Modulation of Neurotransmitter Pathways and Associated Metabolites by Systemic Silencing of Gut Genes in C. elegans
Abstract
1. Introduction
2. Materials and Methods
2.1. C. elegans Strains and Maintenance
2.2. dsRNA-Induced RNAi Silencing
2.3. Thrashing Assay
2.4. Imaging of Human α-Synuclein Protein Expression
2.5. Aversion Assay
2.6. NMR Sample Preparation
2.7. NMR Experimental Condition
3. Results
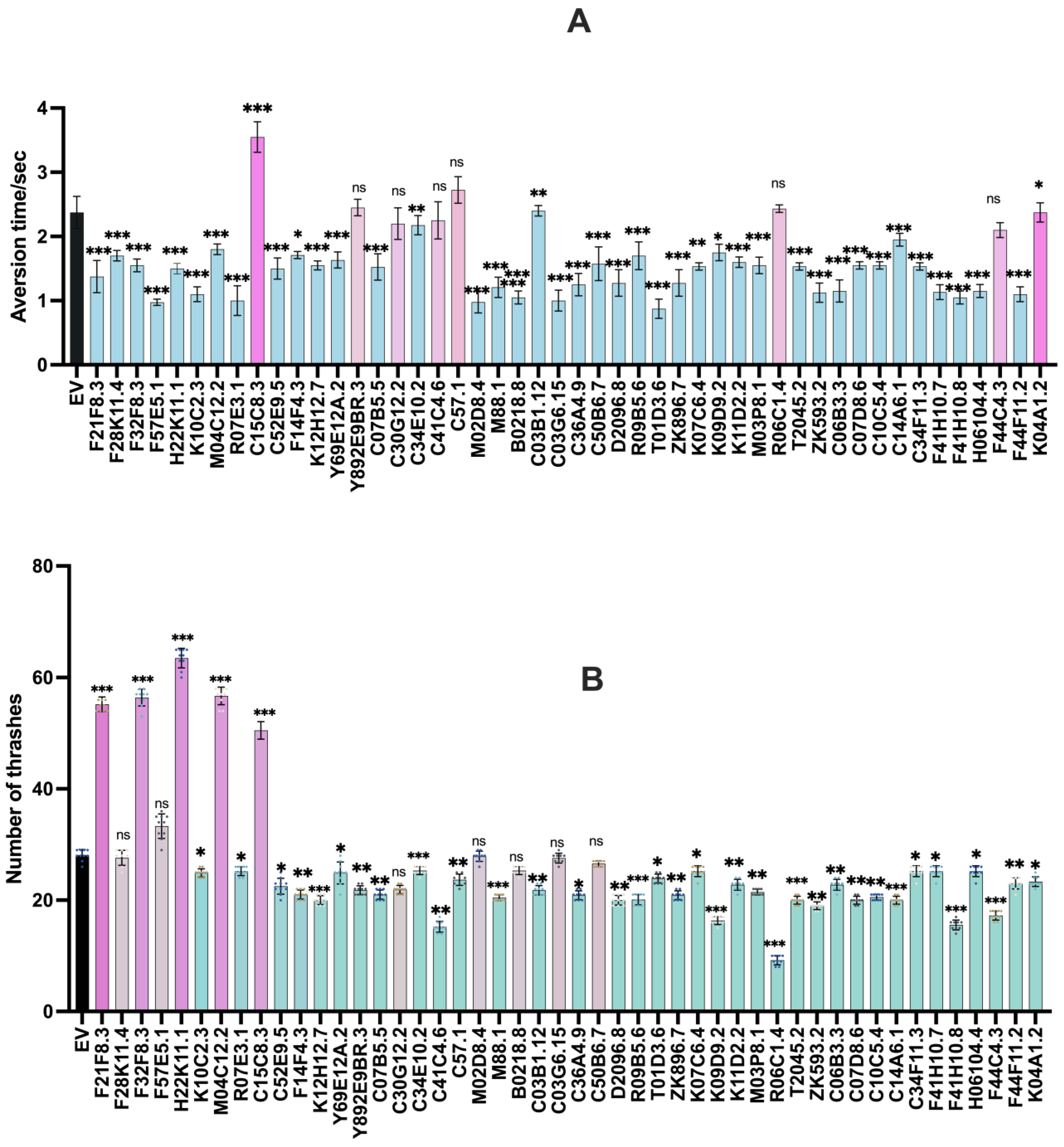
3.1. Screening of 48 Gut Genes on the Basis of Behavioural Assays That are Regulated by Dopamine and ACh
3.1.1. Aversion Assay
3.1.2. Thrashing Assay
3.2. dsRNA-Induced RNAi Systemic Silencing of Gut Genes in Transgenic Model of C. elegans Altered α-Synuclein Expression
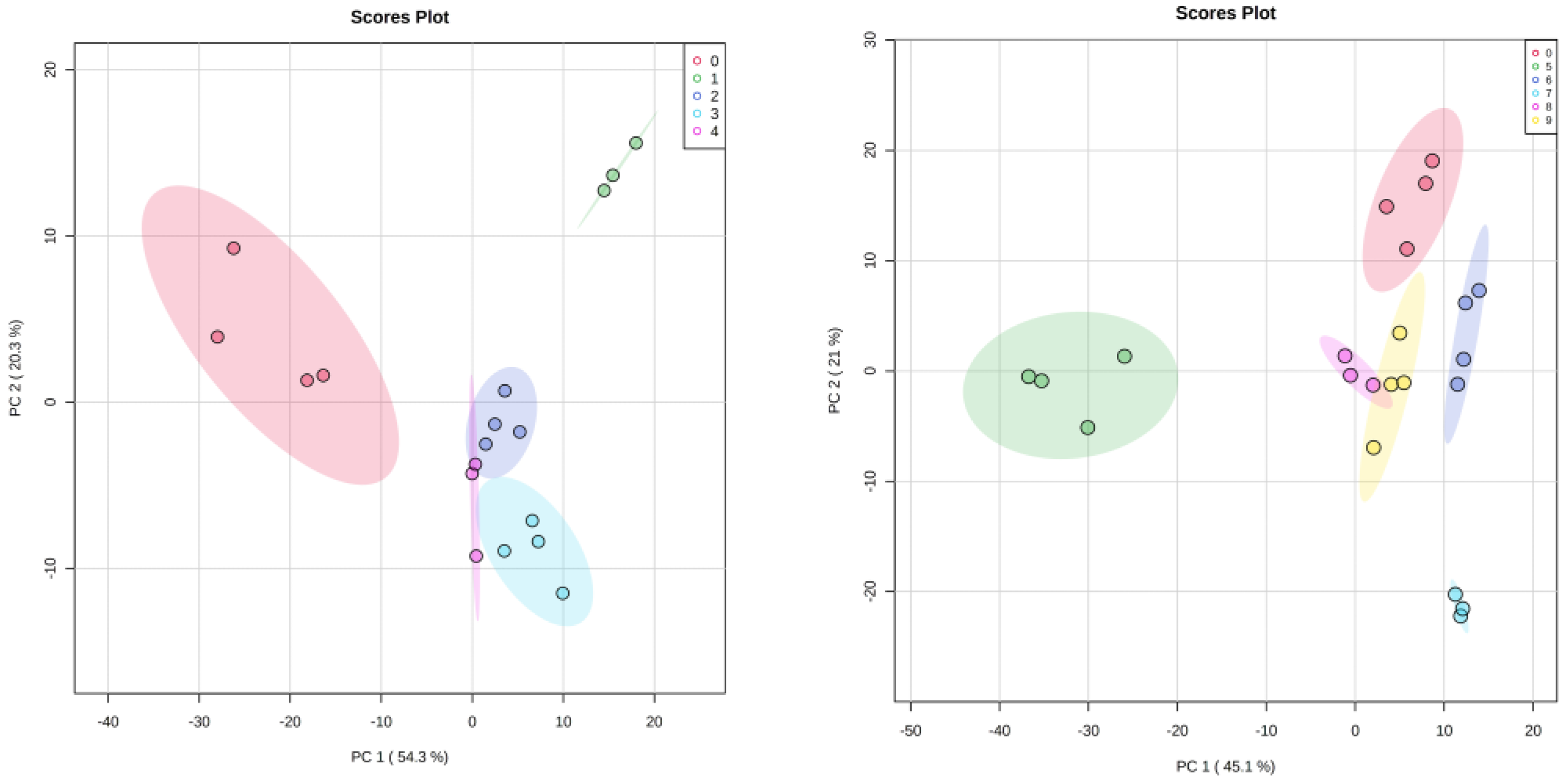
3.3. Characterization of Metabolites
3.4. Statistical Analysis
3.5. HR-MAS NMR Spectroscopy Based Metabolome Profiling Strongly Reflects the Reciprocity of Gut Homeostasis and Neuronal Health
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Przedborski, S.; Vila, M.; Jackson-Lewis, V. Series Introduction: Neurodegeneration: What Is It and Where Are We? J. Clin. Investig. 2003, 111, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Collins, S.M.; Surette, M.; Bercik, P. The interplay between the intestinal microbiota and the brain. Nat. Rev. Microbiol. 2012, 10, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Bonaz, B.; Bazin, T.; Pellissier, S. The Vagus Nerve at the Interface of the Microbiota-Gut-Brain Axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef]
- Badman, M.K.; Flier, J.S. The Gut and Energy Balance: Visceral Allies in the Obesity Wars. Science 2005, 307, 1909–1914. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Wu, W.; Li, Y.; Li, L. Influences of the Gut Microbiota on DNA Methylation and Histone Modification. Dig. Dis. Sci. 2017, 62, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Knauf, C. How gut microbes talk to organs: The role of endocrine and nervous routes. Mol. Metab. 2016, 5, 743–752. [Google Scholar] [CrossRef]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef]
- Koutzoumis, D.N.; Vergara, M.; Pino, J.; Buddendorff, J.; Khoshbouei, H.; Mandel, R.J.; Torres, G.E. Alterations of the gut microbiota with antibiotics protects dopamine neuron loss and improve motor deficits in a pharmacological rodent model of Parkinson’s disease. Exp. Neurol. 2020, 325, 113159. [Google Scholar] [CrossRef]
- Maiuolo, J.; Gliozzi, M.; Musolino, V.; Carresi, C.; Scarano, F.; Nucera, S.; Scicchitano, M.; Oppedisano, F.; Bosco, F.; Ruga, S.; et al. The Contribution of Gut Microbiota–Brain Axis in the Development of Brain Disorders. Front. Neurosci. 2021, 15, 616883. [Google Scholar] [CrossRef]
- Gubert, C.; Kong, G.; Renoir, T.; Hannan, A.J. Exercise, diet and stress as modulators of gut microbiota: Implications for neurodegenerative diseases. Neurobiol. Dis. 2020, 134, 104621. [Google Scholar] [CrossRef]
- Muller, P.A.; Schneeberger, M.; Matheis, F.; Wang, P.; Kerner, Z.; Ilanges, A.; Pellegrino, K.; del Mármol, J.; Castro, T.B.R.; Furuichi, M.; et al. Microbiota modulate sympathetic neurons via a gut–brain circuit. Nature 2020, 583, 441–446. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, K.; Mulak, A. Brain-Gut-Microbiota Axis in Alzheimer’s Disease. J. Neurogastroenterol. Motil. 2019, 25, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Stiernagle, T. Maintenance of C. Elegans. WormBook Online Rev. C. Elegans Biol. 2006, 11, 1–11. [Google Scholar] [CrossRef]
- Sloan, M.A.; Reaves, B.J.; Maclean, M.J.; Storey, B.E.; Wolstenholme, A.J. Expression of nicotinic acetylcholine receptor subunits from parasitic nematodes in Caenorhabditis elegans. Mol. Biochem. Parasitol. 2015, 204, 44–50. [Google Scholar] [CrossRef]
- Sammi, S.R.; Agim, Z.; Cannon, J.R. From the Cover: Harmane-Induced Selective Dopaminergic Neurotoxicity in Caenorhabditis elegans. Toxicol. Sci. 2017, 161, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Blaise, B.J.; Giacomotto, J.; Elena, B.; Dumas, M.-E.; Toulhoat, P.; Ségalat, L.; Emsley, L. Metabotyping of Caenorhabditis elegans reveals latent phenotypes. Proc. Natl. Acad. Sci. USA 2007, 104, 19808–19812. [Google Scholar] [CrossRef]
- Singh, N.; Sharma, G.; Dev, I.; Shukla, S.K.; Ansari, K.M. Study of the metabolic alterations in patulin-induced neoplastic transformation in normal intestinal cells. Toxicol. Res. 2021, 10, 592–600. [Google Scholar] [CrossRef]
- Sethi, N.; Mahar, R.; Shukla, S.K.; Kumar, A.; Sinha, N. A novel approach for testing the teratogenic potential of chemicals on the platform of metabolomics: Studies employing HR-MAS nuclear magnetic resonance spectroscopy. RSC Adv. 2015, 5, 26027–26039. [Google Scholar] [CrossRef]
- McDonald, P.W.; Hardie, S.L.; Jessen, T.N.; Carvelli, L.; Matthies, D.S.; Blakely, R.D. Vigorous Motor Activity in Caenorhabditis elegans Requires Efficient Clearance of Dopamine Mediated by Synaptic Localization of the Dopamine Transporter DAT-1. J. Neurosci. 2007, 27, 14216–14227. [Google Scholar] [CrossRef]
- Pahuja, R.; Seth, K.; Shukla, A.; Shukla, R.K.; Bhatnagar, P.; Chauhan, L.K.S.; Saxena, P.N.; Arun, J.; Chaudhari, B.P.; Patel, D.K.; et al. Trans-Blood Brain Barrier Delivery of Dopamine-Loaded Nanoparticles Reverses Functional Deficits in Parkinsonian Rats. ACS Nano 2015, 9, 4850–4871. [Google Scholar] [CrossRef] [PubMed]
- Nishijima, S.; Maruyama, I.N. Appetitive Olfactory Learning and Long-Term Associative Memory in Caenorhabditis elegans. Front. Behav. Neurosci. 2017, 11, 80. [Google Scholar] [CrossRef] [PubMed]
- Borgohain, R.; Kandadai, R.M.; Ch, M.K.V.; Jabeen, A.; Kanikannan, M.A. Prevalence of Quantitaive Sensory Abnormalities and Correlation with Autonomic Disturbances in Patients with Idiopathic Parkinson’s Disease and Parkinson’s plus Syndromes. In Movement Disorders; Wiley-Blackwell 111 River ST: Hoboken, NJ, USA, 2015; Volume 30, p. S192. [Google Scholar]
- Caputi, V.; Giron, M.C. Microbiome-Gut-Brain Axis and Toll-Like Receptors in Parkinson’s Disease. Int. J. Mol. Sci. 2018, 19, 1689. [Google Scholar] [CrossRef] [PubMed]
- Stolzenberg, E.; Berry, D.; Yang, D.; Lee, E.Y.; Kroemer, A.; Kaufman, S.; Wong, G.C.; Oppenheim, J.J.; Sen, S.; Fishbein, T.; et al. A Role for Neuronal Alpha-Synuclein in Gastrointestinal Immunity. J. Innate Immun. 2017, 9, 456–463. [Google Scholar] [CrossRef]
- Doroszkiewicz, J.; Groblewska, M.; Mroczko, B. The Role of Gut Microbiota and Gut–Brain Interplay in Selected Diseases of the Central Nervous System. Int. J. Mol. Sci. 2021, 22, 10028. [Google Scholar] [CrossRef]
- McGhee, J.D.; Sleumer, M.C.; Bilenky, M.; Wong, K.; McKay, S.J.; Goszczynski, B.; Tian, H.; Krich, N.D.; Khattra, J.; Holt, R.A.; et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2007, 302, 627–645. [Google Scholar] [CrossRef]
- Wong, A.; Li, X.; Molin, L.; Solari, F.; Elena-Herrmann, B.; Sakellariou, D. μHigh Resolution-Magic-Angle Spinning NMR Spectroscopy for Metabolic Phenotyping of Caenorhabditis elegans. Anal. Chem. 2014, 86, 6064–6070. [Google Scholar] [CrossRef]
- Ahmed, S.S.; Santosh, W.; Kumar, S.; Christlet, H.T.T. Metabolic profiling of Parkinson’s disease: Evidence of biomarker from gene expression analysis and rapid neural network detection. J. Biomed. Sci. 2009, 16, 63. [Google Scholar] [CrossRef]
- Andersen, J.V.; Markussen, K.H.; Jakobsen, E.; Schousboe, A.; Waagepetersen, H.S.; Rosenberg, P.A.; Aldana, B.I. Glutamate metabolism and recycling at the excitatory synapse in health and neurodegeneration. Neuropharmacology 2021, 196, 108719. [Google Scholar] [CrossRef]
- Ruhal, P.; Dhingra, D. Inosine improves cognitive function and decreases aging-induced oxidative stress and neuroinflammation in aged female rats. Inflammopharmacology 2018, 26, 1317–1329. [Google Scholar] [CrossRef]
- Banderet, L.E.; Lieberman, H.R. Treatment with tyrosine, a neurotransmitter precursor, reduces environmental stress in humans. Brain Res. Bull. 1989, 22, 759–762. [Google Scholar] [CrossRef]
- Chatterjee, P.; Cheong, Y.; Bhatnagar, A.; Goozee, K.; Wu, Y.; McKay, M.; Martins, I.J.; Lim, W.L.F.; Pedrini, S.; Tegg, M.; et al. Plasma metabolites associated with biomarker evidence of neurodegeneration in cognitively normal older adults. J. Neurochem. 2021, 159, 389–402. [Google Scholar] [CrossRef]
- Stone, T.W.; Darlington, L.G. The kynurenine pathway as a therapeutic target in cognitive and neurodegenerative disorders. Br. J. Pharmacol. 2013, 169, 1211–1227. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Laparra, J.M.; Sanz, Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol. Res. 2010, 61, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Imai, J.; Katagiri, H. Regulation of systemic metabolism by the autonomic nervous system consisting of afferent and efferent innervation. Int. Immunol. 2022, 34, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Lotharius, J.; Brundin, P. Pathogenesis of parkinson’s disease: Dopamine, vesicles and α-synuclein. Nat. Rev. Neurosci. 2002, 3, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Taliyan, R.; Dubey, S.K. Comprehensive Review on Potential Signaling Pathways Involving the Transfer of α-Synuclein from the Gut to the Brain That Leads to Parkinson’s Disease. ACS Chem. Neurosci. 2023, 14, 590–602. [Google Scholar] [CrossRef] [PubMed]


| F41H10.8 | elo(Predicted to enable fatty acid elongase activity) |
| M02D8.4 | asns(Predicted to enable asparagine synthase (glutamine-hydrolyzing) activity) |
| M88.1 | ugt(Predicted to enable glucuronosyltransferase activity) |
| C03G6.15 | cyp-35A2(Predicted to enable heme binding activity; oxidoreductase activity) |
| T01D3.6 | Predicted to enable calcium ion binding activity |
| R07E3.1 | Predicted to enable cysteine-type endopeptidase activity |
| C14A6.1 | clec-48(Predicted to enable signaling receptor activity. |
| K09D9.2 | cyp-35A3(Predicted to enable heme binding activity; oxidoreductase activity) |
| ZK593.2 | Is affected by several genes including daf-2; daf-12; eat-2 |
| S. No. | Metabolites | Concentration (µM) | ||||
|---|---|---|---|---|---|---|
| NL5901 (Mean ± SD) | F41H10.8 (Mean ± SD) | M02D8.4 (Mean ± SD) | M88.1 (Mean ± SD) | C03G6.15 (Mean ± SD) | ||
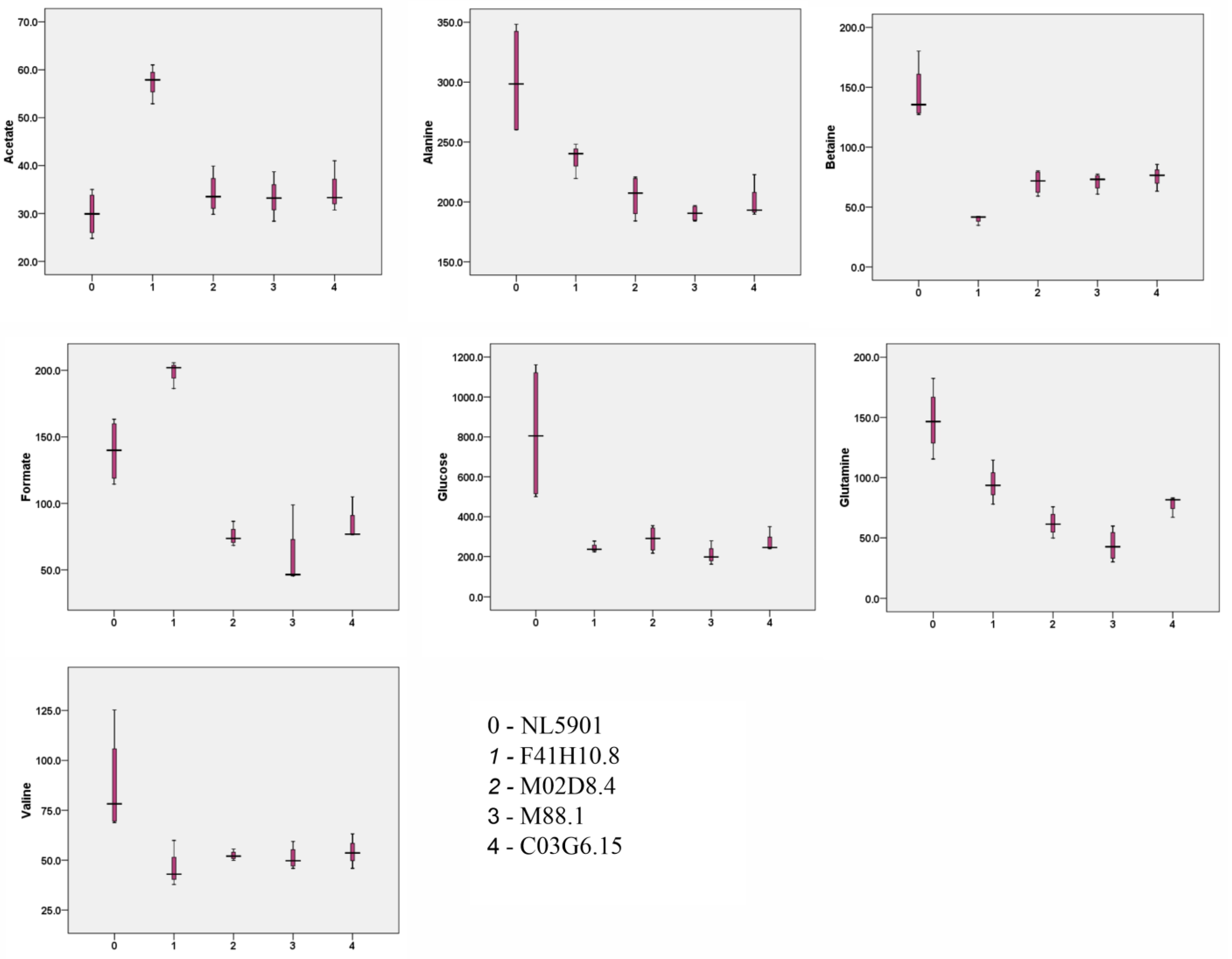
| 1 | Valine | 87.6 ± 26.3 | 46.9 ± 11.6 | 52.4 ± 2.4 | 51.2 ± 5.9 | 54.2 ± 8.7 |
| 2 | Ethanol | 136.5 ± 33.7 | - | - | - | 104.2 ± 14.3 |
| 3 | Leucine | 139.5 ± 75.8 | 83.0 ± 43.6 | 66.5 ± 22.4 | 71.9 ± 17.5 | 85.5 ± 15.2 |
| 4 | Alanine | 301.4 ± 47.6 | 236.0 ± 14.8 | 204.9 ± 17.6 | 190.6 ± 6.6 | 201.9 ± 18.2 |
| 5 | Acetate | 29.9 ± 4.7 | 57.2 ± 4.1 | 34.2 ± 4.3 | 33.4 ± 4.2 | 35.0 ± 5.4 |
| 6 | Glutamate | 276.5 ± 32.0 | 399.5 ± 25.3 | 322.9 ± 62.1 | 327.9 ± 38.3 | 369.1 ± 62.4 |
| 7 | Glutamine | 147.8 ± 27.6 | 95.4 ± 18.4 | 62.1 ± 10.7 | 43.8 ± 13.3 | 77.3 ± 9.0 |
| 8 | Betaine | 144.7 ± 24.6 | 39.6 ± 4.1 | 70.8 ± 10.1 | 71.2 ± 7.4 | 75.1 ± 11.3 |
| 9 | Glucose | 818.2 ± 351.8 | 246.2 ± 28.1 | 288.5 ± 65.8 | 209.4 ± 49.5 | 278.6 ± 61.8 |
| 10 | Trehalose | 133.2 ± 16.1 | - | 74.9 ± 9.7 | 65.7 ± 14.7 | 55.8 ± 22.0 |
| 11 | Inosine | 85.3 ± 20.7 | - | - | - | - |
| 12 | Tyrosine | 62.2 ± 24.6 | - | - | - | - |
| 13 | Phenylalanine | 70.4 ± 20.7 | - | - | - | - |
| 14 | Formate | 139.4 ± 24.0 | 198.0 ± 10.2 | 75.6 ± 7.7 | 59.3 ± 26.3 | 86.1 ± 16.3 |
| 15 | Methanol | 99.4 ± 12.4 | 194.0 ± 11.3 | 96.5 ± 17.0 | 96.9 ± 7.1 | 64.4 ± 20.6 |
| 16 | Glycine | 68.2 ± 28.8 | - | - | 69.2 ± 14.4 | 67.2 ± 14.7 |
| S.No. | Metabolites | Concentration (µM) | ||||
|---|---|---|---|---|---|---|
| T01D3.6 (Mean ± SD) | R07E3.1 (Mean ± SD) | C14A6.1 (Mean ± SD) | K09D9.2 (Mean ± SD) | ZK593.2 (Mean ± SD) | ||
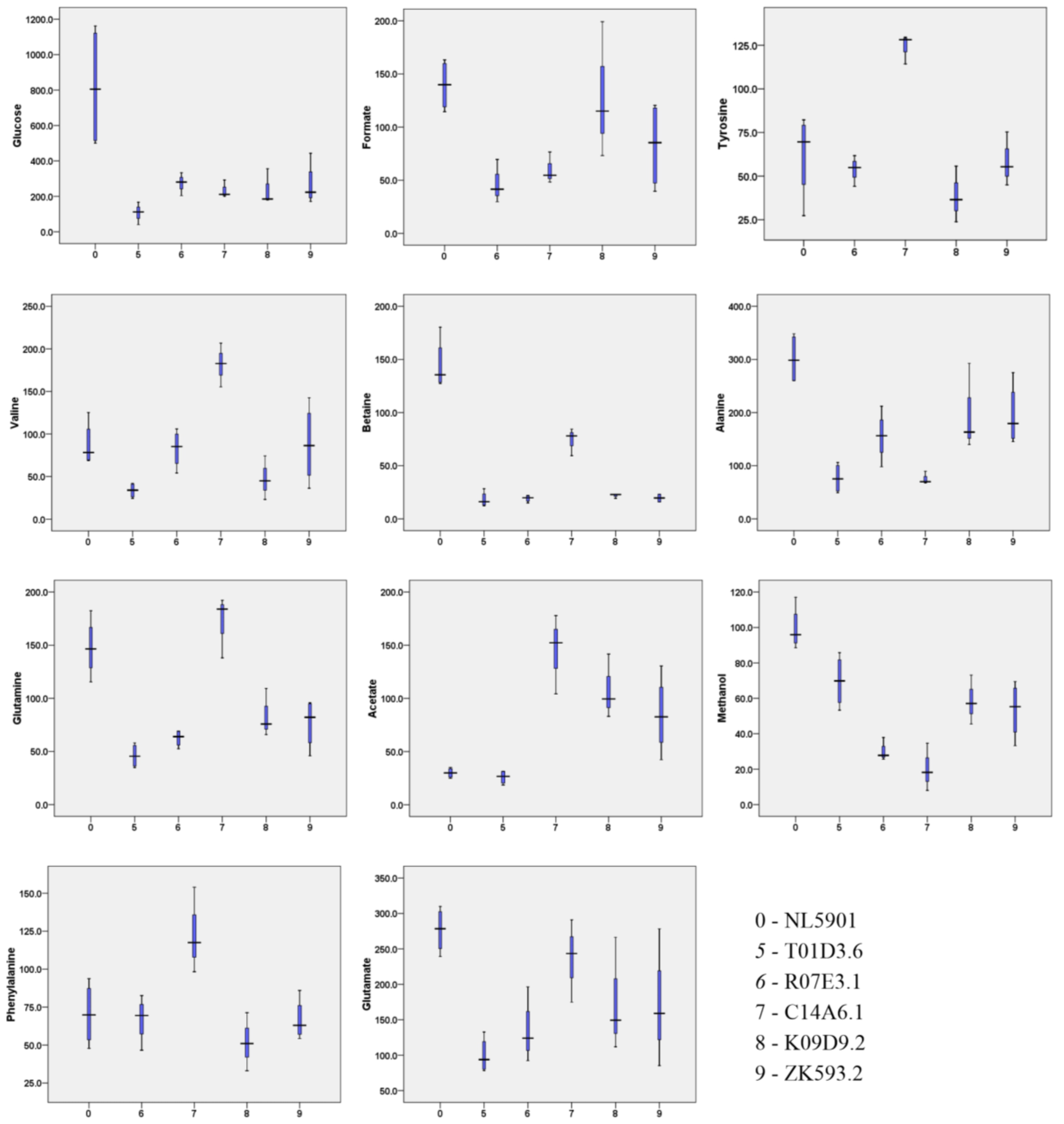
| 1 | Valine | 33.6 ± 8.8 | 82.7 ± 22.5 | 181.7 ± 25.7 | 47.5 ± 25.7 | 88.0 ± 46.3 |
| 2 | Ethanol | 468.2 ± 145.8 | - | - | 163.3 ± 48.3 | 109.2 ± 47.5 |
| 3 | Leucine | 51.3 ± 9.3 | 82.8 ± 18.7 | 187.6 ± 36.2 | 46.0 ± 14.4 | 101.9 ± 20.5 |
| 4 | Alanine | 76.4 ± 28.3 | 155.7 ± 46.6 | 75.8 ± 12.0 | 198.7 ± 82.3 | 194.9 ± 58.7 |
| 5 | Acetate | 25.8 ± 6.4 | - | 144.8 ± 37.4 | 108.1 ± 30.2 | 84.6 ± 36.7 |
| 6 | Glutamate | 99.6 ± 25.2 | 134.2 ± 44.2 | 236.5 ± 58.4 | 175.9 ± 80.6 | 170.5 ± 79.9 |
| 7 | Glutamine | 46.0 ± 11.4 | 62.5 ± 7.9 | 171.5 ± 29.2 | 83.7 ± 22.8 | 76.5 ± 23.4 |
| 8 | Betaine | 18.2 ± 7.2 | 19.2 ± 3.2 | 74.0 ± 12.9 | 21.8 ± 2.1 | 19.6 ± 4.0 |
| 9 | Glucose | 107.7 ± 52.0 | 274.7 ± 53.2 | 234.8 ± 50.4 | 240.3 ± 100.1 | 265.3 ± 121.4 |
| 10 | Trehalose | - | - | 40.0 ± 17.0 | - | - |
| 11 | Inosine | - | 61.2 ± 18.6 | 42.5 ± 3.5 | 46.1 ± 26.4 | 33.3 ± 13.8 |
| 12 | Tyrosine | - | 53.9 ± 7.3 | 124.0 ± 8.5 | 38.7 ± 16.1 | 57.8 ± 12.78 |
| 13 | Phenylalanine | - | 67.0 ± 15.0 | 123.3 ± 28.3 | 51.8 ± 19.1 | 66.6 ± 13.8 |
| 14 | Formate | - | 45.6 ± 16.9 | 59.9 ± 14.8 | 129.2 ± 64.2 | 82.7 ± 41.3 |
| 15 | Methanol | 69.7 ± 14.7 | 29.8 ± 5.5 | 20.3 ± 13.4 | 58.6 ± 13.9 | 53.4 ± 15.9 |
| 16 | Glycine | 39.4 ± 13.0 | 84.0 ± 18.9 | 126.8 ± 47.0 | 150.0 ± 144.6 | 99.5 ± 25.1 |
| 17 | Uridine | - | 76.7 ± 36.8 | - | 59.5 ± 35.1 | 46.2 ± 9.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shukla, S.; Saxena, A.; Shukla, S.K.; Nazir, A. Modulation of Neurotransmitter Pathways and Associated Metabolites by Systemic Silencing of Gut Genes in C. elegans. Diagnostics 2023, 13, 2322. https://doi.org/10.3390/diagnostics13142322
Shukla S, Saxena A, Shukla SK, Nazir A. Modulation of Neurotransmitter Pathways and Associated Metabolites by Systemic Silencing of Gut Genes in C. elegans. Diagnostics. 2023; 13(14):2322. https://doi.org/10.3390/diagnostics13142322
Chicago/Turabian StyleShukla, Shikha, Ankit Saxena, Sanjeev K. Shukla, and Aamir Nazir. 2023. "Modulation of Neurotransmitter Pathways and Associated Metabolites by Systemic Silencing of Gut Genes in C. elegans" Diagnostics 13, no. 14: 2322. https://doi.org/10.3390/diagnostics13142322
APA StyleShukla, S., Saxena, A., Shukla, S. K., & Nazir, A. (2023). Modulation of Neurotransmitter Pathways and Associated Metabolites by Systemic Silencing of Gut Genes in C. elegans. Diagnostics, 13(14), 2322. https://doi.org/10.3390/diagnostics13142322






