.7415 | 17.0362 | 21.4753 | 21.7739 | 21.6554 | 23.0862 | |
| 5 | 22.2005 | 20.0964 | 23.0482 | 21.4175 | 22.7238 | 22.4057 | 25.2347 | |
| Img9 | 2 | 18.5839 | 18.5990 | 15.9292 | 18.9314 | 18.5884 | 18.6395 | 18.8499 |
| 3 | 20.7089 | 16.8991 | 16.5721 | 19.5291 | 20.3260 | 21.0314 | 21.2396 | |
| 4 | 21.4919 | 16.5244 | 19.9028 | 21.0434 | 20.3115 | 21.9869 | 22.6152 | |
| 5 | 15.8094 | 15.2637 | 20.4012 | 21.9074 | 21.5602 | 22.3567 | 23.8510 | |
| Img10 | 2 | 20.5999 | 15.2959 | 18.0531 | 20.6405 | 20.4843 | 20.5994 | 20.8147 |
| 3 | 23.2996 | 12.6441 | 19.8027 | 23.1252 | 23.9542 | 23.0295 | 23.9158 | |
| 4 | 22.7509 | 12.7175 | 18.8998 | 24.1067 | 23.5074 | 24.2329 | 24.9056 | |
| 5 | 15.2400 | 14.9047 | 18.5323 | 22.5299 | 23.9037 | 25.0195 | 25.2335 |
| Image | Th | AOA | SCA | RSA | FPA | SOA | GTO | COVIDOA |
|---|---|---|---|---|---|---|---|---|
| Img1 | 2 | 0.7378 | 0.7389 | 0.7359 | 0.7382 | 0.7230 | 0.7387 | 0.7387 |
| 3 | 0.7586 | 0.7628 | 0.7356 | 0.7627 | 0.7227 | 0.7667 | 0.7654 | |
| 4 | 0.7506 | 0.7814 | 0.7649 | 0.7734 | 0.7707 | 0.7982 | 0.7999 | |
| 5 | 0.7365 | 0.7861 | 0.7556 | 0.8125 | 0.7608 | 0.8159 | 0.8179 | |
| Img2 | 2 | 0.6497 | 0.6501 | 0.6586 | 0.6503 | 0.6339 | 0.6503 | 0.6503 |
| 3 | 0.6696 | 0.6666 | 0.6610 | 0.6685 | 0.6658 | 0.6683 | 0.6688 | |
| 4 | 0.6967 | 0.6892 | 0.6825 | 0.6892 | 0.6724 | 0.7002 | 0.7018 | |
| 5 | 0.6284 | 0.7244 | 0.6993 | 0.7168 | 0.7141 | 0.7205 | 0.7244 | |
| Img3 | 2 | 0.6684 | 0.6675 | 0.6716 | 0.6690 | 0.6524 | 0.6689 | 0.6612 |
| 3 | 0.6919 | 0.6560 | 0.6823 | 0.6832 | 0.6906 | 0.6894 | 0.6870 | |
| 4 | 0.6873 | 0.6975 | 0.7124 | 0.7156 | 0.7134 | 0.7134 | 0.7159 | |
| 5 | 0.6762 | 0.7130 | 0.7365 | 0.7457 | 0.7194 | 0.7432 | 0.7308 | |
| Img4 | 2 | 0.6588 | 0.6618 | 0.6549 | 0.6623 | 0.6629 | 0.6623 | 0.6717 |
| 3 | 0.7117 | 0.7060 | 0.7022 | 0.6990 | 0.6835 | 0.6936 | 0.7169 | |
| 4 | 0.7258 | 0.6906 | 0.7113 | 0.7378 | 0.7113 | 0.7151 | 0.7359 | |
| 5 | 0.6525 | 0.7331 | 0.7326 | 0.7312 | 0.7375 | 0.7521 | 0.7643 | |
| Img5 | 2 | 0.6807 | 0.6828 | 0.6745 | 0.6830 | 0.6827 | 0.6827 | 0.6830 |
| 3 | 0.7324 | 0.6673 | 0.6870 | 0.7285 | 0.6942 | 0.7296 | 0.7141 | |
| 4 | 0.7255 | 0.7066 | 0.7574 | 0.7766 | 0.7721 | 0.7512 | 0.7572 | |
| 5 | 0.6582 | 0.7875 | 0.7831 | 0.8198 | 0.8117 | 0.8100 | 0.8277 | |
| Img6 | 2 | 0.6680 | 0.6690 | 0.6635 | 0.6716 | 0.6708 | 0.6716 | 0.6681 |
| 3 | 0.7140 | 0.6868 | 0.7054 | 0.7161 | 0.7156 | 0.7013 | 0.7161 | |
| 4 | 0.7397 | 0.7382 | 0.7676 | 0.7423 | 0.7435 | 0.7469 | 0.7464 | |
| 5 | 0.6986 | 0.7468 | 0.7508 | 0.7617 | 0.7722 | 0.7961 | 0.7801 | |
| Img7 | 2 | 0.7007 | 0.6736 | 0.6888 | 0.7054 | 0.7032 | 0.7051 | 0.7096 |
| 3 | 0.7142 | 0.6890 | 0.7097 | 0.7138 | 0.7120 | 0.7112 | 0.7200 | |
| 4 | 0.7320 | 0.7407 | 0.7005 | 0.7415 | 0.7250 | 0.7253 | 0.7731 | |
| 5 | 0.7088 | 0.7406 | 0.7087 | 0.7814 | 0.7403 | 0.7821 | 0.8041 | |
| Img8 | 2 | 0.6745 | 0.5882 | 0.6317 | 0.6726 | 0.6881 | 0.6780 | 0.6806 |
| 3 | 0.7562 | 0.6674 | 0.6525 | 0.7459 | 0.7359 | 0.7532 | 0.7739 | |
| 4 | 0.8245 | 0.6624 | 0.7052 | 0.7736 | 0.7860 | 0.7801 | 0.8464 | |
| 5 | 0.8077 | 0.7626 | 0.8324 | 0.6869 | 0.8106 | 0.8061 | 0.8712 | |
| Img9 | 2 | 0.6925 | 0.6916 | 0.6740 | 0.6903 | 0.6869 | 0.6928 | 0.6962 |
| 3 | 0.7320 | 0.6903 | 0.7999 | 0.7055 | 0.7234 | 0.7564 | 0.7444 | |
| 4 | 0.7516 | 0.6767 | 0.7241 | 0.7386 | 0.7219 | 0.7801 | 0.7872 | |
| 5 | 0.6561 | 0.6966 | 0.7493 | 0.7728 | 0.7545 | 0.8210 | 0.8225 | |
| Img10 | 2 | 0.7364 | 0.7254 | 0.7298 | 0.7374 | 0.7362 | 0.7369 | 0.7381 |
| 3 | 0.7437 | 0.7188 | 0.7245 | 0.7454 | 0.7439 | 0.7437 | 0.7617 | |
| 4 | 0.7687 | 0.7198 | 0.7377 | 0.7507 | 0.7507 | 0.7522 | 0.7672 | |
| 5 | 0.7212 | 0.6696 | 0.7377 | 0.7490 | 0.7844 | 0.7785 | 0.7814 |
| Image | Th | AOA | SCA | RSA | FPA | SOA | GTO | COVIDOA |
|---|---|---|---|---|---|---|---|---|
| Img1 | 2 | 0.9150 | 0.91607 | 0.9155 | 0.9142 | 0.9125 | 0.9160 | 0.9152 |
| 3 | 0.9105 | 0.9197 | 0.9080 | 0.9231 | 0.9148 | 0.9232 | 0.9238 | |
| 4 | 0.3242 | 0.9372 | 0.9280 | 0.9271 | 0.8974 | 0.9312 | 0.9372 | |
| 5 | 0.9138 | 0.9420 | 0.8947 | 0.9404 | 0.9417 | 0.9651 | 0.9523 | |
| Img2 | 2 | 0.9628 | 0.9624 | 0.9504 | 0.9627 | 0.9497 | 0.9628 | 0.9627 |
| 3 | 0.9778 | 0.9777 | 0.9785 | 0.9787 | 0.9719 | 0.9788 | 0.9808 | |
| 4 | 0.9812 | 0.9815 | 0.9794 | 0.9841 | 0.9778 | 0.9821 | 0.9852 | |
| 5 | 0.9483 | 0.9840 | 0.9746 | 0.9872 | 0.9700 | 0.9881 | 0.9879 | |
| Img3 | 2 | 0.9055 | 0.9060 | 0.9058 | 0.9063 | 0.9064 | 0.9063 | 0.9146 |
| 3 | 0.9272 | 0.7407 | 0.9359 | 0.9276 | 0.9215 | 0.9332 | 0.9332 | |
| 4 | 0.9031 | 0.9299 | 0.9326 | 0.9585 | 0.9515 | 0.9573 | 0.9542 | |
| 5 | 0.9030 | 0.9163 | 0.9452 | 0.9668 | 0.9519 | 0.9728 | 0.9684 | |
| Img4 | 2 | 0.9162 | 0.9172 | 0.9103 | 0.9174 | 0.9176 | 0.9174 | 0.9292 |
| 3 | 0.9550 | 0.9532 | 0.9499 | 0.9535 | 0.9509 | 0.9364 | 0.9622 | |
| 4 | 0.9579 | 0.9394 | 0.9201 | 0.9677 | 0.9629 | 0.9578 | 0.9693 | |
| 5 | 0.8117 | 0.9676 | 0.9527 | 0.9757 | 0.9715 | 0.6756 | 0.9763 | |
| Img5 | 2 | 0.9655 | 0.9641 | 0.9550 | 0.9648 | 0.9627 | 0.9646 | 0.9658 |
| 3 | 0.9598 | 0.9351 | 0.9521 | 0.9686 | 0.9664 | 0.9684 | 0.9506 | |
| 4 | 0.9620 | 0.9117 | 0.9625 | 0.9656 | 0.9674 | 0.9675 | 0.9692 | |
| 5 | 0.9476 | 0.9710 | 0.9682 | 0.9715 | 0.9762 | 0.9754 | 0.9802 | |
| Img6 | 2 | 0.9459 | 0.9478 | 0.9269 | 0.9478 | 0.9481 | 0.9478 | 0.9480 |
| 3 | 0.9773 | 0.9783 | 0.9612 | 0.9796 | 0.9797 | 0.9634 | 0.9800 | |
| 4 | 0.9820 | 0.9673 | 0.9808 | 0.9850 | 0.9819 | 0.9805 | 0.9844 | |
| 5 | 0.9499 | 0.9498 | 0.9739 | 0.9859 | 0.9854 | 0.9900 | 0.9884 | |
| Img7 | 2 | 0.9384 | 0.7331 | 0.9254 | 0.9407 | 0.9379 | 0.9408 | 0.9415 |
| 3 | 0.9589 | 0.9295 | 0.9456 | 0.9576 | 0.9578 | 0.9521 | 0.9589 | |
| 4 | 0.9636 | 0.9318 | 0.9127 | 0.9629 | 0.9647 | 0.9643 | 0.9680 | |
| 5 | 0.9180 | 0.9506 | 0.9492 | 0.9657 | 0.9606 | 0.9621 | 0.9688 | |
| Img8 | 2 | 0.8210 | 0.7439 | 0.8228 | 0.8193 | 0.8220 | 0.8159 | 0.8245 |
| 3 | 0.8635 | 0.8379 | 0.7984 | 0.8593 | 0.8572 | 0.8501 | 0.8704 | |
| 4 | 0.9032 | 0.7676 | 0.7643 | 0.8732 | 0.8812 | 0.8784 | 0.9064 | |
| 5 | 0.8846 | 0.8582 | 0.8829 | 0.8790 | 0.8930 | 0.8897 | 0.9176 | |
| Img9 | 2 | 0.9569 | 0.9574 | 0.9473 | 0.9550 | 0.9566 | 0.9574 | 0.9583 |
| 3 | 0.9703 | 0.9285 | 0.9468 | 0.9659 | 0.9694 | 0.9713 | 0.9729 | |
| 4 | 0.9727 | 0.9261 | 0.9530 | 0.9665 | 0.9691 | 0.9790 | 0.9785 | |
| 5 | 0.9255 | 0.8852 | 0.9727 | 0.9705 | 0.9728 | 0.9799 | 0.9830 | |
| Img10 | 2 | 0.9402 | 0.8591 | 0.9199 | 0.9406 | 0.9405 | 0.9405 | 0.9470 |
| 3 | 0.9579 | 0.8472 | 0.9350 | 0.9500 | 0.9642 | 0.9648 | 0.9582 | |
| 4 | 0.9552 | 0.8387 | 0.8822 | 0.9629 | 0.9679 | 0.9694 | 0.9665 | |
| 5 | 0.8850 | 0.8219 | 0.9426 | 0.9677 | 0.9420 | 0.9672 | 0.9543 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsahafi, Y.S.; Elshora, D.S.; Mohamed, E.R.; Hosny, K.M. Multilevel Threshold Segmentation of Skin Lesions in Color Images Using Coronavirus Optimization Algorithm. Diagnostics 2023, 13, 2958. https://doi.org/10.3390/diagnostics13182958
Alsahafi YS, Elshora DS, Mohamed ER, Hosny KM. Multilevel Threshold Segmentation of Skin Lesions in Color Images Using Coronavirus Optimization Algorithm. Diagnostics. 2023; 13(18):2958. https://doi.org/10.3390/diagnostics13182958
Chicago/Turabian StyleAlsahafi, Yousef S., Doaa S. Elshora, Ehab R. Mohamed, and Khalid M. Hosny. 2023. "Multilevel Threshold Segmentation of Skin Lesions in Color Images Using Coronavirus Optimization Algorithm" Diagnostics 13, no. 18: 2958. https://doi.org/10.3390/diagnostics13182958
APA StyleAlsahafi, Y. S., Elshora, D. S., Mohamed, E. R., & Hosny, K. M. (2023). Multilevel Threshold Segmentation of Skin Lesions in Color Images Using Coronavirus Optimization Algorithm. Diagnostics, 13(18), 2958. https://doi.org/10.3390/diagnostics13182958






