DSCNet: Deep Skip Connections-Based Dense Network for ALL Diagnosis Using Peripheral Blood Smear Images
Abstract
:1. Introduction
- 1.
- Deep Skip Connections-Based Dense Network (DSCNet): This paper proposes a novel architecture, DSCNet, specifically tailored for the diagnosis of ALL using peripheral blood smear images. DSCNet utilizes skip connections, custom image filtering, and dense blocks to capture long-range dependencies and enhance feature extraction.
- 2.
- Custom Image Filtering: A custom image filter is used as a pre-processing step to enhance input images and highlight relevant features. This filtering process is used to improve the quality of input data and aids the model in detecting intricate patterns associated with different stages of ALL.
- 3.
- KL Divergence Loss Optimization: KL divergence loss is used as an objective function for model optimization. By minimizing KL divergence between predicted and ground truth distributions, the model learns to make accurate predictions, enhancing its diagnostic accuracy.
- 4.
- Dropout Regularization: Dropout regularization is used to prevent overfitting during model training. This technique is used to enhance the robustness of feature representations and improve the way DSCNet generalizes unseen data.
2. Related Work
3. Deep Skip Connections-Based Dense Network (DSCNet) for ALL Diagnosis
| Algorithm 1 Deep Skip Connections-Based Dense Network (DSCNet) for ALL diagnosis |
|
3.1. Training Process of DSCNet
| Algorithm 2 Training the Deep Skip Connections-Based Dense Network (DSCNet) |
|
3.2. Hyperparameters of DSCNet
- 1.
- Learning Rate: The learning rate controls the step size during model parameter updates. An appropriate learning rate is crucial for successful training without overshooting or becoming stuck in local minima. Learning Rate = 0.001.
- 2.
- Number of Epochs: The number of epochs determines how many times the entire dataset is passed through the model during training. Number of Epochs = 50.
- 3.
- Batch Size: The batch size specifies the number of training examples in each mini-batch. Larger batch sizes may increase training speed, but too large a batch can lead to memory issues. Batch Size = 32.
- 4.
- Regularization Parameter (): The regularization parameter controls the strength of regularization, preventing overfitting by penalizing large weights. = 0.01.
- 5.
- Dropout Rate: The dropout rate determines the fraction of neurons dropped during training, promoting robustness. We have used two dropout rates—0.3 and 0.2, respectively.
4. Performance Analysis
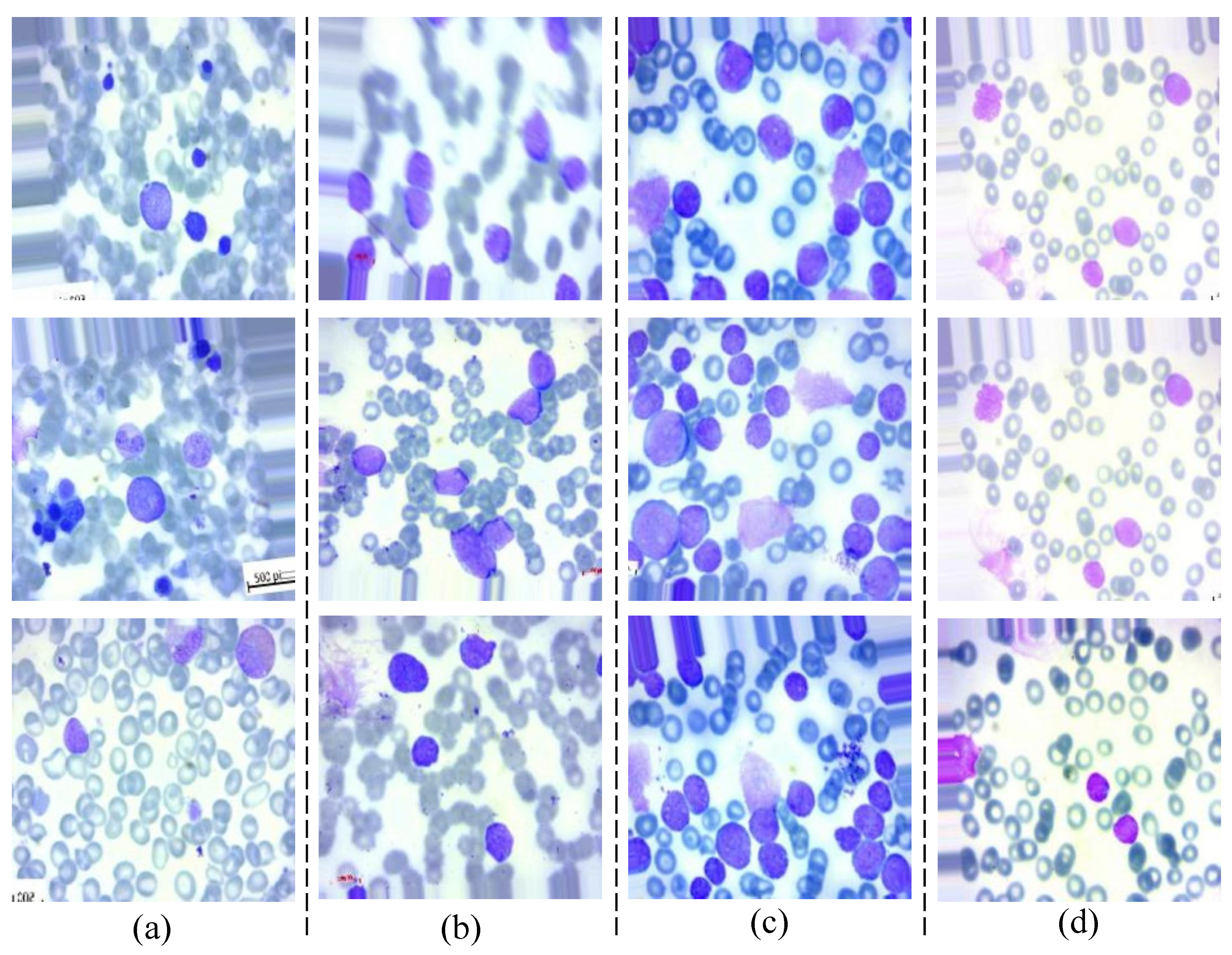
4.1. Dataset
- 1.
- ALL_benign: Benign—Represents images of peripheral blood smears that are classified as benign, meaning there are no signs of leukemia.
- 2.
- ALL_early: Early—Contains images representing the early stages of ALL.
- 3.
- ALL_pre: Pre—Includes images of peripheral blood smears from patients in the pre-ALL stage, indicating a progression towards leukemia.
- 4.
- ALL_pro: Pro—Comprises images from patients in the pro-ALL stage, representing a more advanced state of ALL.
4.2. Data Augmentation
- 1.
- Horizontal and Vertical Flips: This data augmentation technique involves flipping the image horizontally or vertically. By doing so, the model is exposed to different orientations of objects in the image, which enhances its robustness to variations in object direction.
- 2.
- Random Rotations: Random rotations are applied to the image by rotating it by a random angle. This approach allows the model to learn from images with various angles, making it more capable of handling rotated images during inference. By augmenting the dataset with rotated versions of the original images, the model gains the ability to recognize objects and patterns from different perspectives.
- 3.
- Random Crop and Resize: With random crop and resize, a portion of the image is randomly cropped and then resized back to the original size. This technique enables the model to focus on different regions of the image during training, promoting robustness and reducing sensitivity to the precise object location. By training on diverse crops, DSCNet learns to recognize important features that may appear in different parts of the image, improving its generalization performance on unseen data.
- 4.
- Color Jittering: Color jittering involves randomly modifying the color channels of the image, including altering the hue, saturation, and brightness. This augmentation introduces variations in color, making the model more resilient to changes in lighting conditions and color distributions in the dataset. By simulating different lighting conditions and color shifts, the model becomes more adaptable to real-world scenarios where images may have varying color casts or brightness levels.
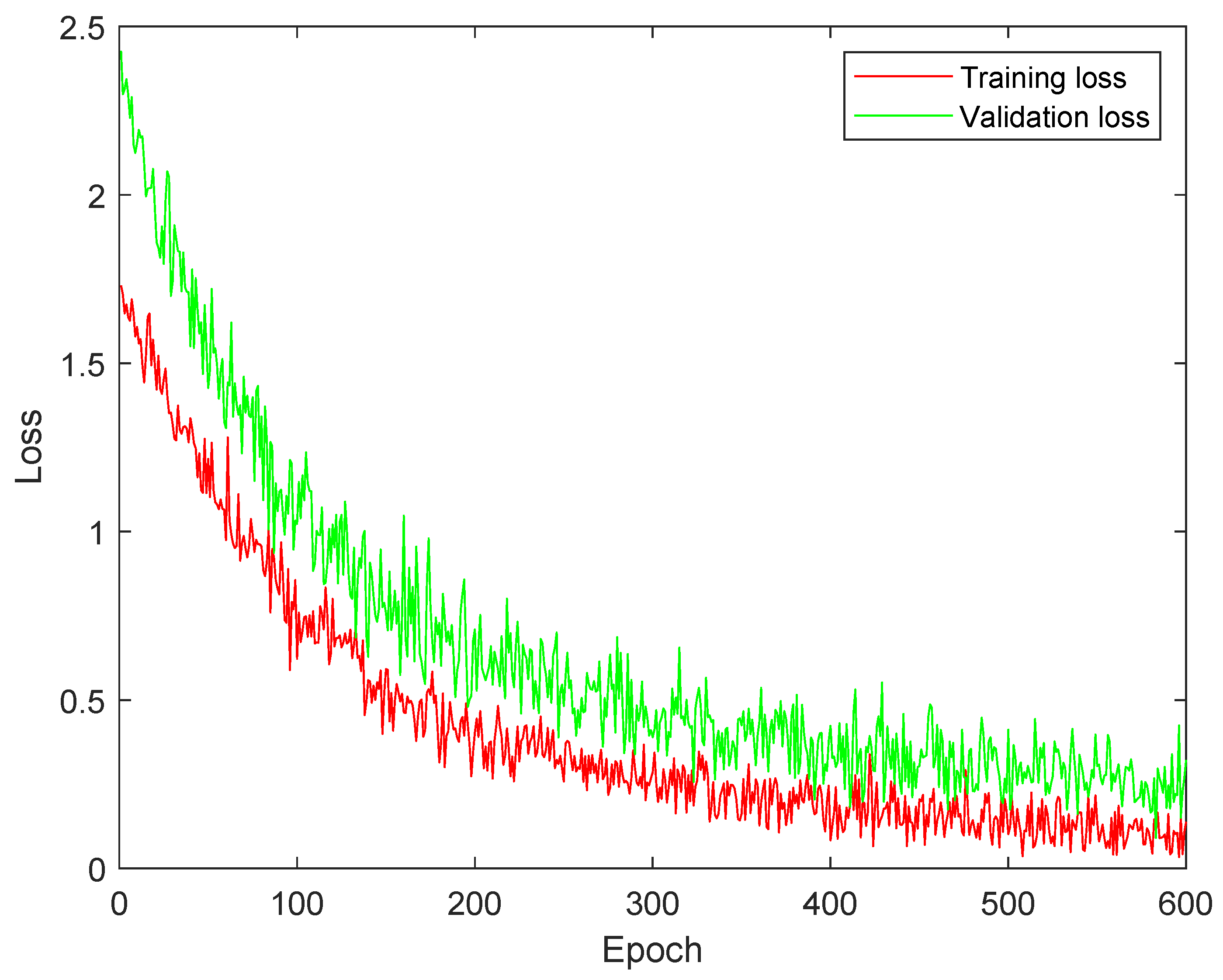
4.3. Training and Validation Loss Analysis
4.4. Comparative Analysis
4.4.1. Without Augmented Dataset
4.4.2. With Augmented Dataset
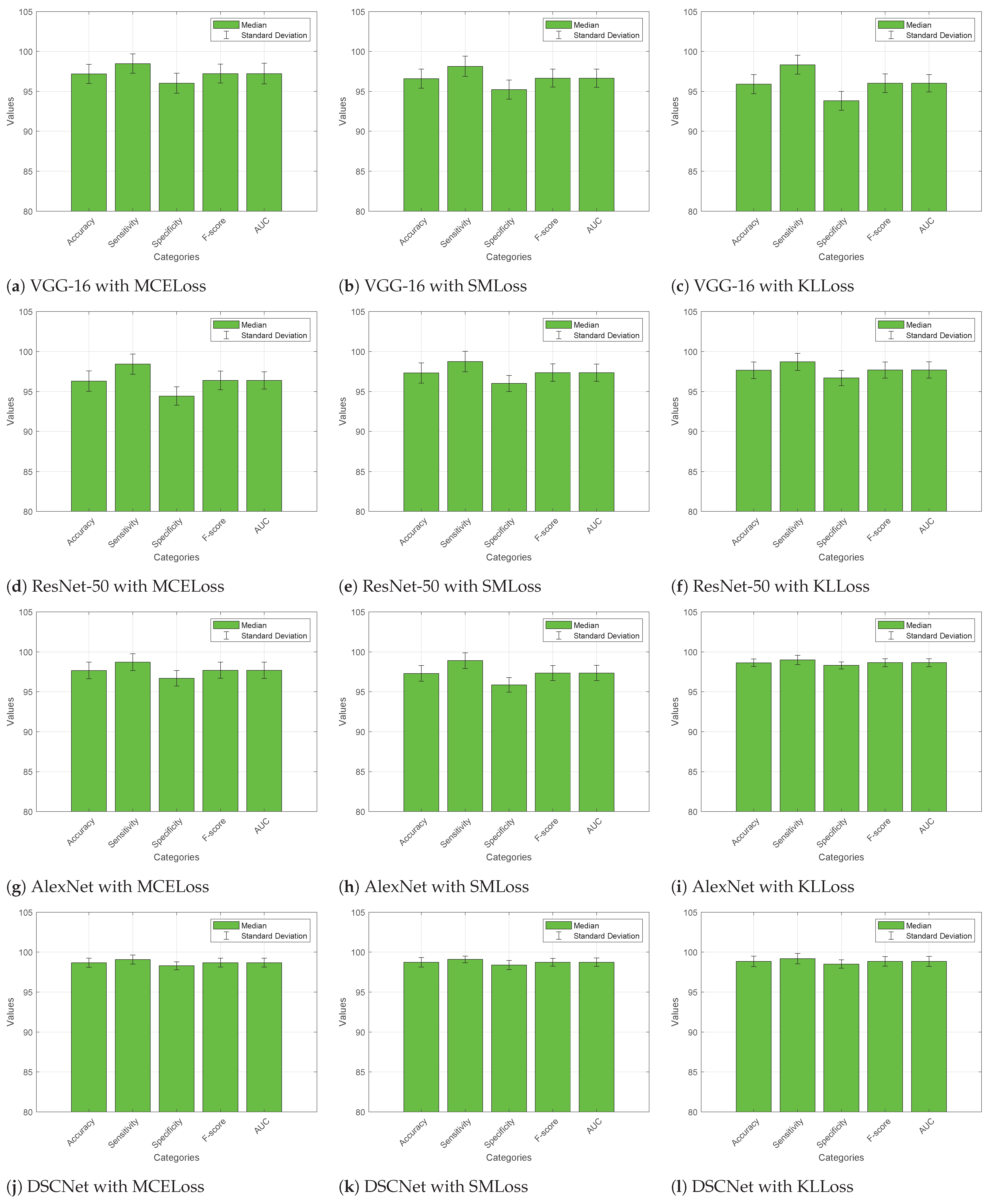
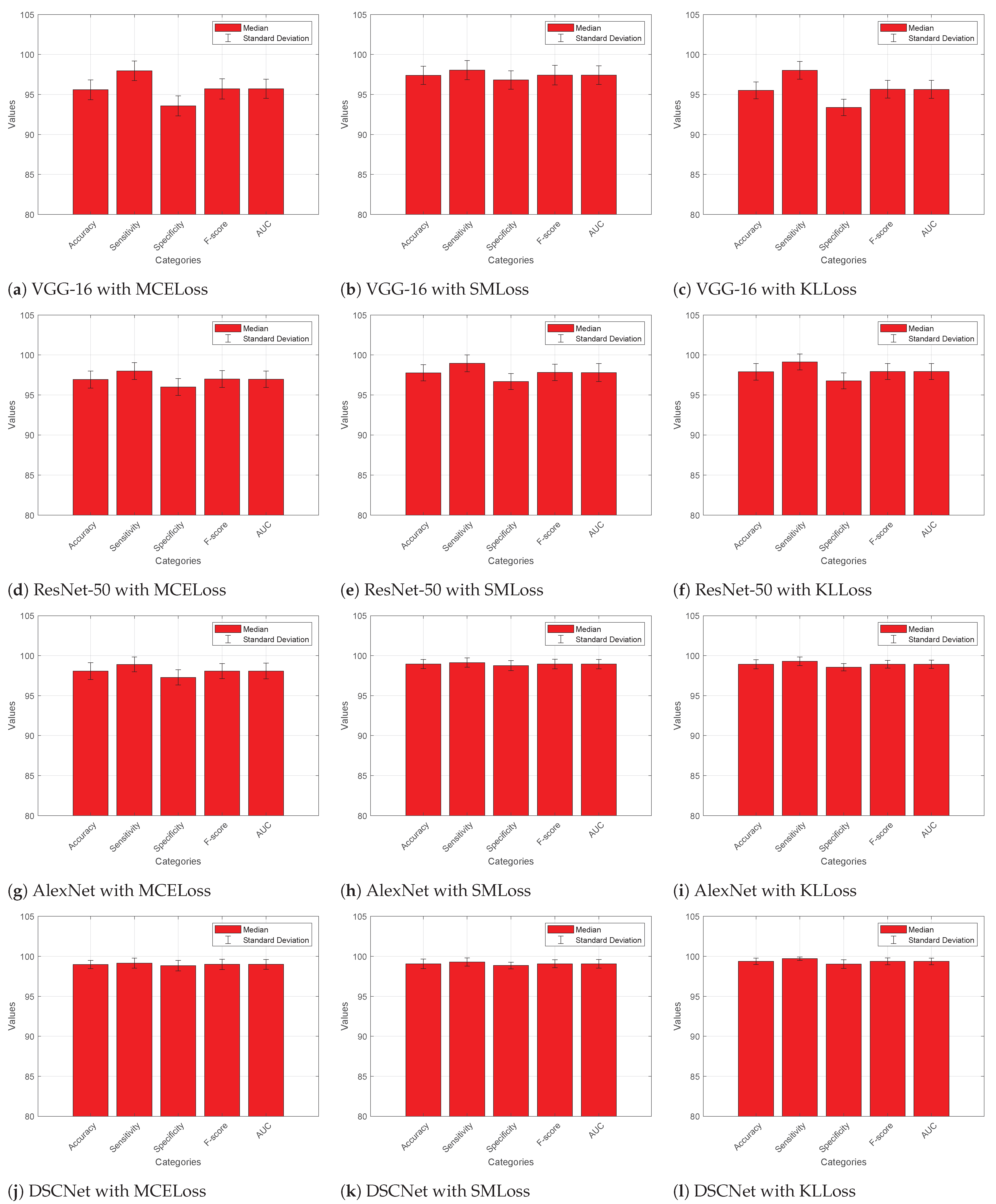
4.5. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atteia, G.E. Latent Space Representational Learning of Deep Features for Acute Lymphoblastic Leukemia Diagnosis. Comput. Syst. Sci. Eng. 2023, 45, 361–376. [Google Scholar] [CrossRef]
- Mallikarjuna, S.B.; Shivakumara, P.; Khare, V.; Basavanna, M.; Pal, U.; Poornima, B. Multi-gradient-direction based deep learning model for arecanut disease identification. CAAI Trans. Intell. Technol. 2022, 7, 156–166. [Google Scholar] [CrossRef]
- Kumar, S.; Rani, S.; Jain, A.; Verma, C.; Raboaca, M.S.; Illés, Z.; Neagu, B.C. Face spoofing, age, gender and facial expression recognition using advance neural network architecture-based biometric system. Sensors 2022, 22, 5160. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhi, K.; Zeng, J.; Tian, C.; You, L. Multi-gradient-direction based deep learning model for arecanut disease identification. J. Artif. Intell. Technol. 2022, 2, 93–99. [Google Scholar]
- Chand, S.; Vishwakarma, V.P. A novel Deep Learning Framework (DLF) for classification of Acute Lymphoblastic Leukemia. Multimed. Tools Appl. 2022, 81, 37243–37262. [Google Scholar] [CrossRef]
- Masoudi, B. VKCS: A pre-trained deep network with attention mechanism to diagnose acute lymphoblastic leukemia. Multimed. Tools Appl. 2023, 82, 18967–18983. [Google Scholar] [CrossRef]
- Das, P.K.; Meher, S. An efficient deep Convolutional Neural Network based detection and classification of Acute Lymphoblastic Leukemia. Expert Syst. Appl. 2021, 183, 115311. [Google Scholar] [CrossRef]
- Kumar, S.; Choudhary, S.; Jain, A.; Singh, K.; Ahmadian, A.; Bajuri, M.Y. Brain Tumor Classification Using Deep Neural Network and Transfer Learning. Brain Topogr. 2023, 36, 305–318. [Google Scholar] [CrossRef]
- Meng, J.; Li, Y.; Liang, H.; Ma, Y. Single-image dehazing based on two-stream convolutional neural network. J. Artif. Intell. Technol. 2022, 2, 100–110. [Google Scholar] [CrossRef]
- Wu, Z.; Luo, G.; Yang, Z.; Guo, Y.; Li, K.; Xue, Y. A comprehensive review on deep learning approaches in wind forecasting applications. CAAI Trans. Intell. Technol. 2022, 7, 129–143. [Google Scholar] [CrossRef]
- Genovese, A.; Hosseini, M.S.; Piuri, V.; Plataniotis, K.N.; Scotti, F. Acute lymphoblastic leukemia detection based on adaptive unsharpening and deep learning. In Proceedings of the 2021 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP 2021), Toronto, ON, Canada, 6–11 June 2021; pp. 1205–1209. [Google Scholar] [CrossRef]
- Rezayi, S.; Mohammadzadeh, N.; Bouraghi, H.; Saeedi, S.; Mohammadpour, A. Timely Diagnosis of Acute Lymphoblastic Leukemia Using Artificial Intelligence-Oriented Deep Learning Methods. Comput. Intell. Neurosci. 2021, 2021, 5478157. [Google Scholar] [CrossRef] [PubMed]
- Abunadi, I.; Senan, E.M. Multi-Method Diagnosis of Blood Microscopic Sample for Early Detection of Acute Lymphoblastic Leukemia Based on Deep Learning and Hybrid Techniques. Sensors 2022, 22, 1629. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.J.; Nayak, P. Detection of Lymphoblastic Leukemia Using VGG19 Model. In Proceedings of the 2021 Fifth International Conference on I-SMAC (IOT in Social, Mobile, Analytics and Cloud) (I-SMAC 2021), Palladam, India, 11–13 November 2021; pp. 716–723. [Google Scholar] [CrossRef]
- Ansari, S.; Navin, A.H.; Sangar, A.B.; Gharamaleki, J.V.; Danishvar, S. A Customized Efficient Deep Learning Model for the Diagnosis of Acute Leukemia Cells Based on Lymphocyte and Monocyte Images. Electronics 2023, 12, 322. [Google Scholar] [CrossRef]
- Das, P.K.; Meher, S. Transfer Learning-Based Automatic Detection of Acute Lymphocytic Leukemia. In Proceedings of the 2021 National Conference on Communications (NCC), Virtual, 27–30 July 2021; pp. 386–391. [Google Scholar] [CrossRef]
- Genovese, A.; Hosseini, M.S.; Piuri, V.; Plataniotis, K.N.; Scotti, F. Histopathological Transfer Learning for Acute Lymphoblastic Leukemia Detection. In Proceedings of the 2021 IEEE International Conference on Computational Intelligence and Virtual Environments for Measurement Systems and Applications (IEEE CIVEMSA 2021), Hong Kong, China, 18–19 June 2021. [Google Scholar] [CrossRef]
- Jawahar, M.; Sharen, H.; Anbarasi, J.L.; Gandomi, A.H. ALNett: A cluster layer deep convolutional neural network for acute lymphoblastic leukemia classification. Comput. Biol. Med. 2022, 148, 105894. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Sahoo, B.; Meher, S. An Efficient Detection and Classification of Acute Leukemia Using Transfer Learning and Orthogonal Softmax Layer-Based Model. IEEE ACM Trans. Comput. Biol. Bioinform. 2023, 20, 1817–1828. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Singh, S.; Sheet, D. Simultaneous Localization and Classification of Acute Lymphoblastic Leukemic Cells in Peripheral Blood Smears Using a Deep Convolutional Network with Average Pooling Layer. In Proceedings of the 2017 IEEE International Conference on Industrial and Information Systems (ICIIS), Peradeniya, Sri Lanka, 15–16 December 2017; pp. 529–534. [Google Scholar]
- Atteia, G.; Alhussan, A.A.; Samee, N.A. BO-ALLCNN: Bayesian-Based Optimized CNN for Acute Lymphoblastic Leukemia Detection in Microscopic Blood Smear Images. Sensors 2022, 22, 5520. [Google Scholar] [CrossRef] [PubMed]
- Ghaderzadeh, M.; Aria, M.; Hosseini, A.; Asadi, F.; Bashash, D.; Abolghasemi, H. A fast and efficient CNN model for BALL diagnosis and its subtypes classification using peripheral blood smear images. Int. J. Intell. Syst. 2022, 37, 5113–5133. [Google Scholar] [CrossRef]
- Gehlot, S.; Gupta, A.; Gupta, R. SDCT-AuxNet(theta): DCT augmented stain deconvolutional CNN with auxiliary classifier for cancer diagnosis. Med. Image Anal. 2020, 61, 101661. [Google Scholar] [CrossRef]
- Mohammed, K.K.; Hassanien, A.E.; Afify, H.M. Refinement of ensemble strategy for acute lymphoblastic leukemia microscopic images using hybrid CNN-GRU-BiLSTM and MSVM classifier. Neural Comput. Appl. 2023, 35, 17415–17427. [Google Scholar] [CrossRef]
- Hui, L.W.; Mashor, M.Y.; Abd Halim, N.H.; Hassan, R. An Acute Leukemia Subtype Classification Procedure Based on Blood Sample Images. In Proceedings of the International Conference on Biomedical Engineering, ICOBE 2021, Perlis, Malaysia, 14–15 September 2021; Volume 2562. [Google Scholar] [CrossRef]
- Billah, M.E.; Javed, F. Bayesian Convolutional Neural Network-based Models for Diagnosis of Blood Cancer. Appl. Artif. Intell. 2022, 36, 2011688. [Google Scholar] [CrossRef]
- Jha, K.K.; Dutta, H.S. Mutual information based hybrid model and deep learning for acute lymphocytic leukemia detection in single cell blood smear images. Comput. Methods Programs Biomed. 2019, 179, 104987. [Google Scholar] [CrossRef] [PubMed]
- Genovese, A.; Piuri, V.; Plataniotis, K.N.; Scotti, F. DL4ALL: Multi-Task Cross-Dataset Transfer Learning for Acute Lymphoblastic Leukemia Detection. IEEE Access 2023, 2023, 65222–65237. [Google Scholar] [CrossRef]
- Ullah, Z.M.; Zheng, Y.; Song, J.; Aslam, S.; Xu, C.; Kiazolu, G.D.; Wang, L. An attention-based convolutional neural network for acute lymphoblastic leukemia classification. Appl. Sci. 2021, 11, 10662. [Google Scholar] [CrossRef]
- Mirmohammadi, P.; Ameri, M.; Shalbaf, A. Recognition of acute lymphoblastic leukemia and lymphocytes cell subtypes in microscopic images using random forest classifier. Phys. Eng. Sci. Med. 2021, 44, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Abhishek, A.; Jha, R.K.; Sinha, R.; Jha, K. Automated classification of acute leukemia on a heterogeneous dataset using machine learning and deep learning techniques. Biomed. Signal Process. Control 2022, 72, 103341. [Google Scholar] [CrossRef]
- Devi, T.G.; Patil, N.; Rai, S.; Philipose, C.S. Gaussian blurring technique for detecting and classifying acute lymphoblastic leukemia cancer cells from microscopic biopsy images. Life 2023, 13, 348. [Google Scholar] [CrossRef] [PubMed]
- Khandekar, R.; Shastry, P.; Jaishankar, S.; Faust, O.; Sampathila, N. Automated blast cell detection for Acute Lymphoblastic Leukemia diagnosis. Biomed. Signal Process. Control 2021, 68, 102690. [Google Scholar] [CrossRef]
- Sampathila, N.; Chadaga, K.; Goswami, N.; Chadaga, R.P.; Pandya, M.; Prabhu, S.; Bairy, M.G.; Katta, S.S.; Bhat, D.; Upadya, S.P. Customized deep learning classifier for detection of acute lymphoblastic leukemia using blood smear images. InHealthcare 2022, 10, 1812. [Google Scholar] [CrossRef]
- Ahmed, I.A.; Senan, E.M.; Shatnawi, H.S.; Alkhraisha, Z.M.; Al-Azzam, M.M. Hybrid techniques for the diagnosis of acute lymphoblastic leukemia based on fusion of CNN features. Diagnostics 2023, 13, 1026. [Google Scholar] [CrossRef]
- Jiang, Z.; Dong, Z.; Wang, L.; Jiang, W. Method for diagnosis of acute lymphoblastic leukemia based on ViT-CNN ensemble model. Comput. Intell. Neurosci. 2021, 2021, 7529893. [Google Scholar] [CrossRef]
- Saeed, A.; Shoukat, S.; Shehzad, K.; Ahmad, I.; Eshmawi, A.A.; Amin, A.H.; Tag-Eldin, E. A deep learning-based approach for the diagnosis of acute lymphoblastic leukemia. Electronics 2022, 11, 3168. [Google Scholar] [CrossRef]
- Hamza, M.A.; Albraikan, A.A.; Alzahrani, J.S.; Dhahbi, S.; Al-Turaiki, I.; Al Duhayyim, M.; Yaseen, I.; Eldesouki, M.I. Optimal deep transfer learning-based human-centric biomedical diagnosis for acute lymphoblastic leukemia detection. Comput. Intell. Neurosci. 2022, 2022, 7954111. [Google Scholar] [CrossRef] [PubMed]
- Ghaderzadeh, M.; Hosseini, A.; Asadi, F.; Abolghasemi, H.; Bashash, D.; Roshanpoor, A. Automated Detection Model in Classification of B-Lymphoblast Cells from Normal B-Lymphoid Precursors in Blood Smear Microscopic Images Based on the Majority Voting Technique. Sci. Program. 2022, 2022, 4801671. [Google Scholar] [CrossRef]
- Gao, S.; Li, S. Bloody Mahjong playing strategy based on the integration of deep learning and XGBoost. CAAI Trans. Intell. Technol. 2022, 7, 95–106. [Google Scholar] [CrossRef]
- Kumar, S.; Jain, A.; Rani, S.; Alshazly, H.; Idris, S.A.; Bourouis, S. Deep Neural Network Based Vehicle Detection and Classification of Aerial Images. Intell. Autom. Soft Comput. 2022, 34, 119–131. [Google Scholar] [CrossRef]
- Liu, X. Real-world data for the drug development in the digital era. J. Artif. Intell. Technol. 2022, 2, 42–46. [Google Scholar] [CrossRef]
- Aria, M.; Ghaderzadeh, M.; Bashash, D.; Abolghasemi, H.; Asadi, F.; Hosseini, A. Acute Lymphoblastic Leukemia (ALL) Image Dataset. 2021. Available online: https://www.kaggle.com/datasets/obulisainaren/multi-cancer (accessed on 13 February 2023). [CrossRef]


| Ref. | Year | Model Name | Benefits | Unsolved Challenges |
|---|---|---|---|---|
| [20] | 2017 | DCNN | Simultaneous localization and classification of ALL in peripheral blood smear images | Limitations in detecting all ALL lymphocytes |
| [21] | 2020 | BO-ALLCNN | Improved performance in ALL image classification | Limitations or trade-offs in employing BO not mentioned |
| [23] | 2020 | SDCT-AuxNet | Improved performance with stain deconvolved quantity images and dual-classifier approach | Generalizability to other cancer types or datasets not addressed |
| [16] | 2021 | SqueezeNet | Highly computationally efficient approach with superior performance | Limited depth and sensitivity to noisy data |
| [17] | 2021 | HTL | Promising results in ALL detection after fine-tuning on ALL database | May perform poorly for images with poor visibility |
| [19] | 2021 | TLOS-Net | Efficient ALL detection on small medical datasets using ResNet18 | Potential computational overhead, requirement of optimal tuning, and sensitivity to certain hyperparameters |
| [14] | 2021 | VGG-19 | Improved accuracy and speed of leukemia image classification | High computational complexity and memory requirements |
| [12] | 2021 | ResNet-50 & VGG-16 | Promising results for ALL classification and potential clinical usage in leukemia diagnosis | Need for further validation in clinical settings not addressed, high computational complexity and memory requirements |
| [22] | 2022 | FCNN | High accuracy, sensitivity, and specificity in distinguishing ALL from benign cases | Potential computational overhead, requirement of optimal tuning, and sensitivity to certain hyperparameters |
| [39] | 2022 | CNN | High accuracy and specificity in distinguishing ALL from benign cases | May lead to overfitting, requirement of optimal tuning, and sensitivity to certain hyperparameters |
| [11] | 2021 | AU-CNN | Enhancement of blood sample images, improved sharpness, and potential for accurate ALL detection | Need for further validation in clinical settings not addressed, high computational complexity and memory requirements |
| [26] | 2022 | BCNN | High accuracy in classifying cancerous and noncancerous lymphocyte cells | Requirement of optimal tuning and sensitivity to certain hyperparameters |
| [13] | 2022 | AlexNet, GoogLeNet, & ResNet18 | High accuracies in ALL image classification, contributing to efficient diagnostic systems | High computational complexity, memory requirements, and challenges with noisy or low-quality input images |
| [15] | 2023 | CDL | Contribution to medical research with GAN-generated dataset | Biases in the generated data and potential lack of diversity in samples |
| [24] | 2023 | CNN–GRU–BiLSTM | High accuracy and sensitivity in ALL cell detection | Sensitivity to specific hyperparameters and the need for careful tuning |
| [18] | 2023 | ALNett | Robust local and global feature extraction for accurate ALL prediction | Challenges with noisy or low-quality input images |
| [25] | 2023 | AlexNet & GoogLeNet | High accuracy in classifying white blood cells for ALL detection | GoogLeNet’s complex architecture leads to high memory consumption and makes fine-tuning challenging |
| [29] | 2021 | VGG-16 | Provides better robustness and adaptability | High computational complexity and memory requirements |
| [36] | 2021 | ViT-CNN | Improved accuracy by using two different feature extraction methods simultaneously | High computational complexity and memory requirements |
| [34] | 2022 | ALLNET | Can be used during peripheral or complete blood count test | High computational complexity and memory requirements |
| [31] | 2022 | CNN model | Heterogeneous dataset utilized for binary and three-class classification | Potential lack of diversity in data |
| [38] | 2022 | ODLHBD-ALLD | Better accuracy by incorporating several state-of-the-art techniques | Potential computational overhead, trained & tested on a small dataset |
| [35] | 2023 | Hybrid CNN | Improved accuracy and fusion of different CNN models | Potential computational overhead, requirement of optimal tuning |
| [32] | 2023 | GBHSV-Leuk | Improved prediction accuracy | May lead to overfitting and sensitivity to certain hyperparameters |
| [28] | 2023 | DL4ALL | Detects ALL even if manual labels are not used for the source domain | Computationally expensive |
| Ref. | Method | Accuracy | Sensitivity | Specificity | F-Score | Dataset |
|---|---|---|---|---|---|---|
| [27] | SCA-based deep CNN | 98.7 | - | - | - | ALL-IDB2 |
| [28] | DL4ALL | 97.85 | 95.81 | 98.79 | - | ADP and C_NMC_2019 |
| [29] | VGG-16 | 91.1 | 92.31 | 90.25 | 90.65 | C_NMC_2019 |
| [30] | RF Classifier | 98.22 | - | - | - | Isfahan Univ. of Med. Sci. |
| [31] | VGG-16 DenseNet & SVM | 97 | - | - | - | Novel dataset and ALL-IDB |
| [32] | GBHSV-Leuk | 95.41 | 87.75 | 95.81 | 91.61 | Private dataset and ALL-IDB1 |
| [33] | YOLOv4 | 92 | 96 | 91 | 92 | ALL_IDB1 and C_NMC_2019 |
| [34] | ALL-NET | 95.54 | 95.91 | 95.81 | 95.43 | ALL and C_NMC_2019 |
| [38] | ODLHBD-ALLD | 96.97 | 96.88 | 96.88 | 96.96 | ALL_IDB1 |
| Proposed | DSCNet | 99.37 ± 0.40 | 99.71 ± 0.21 | 99.03 ± 0.55 | 99.37 ± 0.43 | ALL dataset |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, M.; AlZubi, A.A.; Jain, A.; Singh, D.; Yadav, V.; Alkhayyat, A. DSCNet: Deep Skip Connections-Based Dense Network for ALL Diagnosis Using Peripheral Blood Smear Images. Diagnostics 2023, 13, 2752. https://doi.org/10.3390/diagnostics13172752
Kaur M, AlZubi AA, Jain A, Singh D, Yadav V, Alkhayyat A. DSCNet: Deep Skip Connections-Based Dense Network for ALL Diagnosis Using Peripheral Blood Smear Images. Diagnostics. 2023; 13(17):2752. https://doi.org/10.3390/diagnostics13172752
Chicago/Turabian StyleKaur, Manjit, Ahmad Ali AlZubi, Arpit Jain, Dilbag Singh, Vaishali Yadav, and Ahmed Alkhayyat. 2023. "DSCNet: Deep Skip Connections-Based Dense Network for ALL Diagnosis Using Peripheral Blood Smear Images" Diagnostics 13, no. 17: 2752. https://doi.org/10.3390/diagnostics13172752
APA StyleKaur, M., AlZubi, A. A., Jain, A., Singh, D., Yadav, V., & Alkhayyat, A. (2023). DSCNet: Deep Skip Connections-Based Dense Network for ALL Diagnosis Using Peripheral Blood Smear Images. Diagnostics, 13(17), 2752. https://doi.org/10.3390/diagnostics13172752








