Prediction of Distant Metastases in Patients with Kidney Cancer Based on Gene Expression and Methylation Analysis
Abstract
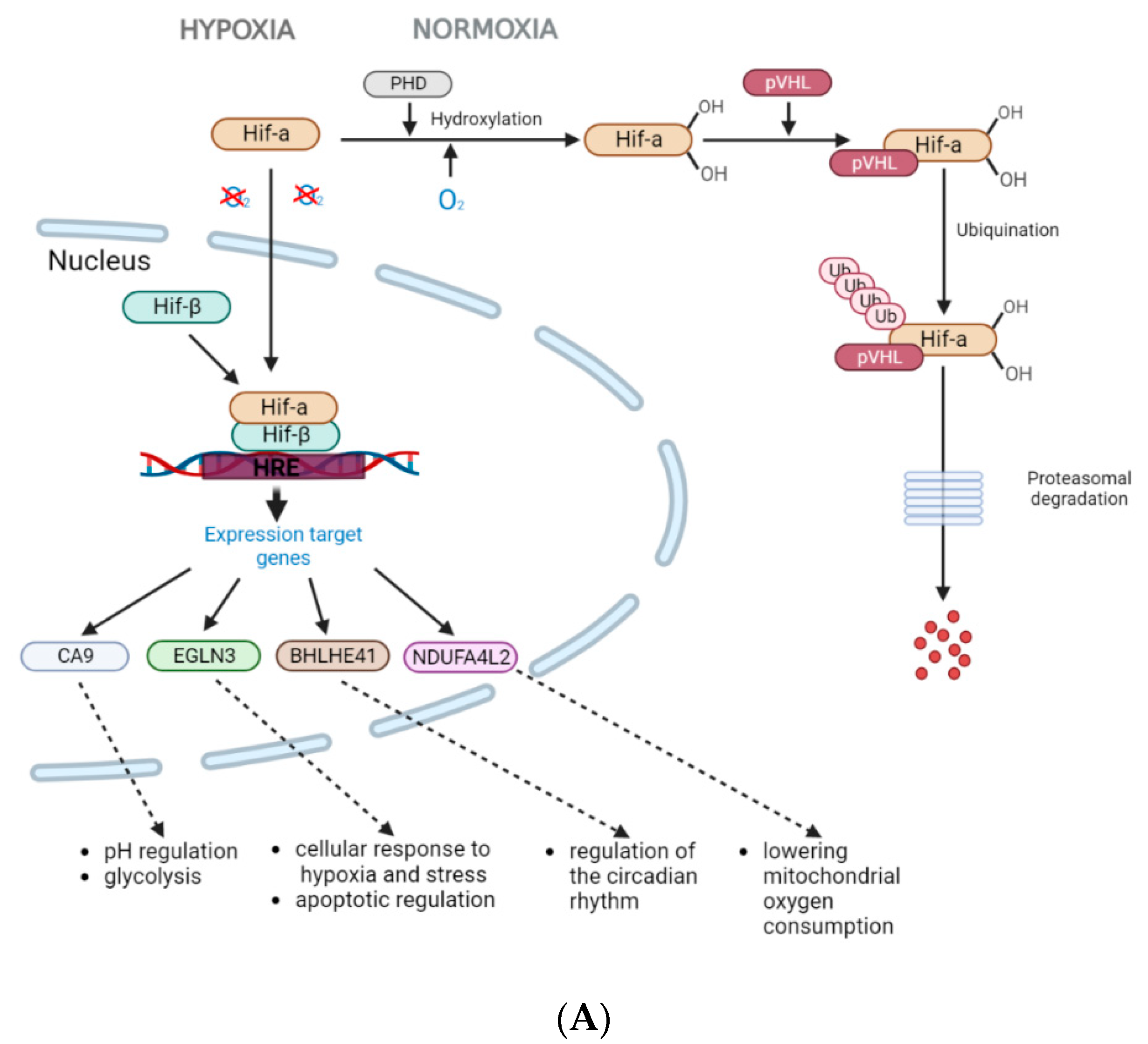
1. Introduction
2. Materials and Methods
3. Results
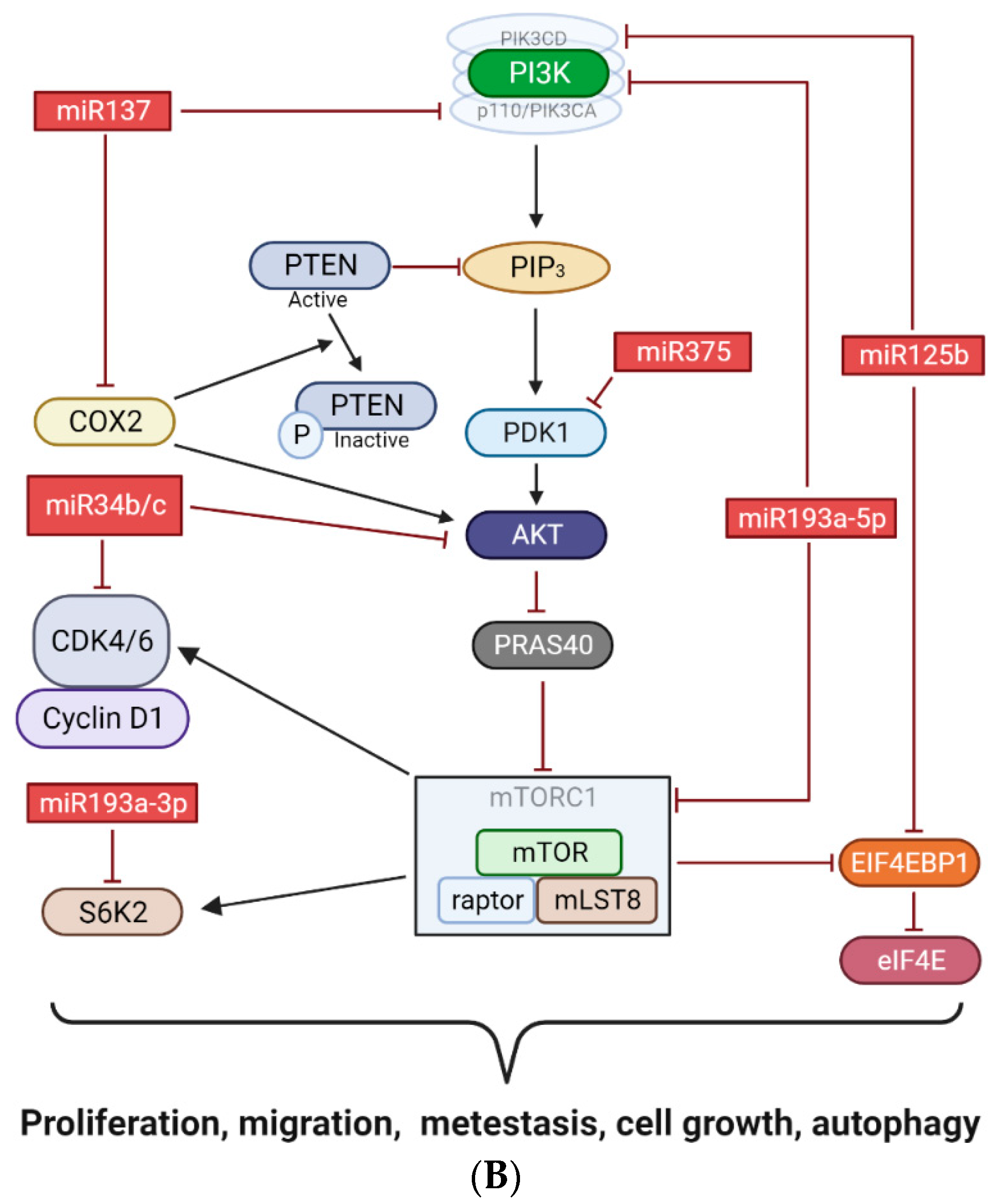
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Makino, T.; Kadomoto, S.; Izumi, K.; Mizokami, A. Epidemiology and Prevention of Renal Cell Carcinoma. Cancers 2022, 14, 4059. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Albiges, L.; Bedke, J.; Bex, A.; Capitanio, U.; Giles, R.H.; Hora, M.; Klatte, T.; Marconi, L.; Powles, T.; et al. EAU Guidelines on Renal Cell Carcinoma; EAU Annual Congress Milan; 2023; ISBN 978-94-92671-19-6. Available online: https://d56bochluxqnz.cloudfront.net/documents/full-guideline/EAU-Guidelines-on-Renal-Cell-Carcinoma-2023.pdf (accessed on 1 March 2023).
- Kaprin, A.; Starinsky, V.; Shakhzadova, A. Malignant Neoplasms in Russia in 2020 (Morbidity and Mortality); Moscow; Russia; 2021; ISBN 978-5-85502-268-1. Available online: https://oncology-association.ru/wp-content/uploads/2021/11/zis-2020-elektronnaya-versiya.pdf (accessed on 1 March 2021).
- Merabishvili, V.M.; Poltorackiy, A.N.; Nosov, A.K.; Artem’Eva, A.S.; Merabishvili, E.N. The State of Oncology Care in Russia. Kidney Cancer (Morbidity, Mortality, Index of Accuracy, One-Year and Year-by-Year Mortality, Histological Structure). Part 1. Onkourologiya 2021, 17, 182–194. [Google Scholar] [CrossRef]
- Athanazio, D.A.; Amorim, L.S.; da Cunha, I.W.; Leite, K.R.M.; da Paz, A.R.; de Paula Xavier Gomes, R.; Tavora, F.R.F.; Faraj, S.F.; Cavalcanti, M.S.; Bezerra, S.M. Classification of Renal Cell Tumors—Current concepts and use of ancillary tests: Recommendations of the Brazilian Society of Pathology. Surg. Exp. Pathol. 2021, 4, 4. [Google Scholar] [CrossRef]
- Mollica, V.; Di Nunno, V.; Gatto, L.; Santoni, M.; Scarpelli, M.; Cimadamore, A.; Lopez-Beltran, A.; Cheng, L.; Battelli, N.; Montironi, R.; et al. Resistance to Systemic Agents in Renal Cell Carcinoma Predict and Overcome Genomic Strategies Adopted by Tumor. Cancers 2019, 11, 830. [Google Scholar] [CrossRef]
- Ma, C.G.; Xu, W.H.; Xu, Y.; Wang, J.; Liu, W.R.; Cao, D.L.; Wang, H.K.; Shi, G.H.; Zhu, Y.P.; Qu, Y.Y.; et al. Identification and validation of novel metastasis-related signatures of clear cell renal cell carcinoma using gene expression databases. Am. J. Transl. Res. 2020, 12, 4108–4126. [Google Scholar] [PubMed]
- Wan, B.; Yang, Y.; Zhang, Z. Identification of Differentially Methylated Genes Associated with Clear Cell Renal Cell Carcinoma and Their Prognostic Values. J. Environ. Public Health 2023, 2023, 8405945. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Jiang, Z.; Wang, X.; Wang, H.; Song, M.; Chen, W.; Yang, S. Key genes associated with prognosis and metastasis of clear cell renal cell carcinoma. PeerJ 2022, 10, e12493. [Google Scholar] [CrossRef]
- Outeiro-Pinho, G.; Barros-Silva, D.; Aznar, E.; Sousa, A.-I.; Vieira-Coimbra, M.; Oliveira, J.; Gonçalves, C.S.; Costa, B.M.; Junker, K.; Henrique, R.; et al. MicroRNA-30a-5pme: A novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J. Exp. Clin. Cancer Res. 2020, 39, 98. [Google Scholar] [CrossRef]
- Grammatikaki, S.; Katifelis, H.; Farooqi, A.A.; Stravodimos, K.; Karamouzis, M.V.; Souliotis, K.; Varvaras, D.; Gazouli, M. An Overview of Epigenetics in Clear Cell Renal Cell Carcinoma. In Vivo 2023, 37, 1–10. [Google Scholar] [CrossRef]
- Patil, N.; Abba, M.L.; Zhou, C.; Chang, S.; Gaiser, T.; Leupold, J.H.; Allgayer, H. Changes in Methylation across Structural and MicroRNA Genes Relevant for Progression and Metastasis in Colorectal Cancer. Cancers 2021, 13, 5951. [Google Scholar] [CrossRef]
- Loginov, V.I.; Beresneva, E.V.; Kazubskaya, T.R.; Braga, E.A.; Karpukhin, A.V. Methylation of 10 miRNA genes in clear cell renal cell carcinoma and their diagnostic value. Onkourologiya 2017, 13, 27–33. [Google Scholar] [CrossRef]
- Grimberg, J.; Nawoschik, S.; Belluscio, L.; McKee, R.; Turck, A.; Eisenberg, A. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucleic Acids Res. 1989, 17, 8390. [Google Scholar] [CrossRef]
- Hattermann, K.; Mehdorn, H.M.; Mentlein, R.; Schultka, S.; Held-Feindt, J. A methylation-specific and SYBR-green-based quantitative polymerase chain reaction technique for O6-methylguanine DNA methyltransferase promoter methylation analysis. Anal. Biochem. 2008, 377, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Loginov, V.I.; Burdennyy, A.M.; Filippova, E.A.; Pronina, I.V.; Lukina, S.S.; Kazubskaya, T.P.; Karpukhin, A.V.; Khodyrev, D.S.; Braga, E.A. Aberrant Methylation of 21 MicroRNA Genes in Breast Cancer: Sets of Genes Associated with Progression and a System of Markers for Predicting Metastasis. Bull. Exp. Biol. Med. 2021, 172, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Loginov, V.I.; Burdennyy, A.M.; Filippova, E.A.; Pronina, I.V.; Kazubskaya, T.P.; Kushlinsky, D.N.; Ermilova, V.D.; Rykov, S.V.; Khodyrev, D.S.; Braga, E.A. Hypermethylation of miR-107, miR-130b, miR-203a, miR-1258 Genes Associated with Ovarian Cancer Development and Metastasis. Mol. Biol. 2018, 52, 693–700. [Google Scholar] [CrossRef]
- van Hoesel, A.Q.; Sato, Y.; Elashoff, D.A.; Turner, R.R.; Giuliano, A.E.; Shamonki, J.M.; Kuppen, P.J.K.; van de Velde, C.J.H.; Hoon, D.S.B. Assessment of DNA methylation status in early stages of breast cancer development. Br. J. Cancer 2013, 108, 2033–2038. [Google Scholar] [CrossRef]
- MedCalc Statistical Software; Ostend, Belgium; 2023. Available online: https://www.medcalc.org/calc/ (accessed on 1 March 2023).
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Apanovich, N.; Peters, M.; Apanovich, P.; Mansorunov, D.; Markova, A.; Matveev, V.; Karpukhin, A. The Genes—Candidates for Prognostic Markers of Metastasis by Expression Level in Clear Cell Renal Cell Cancer. Diagnostics 2020, 10, 30. [Google Scholar] [CrossRef]
- Powles, T.; Tomczak, P.; Park, S.H.; Venugopal, B.; Ferguson, T.; Symeonides, S.N.; Hajek, J.; Gurney, H.; Chang, Y.-H.; Lee, J.L.; et al. Pembrolizumab versus placebo as post-nephrectomy adjuvant therapy for clear cell renal cell carcinoma (KEYNOTE-564): 30-month follow-up analysis of a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2022, 23, 1133–1144. [Google Scholar] [CrossRef]
- Zatovicova, M.; Jelenska, L.; Hulikova, A.; Csaderova, L.; Ditte, Z.; Ditte, P.; Goliasova, T.; Pastorek, J.; Pastorekova, S. Carbonic Anhydrase IX as an Anticancer Therapy Target: Preclinical Evaluation of Internalizing Monoclonal Antibody Directed to Catalytic Domain. Curr. Pharm. Des. 2010, 16, 3255–3263. [Google Scholar] [CrossRef]
- Fredlund, E.; Ovenberger, M.; Borg, K.; Påhlman, S. Transcriptional adaptation of neuroblastoma cells to hypoxia. Biochem. Biophys. Res. Commun. 2008, 366, 1054–1060. [Google Scholar] [CrossRef]
- Pescador, N.; Cuevas, Y.; Naranjo, S.; Alcaide, M.; Villar, D.; Landázuri, M.O.; del Peso, L. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 2005, 390, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Kawamoto, T.; Tanimoto, K.; Nishiyama, M.; Honda, H.; Kato, Y. Identification of Functional Hypoxia Response Elements in the Promoter Region of the DEC1 and DEC2 Genes. J. Biol. Chem. 2002, 277, 47014–47021. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 2003, 3, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Linehan, W.M.; Ricketts, C.J. The Cancer Genome Atlas of renal cell carcinoma: Findings and clinical implications. Nat. Rev. Urol. 2019, 16, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Semenza, G.L. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J. Clin. Investig. 2013, 123, 3664–3671. [Google Scholar] [CrossRef]
- Tostain, J.; Li, G.; Gentil-Perret, A.; Gigante, M. Carbonic anhydrase 9 in clear cell renal cell carcinoma: A marker for diagnosis, prognosis and treatment. Eur. J. Cancer 2010, 46, 3141–3148. [Google Scholar] [CrossRef]
- Wykoff, C.C.; Beasley, N.J.; Watson, P.H.; Turner, K.J.; Pastorek, J.; Sibtain, A.; Wilson, G.D.; Turley, H.; Talks, K.L.; Maxwell, P.; et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res. 2000, 60, 7075–7083. [Google Scholar]
- Courcier, J.; De La Taille, A.; Nourieh, M.; Leguerney, I.; Lassau, N.; Ingels, A. Carbonic Anhydrase IX in Renal Cell Carcinoma, Implications for Disease Management. Int. J. Mol. Sci. 2020, 21, 7146. [Google Scholar] [CrossRef]
- Bui, M.H.T.; Seligson, D.; Han, K.-R.; Pantuck, A.J.; Dorey, F.J.; Huang, Y.; Horvath, S.; Leibovich, B.C.; Chopra, S.; Liao, S.-Y.; et al. Carbonic anhydrase IX is an independent predictor of survival in advanced renal clear cell carcinoma: Implications for prognosis and therapy. Clin. Cancer Res. 2003, 9, 802–811. [Google Scholar]
- Zhao, Z.; Liao, G.; Li, Y.; Zhou, S.; Zou, H.; Fernando, S. Prognostic Value of Carbonic Anhydrase IX Immunohistochemical Expression in Renal Cell Carcinoma: A Meta-Analysis of the Literature. PLoS ONE 2014, 9, e114096. [Google Scholar] [CrossRef]
- Tello, D.; Balsa, E.; Acosta-Iborra, B.; Fuertes-Yebra, E.; Elorza, A.; Ordóñez, A.; Corral-Escariz, M.; Soro, I.; López-Bernardo, E.; Perales-Clemente, E.; et al. Induction of the Mitochondrial NDUFA4L2 Protein by HIF-1α Decreases Oxygen Consumption by Inhibiting Complex I Activity. Cell Metab. 2011, 14, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Bourke, E.; Eriksson, L.A.; Kerin, M.J. Targeting cancer using KAT inhibitors to mimic lethal knockouts. Biochem. Soc. Trans. 2016, 44, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Apanovich, N.V.; Poyarkov, S.V.; Peters, M.V.; Korotaeva, A.A.; Markova, A.S.; Kamolov, B.S.; Pronina, I.V.; Braga, E.A.; Matveev, V.B.; Karpukhin, A. The Differential Gene Expression in Clear Cell Renal Cell Carcinoma and Biomarker Development. Eur. J. Hum.Gen. 2015, 23, 446. [Google Scholar]
- Apanovich, N.V.; Peters, M.V.; Korotaeva, A.A.; Apanovich, P.V.; Markova, A.S.; Kamolov, B.S.; Matveev, V.B.; Karpukhin, A.V. Molecular genetic diagnostics of clear cell renal cell carcinoma. Onkourologiya 2016, 12, 16–20. [Google Scholar] [CrossRef]
- Minton, D.R.; Fu, L.; Mongan, N.P.; Shevchuk, M.M.; Nanus, D.M.; Gudas, L.J. Role of NADH Dehydrogenase (Ubiquinone) 1 Alpha Subcomplex 4-Like 2 in Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2016, 22, 2791–2801. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Lan, G.; Peng, L.; Xie, X.; Peng, F.; Yu, S.; Wang, Y.; Tang, X. NDUFA4L2 expression predicts poor prognosis in clear cell renal cell carcinoma patients. Ren. Fail. 2016, 38, 8. [Google Scholar] [CrossRef]
- Meng, L.; Yang, X.; Xie, X.; Wang, M. Mitochondrial NDUFA4L2 protein promotes the vitality of lung cancer cells by repressing oxidative stress. Thorac. Cancer 2019, 10, 676–685. [Google Scholar] [CrossRef]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Miikkulainen, P.; Högel, H.; Rantanen, K.; Suomi, T.; Kouvonen, P.; Elo, L.L.; Jaakkola, P.M. HIF prolyl hydroxylase PHD3 regulates translational machinery and glucose metabolism in clear cell renal cell carcinoma. Cancer Metab. 2017, 5, 5. [Google Scholar] [CrossRef]
- Doherty, J.R.; Cleveland, J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Investig. 2013, 123, 3685–3692. [Google Scholar] [CrossRef]
- Tamukong, P.K.; Kuhlmann, P.; You, S.; Su, S.; Wang, Y.; Yoon, S.; Gong, J.; Figlin, R.A.; Janes, J.L.; Freedland, S.J.; et al. Hypoxia-inducible factor pathway genes predict survival in metastatic clear cell renal cell carcinoma. Urol. Oncol. Semin. Orig. Investig. 2022, 40, 495.e1–495.e10. [Google Scholar] [CrossRef]
- Lin, L.; Cai, J. Circular RNA circ-EGLN3 promotes renal cell carcinoma proliferation and aggressiveness via miR-1299-mediated IRF7 activation. J. Cell Biochem. 2020, 121, 4377–4385. [Google Scholar] [CrossRef]
- Zaravinos, A.; Pieri, M.; Mourmouras, N.; Anastasiadou, N.; Zouvani, I.; Delakas, D.; Deltas, C. Altered metabolic pathways in clear cell renal cell carcinoma: A meta-analysis and validation study focused on the deregulated genes and their associated networks. Oncoscience 2014, 1, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Zodro, E.; Jaroszewski, M.; Ida, A.; Wrzesinski, T.; Kwias, Z.; Bluyssen, H.; Wesoly, J. FUT11 as a potential biomarker of clear cell renal cell carcinoma progression based on meta-analysis of gene expression data. Tumor Biol. 2014, 35, 2607–2617. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Jia, Y.F.; Ma, X.L.; Zheng, Y.; Kong, Y.; Zhang, Y.; Zong, S.; Chen, Z.T.; Wang, Y.S. DEC2 Suppresses Tumor Proliferation and Metastasis by Regulating ERK/NF-ΚB Pathway in Gastric Cancer. Am. J. Cancer Res. 2016, 6, 1741–1757. [Google Scholar] [CrossRef]
- Bigot, P.; Colli, L.M.; Machiela, M.J.; Jessop, L.; Myers, T.A.; Carrouget, J.; Wagner, S.; Roberson, D.; Eymerit, C.; Henrion, D.; et al. Functional characterization of the 12p12.1 renal cancer-susceptibility locus implicates BHLHE41. Nat. Commun. 2016, 7, 12098. [Google Scholar] [CrossRef]
- Shen, Z.; Zhu, L.; Zhang, C.; Cui, X.; Lu, J. Overexpression of BHLHE41, correlated with DNA hypomethylation in 3’UTR region, promotes the growth of human clear cell renal cell carcinoma. Oncol. Rep. 2019, 41, 2137–2147. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, N.; Zheng, Q.; Zhang, D.; Liu, Y. BHLHE41 promotes U87 and U251 cell proliferation via ERK/cyclinD1 signaling pathway. Cancer Manag. Res. 2019, 11, 7657–7672. [Google Scholar] [CrossRef] [PubMed]
- Montagner, M.; Enzo, E.; Forcato, M.; Zanconato, F.; Parenti, A.; Rampazzo, E.; Basso, G.; Leo, G.; Rosato, A.; Bicciato, S.; et al. SHARP1 suppresses breast cancer metastasis by promoting degradation of hypoxia-inducible factors. Nature 2012, 487, 380–384. [Google Scholar] [CrossRef]
- Asanoma, K.; Liu, G.; Yamane, T.; Miyanari, Y.; Takao, T.; Yagi, H.; Ohgami, T.; Ichinoe, A.; Sonoda, K.; Wake, N.; et al. Regulation of the Mechanism of TWIST1 Transcription by BHLHE40 and BHLHE41 in Cancer Cells. Mol. Cell Biol. 2015, 35, 4096–4109. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Kawamura, H.; Wu, Y.; Sato, H.; Jin, D.; Bhawal, U.K.; Kawamoto, T.; Fujimoto, K.; Noshiro, M.; Seino, H.; et al. The basic helix-loop-helix transcription factor DEC2 inhibits TGF-β-induced tumor progression in human pancreatic cancer BxPC-3 cells. Int. J. Mol. Med. 2012, 30, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, G.; Jiang, Y. The Emerging Roles of miR-125b in Cancers. Cancer Manag. Res. 2020, 12, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Bu, Q.; You, F.; Pan, G.; Yuan, Q.; Cui, T.; Hao, L.; Zhang, J. MiR-125b inhibits anaplastic thyroid cancer cell migration and invasion by targeting PIK3CD. Biomed. Pharmacother. 2017, 88, 443–448. [Google Scholar] [CrossRef]
- Zhou, J.-N.; Zeng, Q.; Wang, H.-Y.; Zhang, B.; Li, S.-T.; Nan, X.; Cao, N.; Fu, C.-J.; Yan, X.-L.; Jia, Y.-L.; et al. MicroRNA-125b attenuates epithelial-mesenchymal transitions and targets stem-like liver cancer cells through small mothers against decapentaplegic 2 and 4. Hepatology 2015, 62, 801–815. [Google Scholar] [CrossRef]
- Lee, M.; Kim, E.J.; Jeon, M.J. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. Oncotarget 2016, 7, 8726–8742. [Google Scholar] [CrossRef]
- Chen, H.; Xu, Z. Hypermethylation-Associated Silencing of miR-125a and miR-125b: A Potential Marker in Colorectal Cancer. Dis. Markers 2015, 2015, 345080. [Google Scholar] [CrossRef]
- Zhang, Y.; Yan, L.-X.; Wu, Q.-N.; Du, Z.-M.; Chen, J.; Liao, D.-Z.; Huang, M.-Y.; Hou, J.-H.; Wu, Q.-L.; Zeng, M.-S.; et al. miR-125b Is Methylated and Functions as a Tumor Suppressor by Regulating the ETS1 Proto-oncogene in Human Invasive Breast Cancer. Cancer Res 2011, 71, 3552–3562. [Google Scholar] [CrossRef]
- Ferracin, M.; Bassi, C.; Pedriali, M.; Pagotto, S.; D’abundo, L.; Zagatti, B.; Corrà, F.; Musa, G.; Callegari, E.; Lupini, L.; et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 2013, 12, 130. [Google Scholar] [CrossRef]
- Zhang, H.; Li, H. miR-137 inhibits renal cell carcinoma growth in vitro and in vivo. Oncol. Lett. 2016, 12, 715–720. [Google Scholar] [CrossRef]
- Sang, L.; Liu, T.; Liu, H.; Li, H.; Zhang, H.; Jiang, F. MicroRNA-137 Suppresses Cell Migration and Invasion in Renal Cell Carcinoma by Targeting PIK3R3. Int. J. Clin. Exp. Med. 2016, 9, 7160–7167. [Google Scholar]
- Cheng, Y.; Li, Y.; Liu, D.; Zhang, R.; Zhang, J. miR-137 effects on gastric carcinogenesis are mediated by targeting Cox-2-activated PI3K/AKT signaling pathway. FEBS Lett. 2014, 588, 3274–3281. [Google Scholar] [CrossRef]
- Chen, Q.; Chen, X.; Zhang, M.; Fan, Q.; Luo, S.; Cao, X. miR-137 Is Frequently Down-Regulated in Gastric Cancer and Is a Negative Regulator of Cdc42. Dig. Dis. Sci. 2011, 56, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xiong, Y.; Watari, H.; Hanley, S.J.B.; Konno, Y.; Ihira, K.; Yamada, T.; Kudo, M.; Yue, J.; Sakuragi, N. MiR-137 and miR-34a directly target Snail and inhibit EMT, invasion and sphere-forming ability of ovarian cancer cells. J. Exp. Clin. Cancer Res. 2016, 35, 132. [Google Scholar] [CrossRef] [PubMed]
- Varachev, V.; Loginov, V.; Pronina, I.; Khodyrev, D.; Kazubskaya, T.; Morozov, A. Hypermethylated Tumor Suppressor MicroRNAs as Novel Markers of Clear Cell Renal Cell Carcinoma. FEBS Open Bio 2018, 8, 304–305. [Google Scholar]
- Wang, J.; Sun, X. MicroRNA-375 inhibits the proliferation, migration and invasion of kidney cancer cells by triggering apoptosis and modulation of PDK1 expression. Environ. Toxicol. Pharmacol. 2018, 62, 227–233. [Google Scholar] [CrossRef]
- Zhou, J.; Song, S.; He, S.; Zhu, X.; Zhang, Y.; Yi, B.; Zhang, B.; Qin, G.; Li, D. MicroRNA-375 targets PDK1 in pancreatic carcinoma and suppresses cell growth through the Akt signaling pathway. Int. J. Mol. Med. 2014, 33, 950–956. [Google Scholar] [CrossRef]
- Cheng, L.; Zhan, B.; Luo, P.; Wang, B. miRNA-375 regulates the cell survival and apoptosis of human non-small cell carcinoma by targeting HER2. Mol. Med. Rep. 2017, 15, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.-Y.; Zhang, Z.-Z.; Liu, H.; Zhao, E.-H.; Cao, H. miR-375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression. Exp. Ther. Med. 2014, 7, 1757–1761. [Google Scholar] [CrossRef]
- Yang, S.; Yang, R.; Lin, R.; Si, L. MicroRNA-375 inhibits the growth, drug sensitivity and metastasis of human ovarian cancer cells by targeting PAX2. J. Buon 2020, 24, 2341–2346. [Google Scholar]
- Yu, T.; Li, J.; Yan, M.; Liu, L.; Lin, H.; Zhao, F.; Sun, L.; Zhang, Y.; Cui, Y.; Zhang, F.; et al. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene 2015, 34, 413–423. [Google Scholar] [CrossRef]
- Bharambe, H.S.; Joshi, A.; Yogi, K.; Kazi, S.; Shirsat, N.V. Restoration of miR-193a expression is tumor-suppressive in MYC amplified Group 3 medulloblastoma. Acta Neuropathol. Commun. 2020, 8, 70. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, M.X.; Mak, C.S.-L.; Yung, M.M.-H.; Leung, T.H.-Y.; Xu, D.; Ngu, S.-F.; Chan, K.K.-L.; Yang, H.; Ngan, H.Y.-S.; et al. Methylation-associated silencing of miR-193a-3p promotes ovarian cancer aggressiveness by targeting GRB7 and MAPK/ERK pathways. Theranostics 2018, 8, 423–436. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Hu, J.; Lu, M.; Gu, H.; Zhou, X.; Chen, X.; Zen, K.; Zhang, C.-Y.; Zhang, T.; Ge, J.; et al. A panel of five serum miRNAs as a potential diagnostic tool for early-stage renal cell carcinoma. Sci. Rep. 2015, 5, 7610. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhang, G.; Xie, C.; Zhou, Y. MiR-34b regulates cervical cancer cell proliferation and apoptosis. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2042–2047. [Google Scholar] [CrossRef]
- Li, Y.; Jin, L.; Rong, W.; Meng, F.; Wang, X.; Wang, S.; Yu, X. Expression and significance of miR-34 with PI3K, AKT and mTOR proteins in colorectal adenocarcinoma tissues. Cell Mol. Biol. 2022, 68, 57–62. [Google Scholar] [CrossRef]
- Liu, X.; Feng, J.; Tang, L.; Liao, L.; Xu, Q.; Zhu, S. The Regulation and Function of miR-21-FOXO3a-miR-34b/c Signaling in Breast Cancer. Int. J. Mol. Sci. 2015, 16, 3148–3162. [Google Scholar] [CrossRef]
- Majid, S.; Dar, A.A.; Saini, S.; Shahryari, V.; Arora, S.; Zaman, M.S.; Chang, I.; Yamamura, S.; Tanaka, Y.; Chiyomaru, T.; et al. miRNA-34b Inhibits Prostate Cancer through Demethylation, Active Chromatin Modifications, and AKT Pathways. Clin. Cancer Res. 2013, 19, 73–84. [Google Scholar] [CrossRef]
- Xiong, S.; Hu, M.; Li, C.; Zhou, X.; Chen, H. Role of miR-34 in gastric cancer: From bench to bedside (Review). Oncol. Rep. 2019, 42, 1635–1646. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, E.J.; Lee, S.; Tan, X.; Liu, X.; Park, S.; Kang, K.; Yoon, J.-S.; Ko, Y.H.; Kurie, J.M.; et al. MiR-34a and miR-34b/c have distinct effects on the suppression of lung adenocarcinomas. Exp. Mol. Med. 2019, 51, 1–10. [Google Scholar] [CrossRef]
- Pantuck, A.J.; Seligson, D.B.; Klatte, T.; Yu, H.; Leppert, J.T.; Moore, L.; O’Toole, T.; Gibbons, J.; Belldegrun, A.S.; Figlin, R.A. Prognostic relevance of the mTOR pathway in renal cell carcinoma: Implications for Molecular Patient Selection for Targeted Therapy. Cancer 2007, 109, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | Number of Samples | Age | Gender M/F (%M/%F) | |
|---|---|---|---|---|
| TNM stage | I | 20 | 60.7 ± 11.1 | 10/10 (50/50) |
| II | 6 | 59.3 ± 8.2 | 5/1 (83.3/16.7) | |
| III | 23 | 62.3 ± 6.9 | 16/7 (69.6/30.4) | |
| IV | 31 | 59.3 ± 7.7 | 16/15 (51.6/48.4) | |
| The presence of metastases | With distant metastases | 31 | 59.3 ± 7.7 | 16/15 (51.6/48.4) |
| No metastases | 49 | 61.3 ± 8.9 | 31/18 (63.3/36.7) | |
| Localization of distant metastases | Lungs | 19 | 60.4 ± 8.3 | 9/10 (47.4/52.6) |
| Adrenal | 9 | 56.2 ± 8.1 | 5/4 (55.6/44.4) | |
| Bones | 3 | 61.0 ± 6.6 | 1/2 (33.3/66.7) | |
| Other | 4 | 57.3 ± 7.0 | 4/0 (100/0) | |
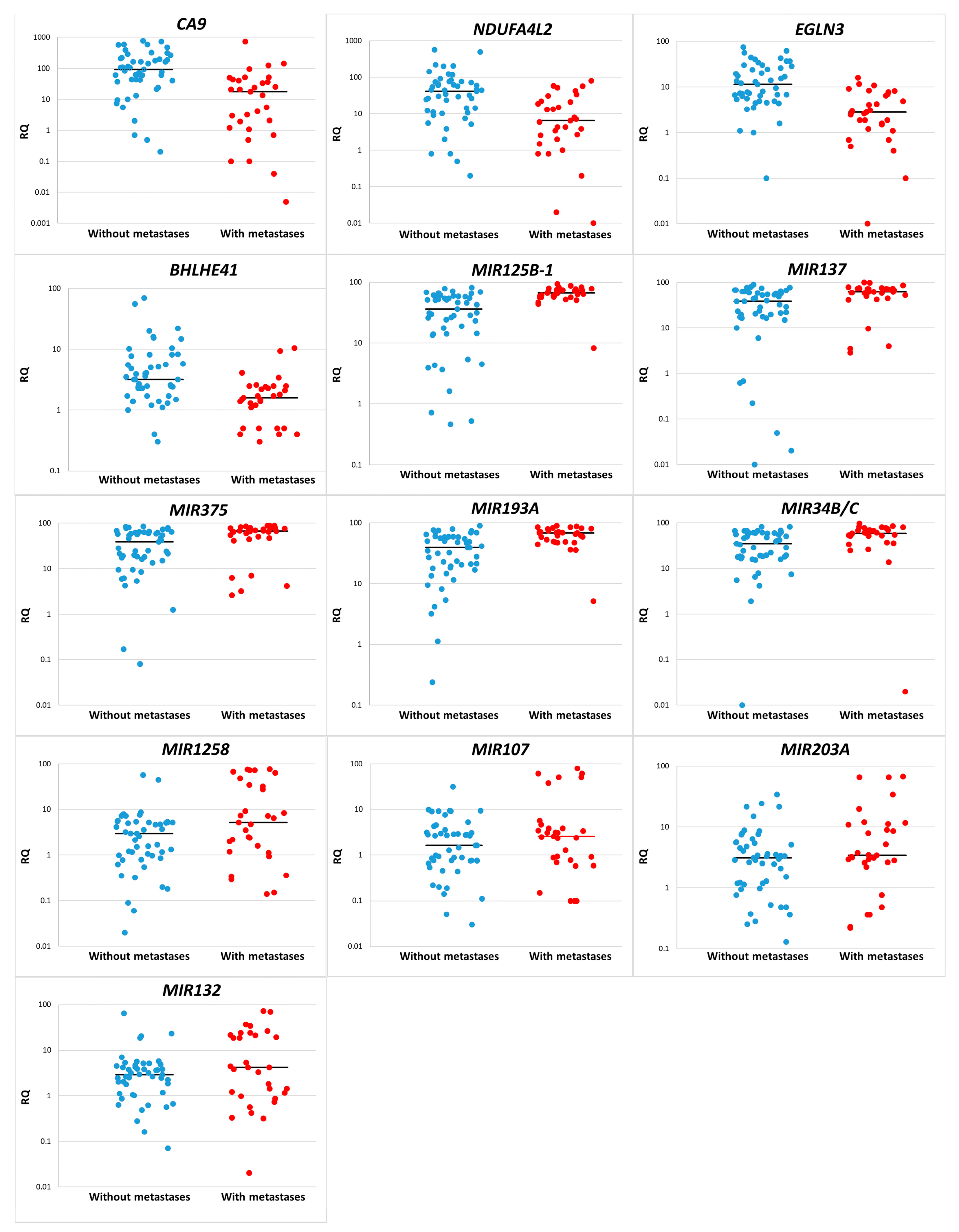
| Gene | The Median Value | (Mann–Whitney U-Test), p = | Logistic Regression, p = | |
|---|---|---|---|---|
| In the Non-Metastasis Group | In the Metastasis Group | |||
| (A) | ||||
| CA9 | 92.7 | 17.8 | <0.001 | 0.022 |
| NDUFA4L2 | 41.1 | 6.5 | <0.001 | 0.007 |
| EGLN3 | 11.4 | 2.8 | <0.001 | 0.004 |
| BHLHE41 | 3.2 | 1.6 | <0.001 | 0.018 |
| (B) | ||||
| MIR125B-1 | 36.27 | 66.34 | <0.001 | 0.001 |
| MIR137 | 38.10 | 61.84 | 0.002 | 0.006 |
| MIR375 | 38.99 | 66.19 | 0.003 | 0.007 |
| MIR193A | 38.99 | 67.58 | <0.001 | 0.001 |
| MIR34B/C | 35.26 | 59.23 | 0.001 | 0.004 |
| MIR1258 | 2.97 | 5.19 | 0.040 | 0.010 |
| MIR107 | 1.62 | 2.57 | 0.252 | 0.036 |
| MIR203A | 3.11 | 3.42 | 0.235 | 0.061 |
| MIR132 | 2.9 | 4.17 | 0.252 | 0.036 |
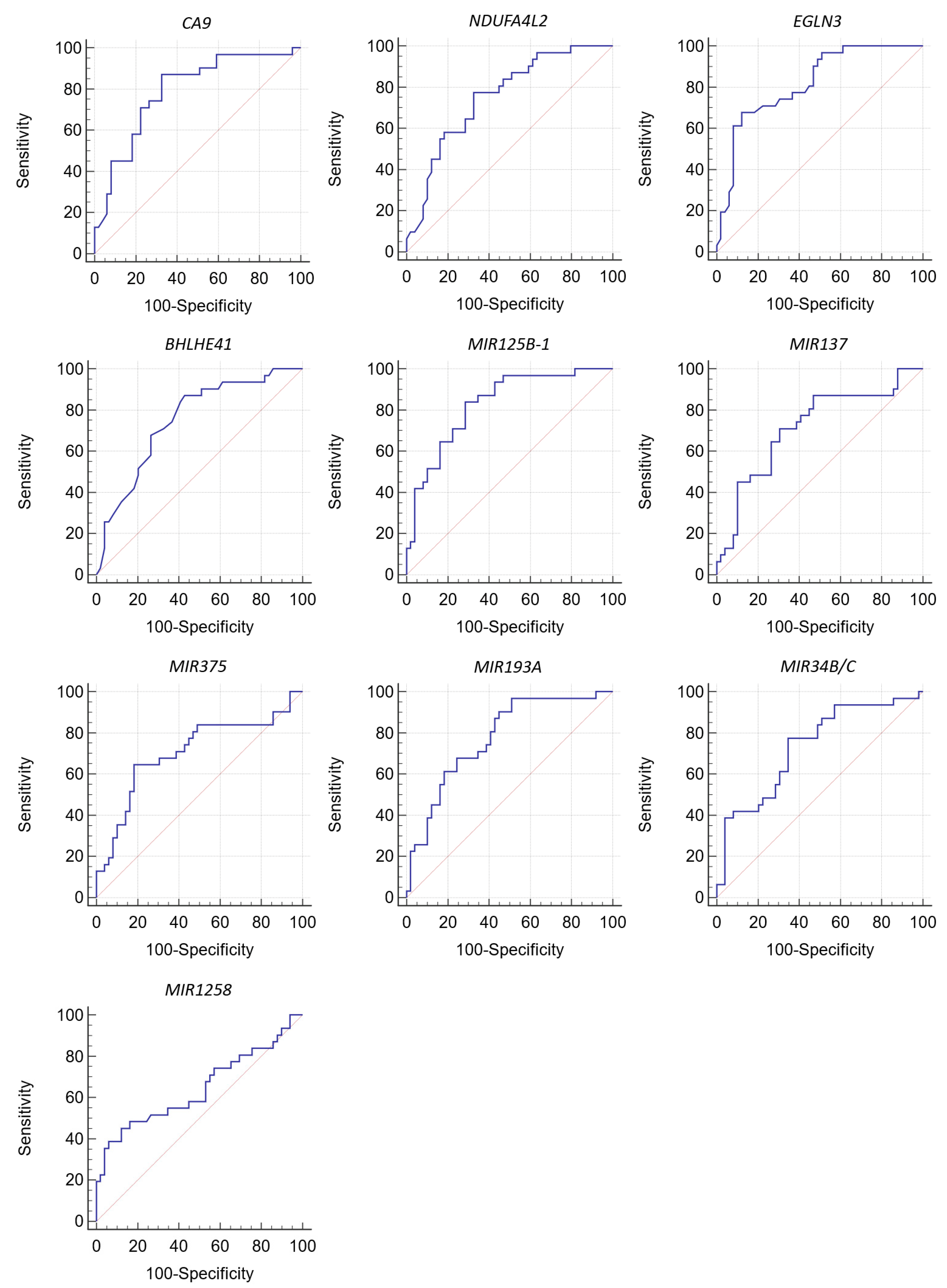
| Gene | Area under ROC Curve (AUC) | 95% CI | Cutoff Value | Significance Level, p (Area = 0.5) | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| CA9 | 0.789 | 0.684–0.873 | ≤51.3 * | <0.001 | 87.10 | 67.35 |
| NDUFA4L2 | 0.753 | 0.644–0.842 | ≤22 * | <0.001 | 77.42 | 67.35 |
| EGLN3 | 0.818 | 0.716–0.895 | ≤4.2 * | <0.001 | 67.74 | 87.76 |
| BHLHE41 | 0.751 | 0.642–0.841 | ≤2.6 * | <0.001 | 87.10 | 57.14 |
| MIR125B-1 | 0.827 | 0.726–0.902 | >55.18 ** | <0.001 | 83.87 | 71.43 |
| MIR137 | 0.716 | 0.604–0.811 | >57.62 ** | 0.001 | 70.97 | 69.39 |
| MIR375 | 0.706 | 0.593–0.802 | >64.29 ** | 0.001 | 64.52 | 81.63 |
| MIR193A | 0.776 | 0.668–0.861 | >34.65 ** | <0.001 | 96.77 | 48.98 |
| MIR34B/C | 0.732 | 0.621–0.825 | >50.35 ** | <0.001 | 77.42 | 65.31 |
| MIR1258 | 0.644 | 0.529–0.748 | >7.15 ** | 0.033 | 45.16 | 87.76 |
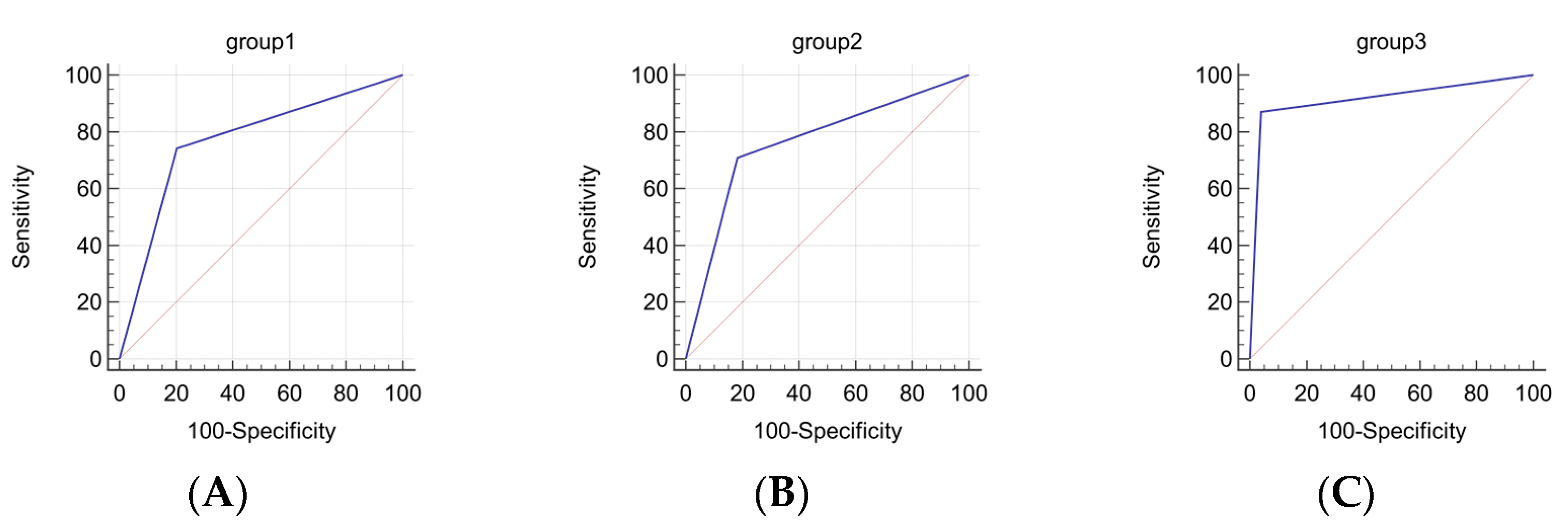
| Gene Group | Sensitivity/ Specificity | Area under ROC Curve (AUC) | Significance Level, p (Area = 0.5) | Negative Predictive Value % (95% CI) | Positive Predictive Value % (95% CI) |
|---|---|---|---|---|---|
| CA9 NDUFA4L2 EGLN3 BHLHE41 | 74.19/79.59 | 0.769 | <0.0001 | 82.98 (69.19–92.35) | 69.70 (51.29–84.41) |
| MIR125B-1 MIR137 MIR375 MIR193A MIR34B/C | 70.97/81.63 | 0.763 | <0.0001 | 81.63 (67.98–91.24) | 70.97 (51.96–85.78) |
| CA9 NDUFA4L2 EGLN3 BHLHE41 MIR125B-1 MIR137 MIR375 MIR193A MIR34B/C | 87.10/95.92 | 0.915 | <0.0001 | 92.16 (81.12–97.82) | 93.10 (77.23–99.15) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apanovich, N.; Matveev, A.; Ivanova, N.; Burdennyy, A.; Apanovich, P.; Pronina, I.; Filippova, E.; Kazubskaya, T.; Loginov, V.; Braga, E.; et al. Prediction of Distant Metastases in Patients with Kidney Cancer Based on Gene Expression and Methylation Analysis. Diagnostics 2023, 13, 2289. https://doi.org/10.3390/diagnostics13132289
Apanovich N, Matveev A, Ivanova N, Burdennyy A, Apanovich P, Pronina I, Filippova E, Kazubskaya T, Loginov V, Braga E, et al. Prediction of Distant Metastases in Patients with Kidney Cancer Based on Gene Expression and Methylation Analysis. Diagnostics. 2023; 13(13):2289. https://doi.org/10.3390/diagnostics13132289
Chicago/Turabian StyleApanovich, Natalya, Alexey Matveev, Natalia Ivanova, Alexey Burdennyy, Pavel Apanovich, Irina Pronina, Elena Filippova, Tatiana Kazubskaya, Vitaly Loginov, Eleonora Braga, and et al. 2023. "Prediction of Distant Metastases in Patients with Kidney Cancer Based on Gene Expression and Methylation Analysis" Diagnostics 13, no. 13: 2289. https://doi.org/10.3390/diagnostics13132289
APA StyleApanovich, N., Matveev, A., Ivanova, N., Burdennyy, A., Apanovich, P., Pronina, I., Filippova, E., Kazubskaya, T., Loginov, V., Braga, E., & Alimov, A. (2023). Prediction of Distant Metastases in Patients with Kidney Cancer Based on Gene Expression and Methylation Analysis. Diagnostics, 13(13), 2289. https://doi.org/10.3390/diagnostics13132289






