Laboratory Automation in Microbiology: Impact on Turnaround Time of Microbiological Samples in COVID Time
Abstract
:1. Introduction
2. Materials and Methods
2.1. Setting
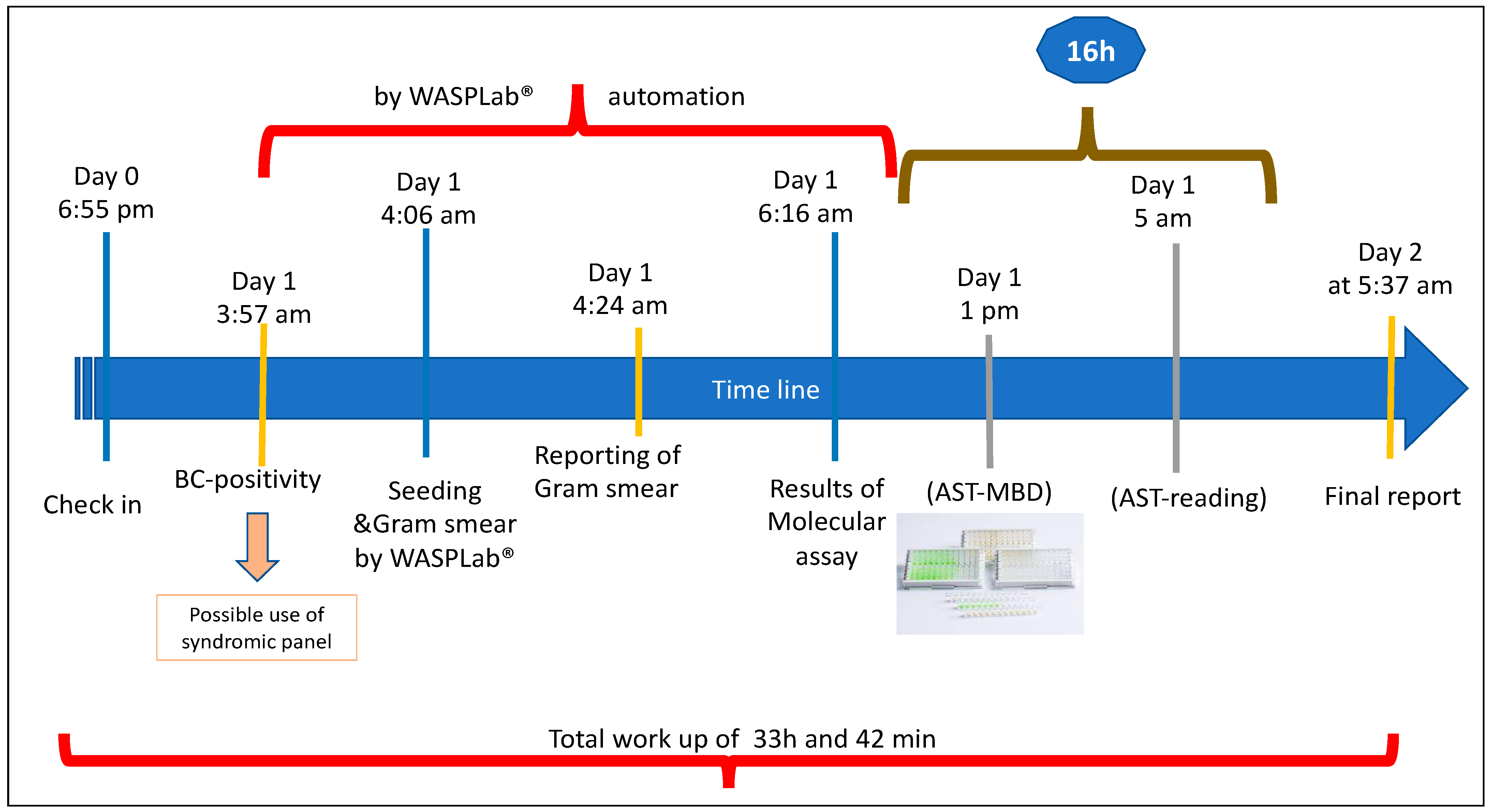
2.2. Protocol for WASPLab®
2.3. TAT Evaluation
2.4. Lab Professionals Involved in the Project
2.5. Statistical Evaluation
2.6. Pathogen Identification, Antimicrobial Susceptibility Testing, and Molecular Assays
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mascart, G.; Martiny, D.; Miendje, Y.; Tummers, G.; Mahadeb, B.; Vandenberg, O.; Hallin, M. Automatisation en bactériologie: Quel avenir? Un exemple concret dans le cadre d’une consolidation de laboratoires universitaires [The future of automation in bacteriology]. Ann. Biol. Clin. 2018, 76, 365–372. [Google Scholar]
- Fontana, C.; Favaro, M.; Favalli, C. How liquid based microbiology can change the workflow in the microbiology laboratories. Adv. Microbiol. 2013, 3, 38450. [Google Scholar] [CrossRef] [Green Version]
- Croxatto, A.; Prod’hom, G.; Faverjon, F.; Rochais, Y.; Greub, G. Laboratory automation in clinical bacteriology: What system to choose? Clin. Microbiol. Infect. 2016, 22, 217–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Da Rin, G.; Zoppelletto, M.; Lippi, G. Integration of Diagnostic Microbiology in a Model of Total Laboratory Automation. Lab. Med. 2016, 47, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Lippi, G.; Da Rin, G. Advantages and limitations of total laboratory automation: A personal overview. Clin. Chem. Lab. Med. 2019, 57, 802–811. [Google Scholar] [CrossRef]
- Naugler, C.; Church, D.L. Automation and artificial intelligence in the clinical laboratory. Crit. Rev. Clin. Lab. Sci. 2019, 56, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, I. Laboratory Automation in Clinical Microbiology. Bioengineering 2018, 5, 102. [Google Scholar] [CrossRef] [Green Version]
- Plebani, M. Laboratory medicine in the COVID-19 era: Six lessons for the future. Chem. Lab. Med. 2021, 59, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Blondeau, M.J. Clinical microbiology laboratories and COVID-19: The calm before the storm. Future Microbiol. 2020, 15, 1419–1424. [Google Scholar] [CrossRef] [PubMed]
- Catalán, P.; Alonso, R.; Alcalá, L.; Marín, M.; Moure, Z.; Pescador, P.; Bouza, E.; Muñoz, P.; Marañón, G. Microbiology-ID COVID 19 Study Group. The challenge of COVID-19 for a Clinical Microbiology Department. Diagn Microbiol. Infect. Dis. 2021, 101, 115426. [Google Scholar] [CrossRef]
- Zimmermann, S. Laboratory Automation in the Microbiology Laboratory: An Ongoing Journey, Not a Tale? J. Clin. Microbiol. 2021, 59, e02592-20. [Google Scholar] [CrossRef] [PubMed]
- Culbreath, K.; Piwonka, H.; Korver, J.; Noorbakhsh, M. Benefits derived from full laboratory automation in microbiology: A tale of four laboratories. J. Clin. Microbiol. 2021, 59, e01969-20. [Google Scholar] [CrossRef]
- Van Eldere, J. Changing needs, opportunities and constraints for the 21st century microbiology laboratory. Clin. Microbiol. Infect. 2005, 11, 15–18. [Google Scholar] [CrossRef] [Green Version]
- Thomson, R.B., Jr.; McElvania, E. Total Laboratory Automation: What Is Gained, What Is Lost, and Who Can Afford It? Clin. Lab. Med. 2019, 39, 371–389. [Google Scholar] [CrossRef] [PubMed]
- Dauwaldwer, O.; Vandenesh, F. Clinical microbiology laboratory: From the Pasteur model to the 24/7 clinical chemistry concept. Clin. Microbiol. Infect. 2014, 20, 593–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenberg, O.; Durand, G.; Hallin, M.; Diefenbach, A.; Gant, V.; Murray, P.; Kozlakidis, Z.; van Belkum, A. Consolidation of clinical microbiology laboratories and introduction of transformative technologies. Clin. Microbiol. Rev. 2020, 33, e00057-19. [Google Scholar] [CrossRef]
- Cherkaoui, A.; Renzi, G.; Martischang, R.; Harbarth, S.; Vuilleumier, N.; Schrenzel, J. Impact of Total Laboratory Automation on Turnaround Times for Urine Cultures and Screening Specimens for MRSA, ESBL, and VRE Carriage: Retrospective Comparison With Manual Workflow. Front. Cell. Infect. Microbiol. 2020, 10, 552122. [Google Scholar] [CrossRef]
- Cherkaoui, A.; Schrenzel, J. Total Laboratory Automation for Rapid Detection and Identification of Microorganisms and Their Antimicrobial Resistance Profiles. Front. Cell. Infect. Microbiol. 2022, 12, 807668. [Google Scholar] [CrossRef]
- Mutters, N.T.; Hodiamont, C.J.; de Jong, M.D.; Overmeijer, H.P.; van den Boogaard, M.; Visser, C.E. Performance of Kiestra total laboratory automation combined with MS in clinical microbiology practice. Ann. Lab. Med. 2014, 34, 111–117. [Google Scholar] [CrossRef]
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. 2022. Available online: http://www.eucast.org/clinical_breakpoints/ (accessed on 1 January 2022).
- van Belkum, A.; Burnham, C.D.; Rossen, J.W.A.; Mallard, F.; Rochas, O.; Dunne, W.M., Jr. Innovative and rapid antimicrobial susceptibility testing systems. Nat. Rev. Microbiol. 2020, 18, 299–311. [Google Scholar] [CrossRef]
- Banerjee, R.; Humphries, R. Rapid Antimicrobial Susceptibility Testing Methods for Blood Cultures and Their Clinical Impact. Front. Med. 2021, 8, 635831. [Google Scholar] [CrossRef] [PubMed]
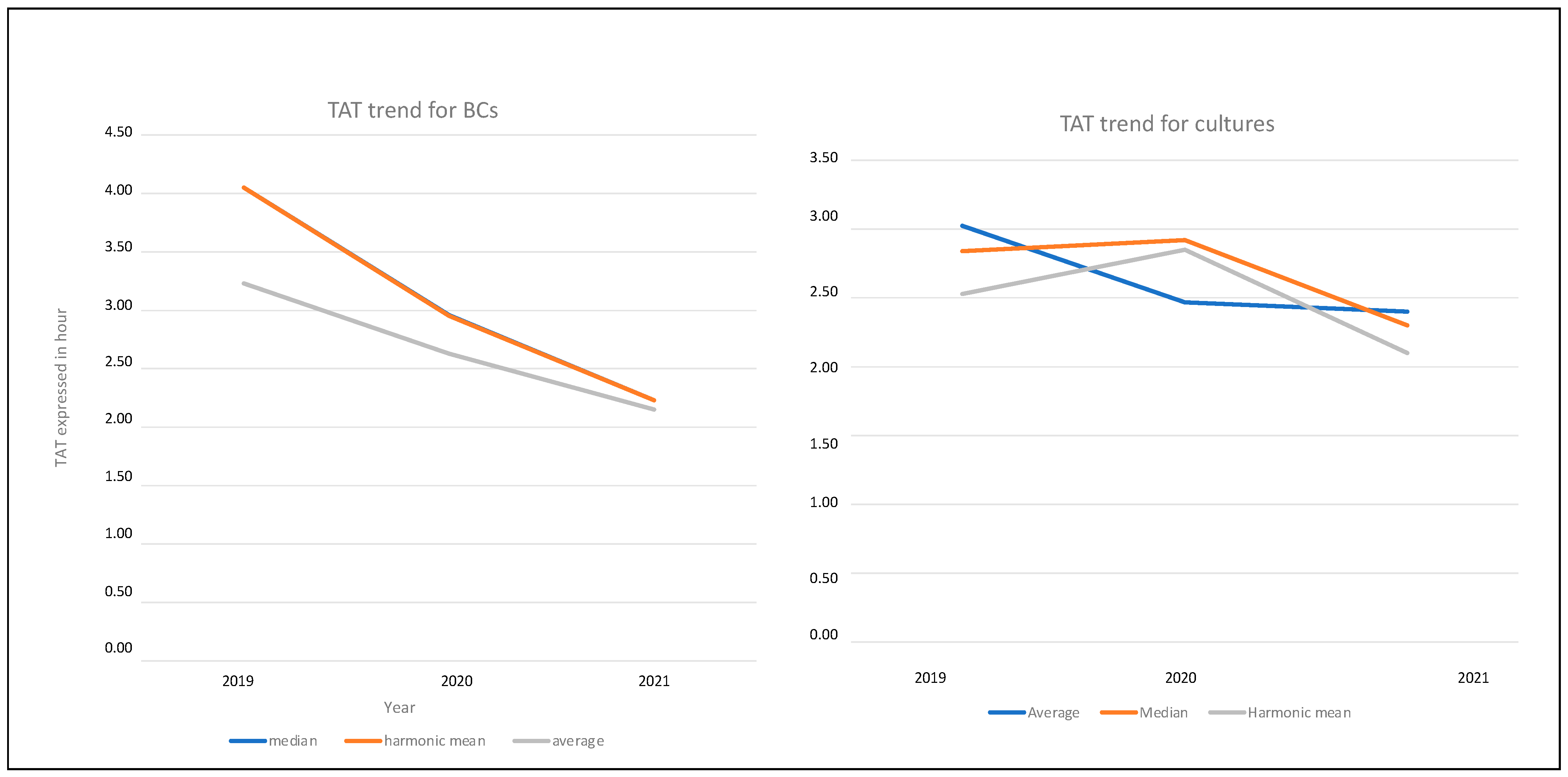
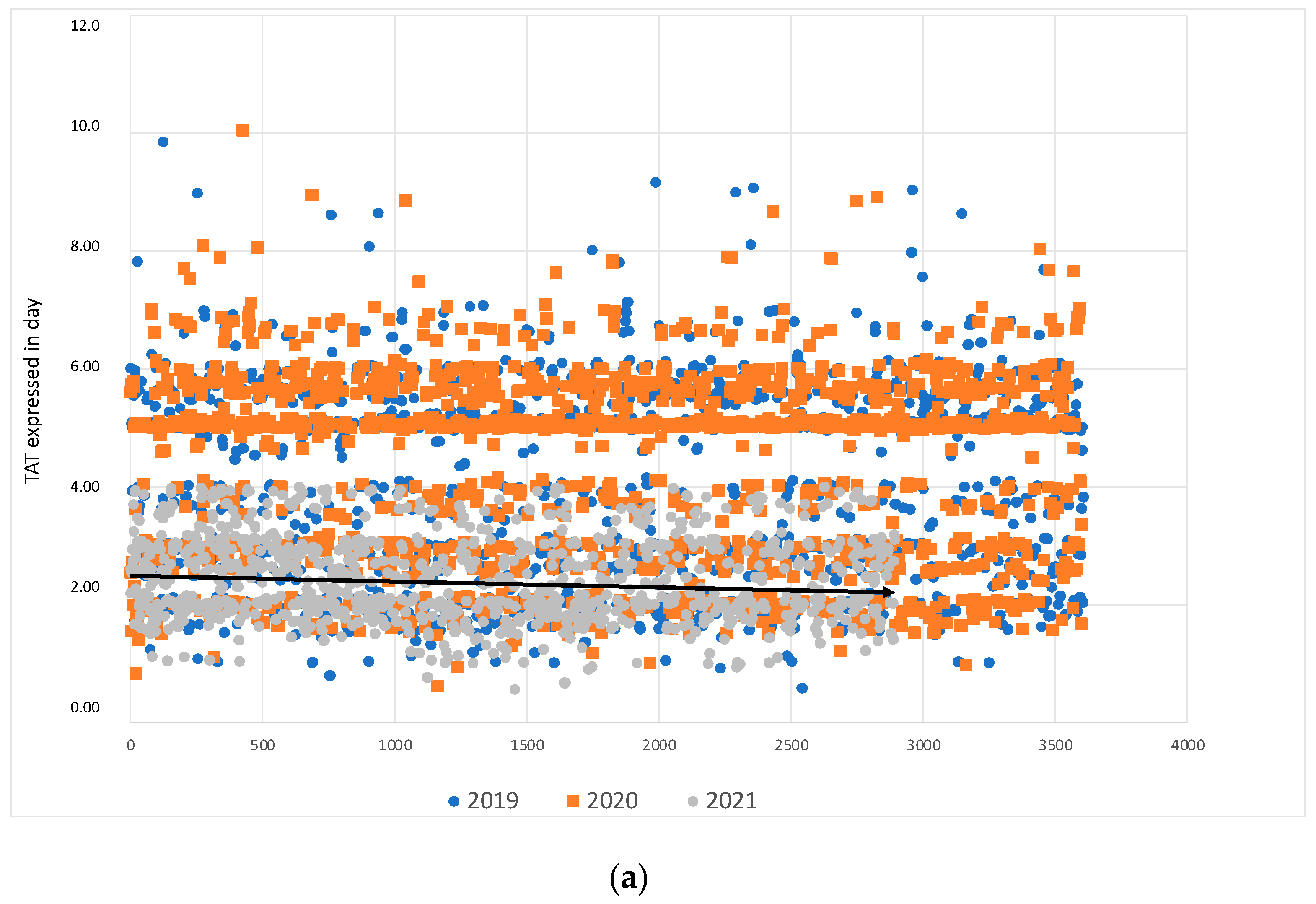

| Statistical Evaluation | 2019 | 2020 | 2021 | |||
|---|---|---|---|---|---|---|
| TAT * 2019 | TAT § 2019 | TAT * 2020 | TAT § 2020 | TAT * 2021 | TAT § 2021 | |
| Average | 4.05 | 97 | 2.96 | 71.04 | 2.23 | 53.52 |
| Median | 4.05 | 97 | 2.95 | 70.80 | 2.23 | 53.52 |
| Harmonic mean | 3.23 | 77 | 2.63 | 63.12 | 2.15 | 51.60 |
| Standard Deviation | 1.56 | 37 | 1.39 | 33.36 | 1.09 | 26.16 |
| No. of positive BCs | 3613 | 2934 | 1732 | |||
| Statistical Evaluation | 2019 | 2020 | 2021 | |||
|---|---|---|---|---|---|---|
| TAT * 2019 | TAT § 2019 | TAT * 2020 | TAT § 2020 | TAT * 2021 | TAT § 2021 | |
| Average | 3.02 | 73 | 2.47 | 59 | 2.40 | 58 |
| Median | 2.84 | 68 | 2.92 | 70 | 2.30 | 55 |
| Harmonic mean | 2.53 | 61 | 2.85 | 68 | 2.10 | 50 |
| Standard Deviation | 1.36 | 33 | 0.45 | 11 | 1.00 | 24 |
| No. of positive cultures | 2299 | 1140 | 2768 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontana, C.; Favaro, M.; Pelliccioni, M.; Minelli, S.; Bossa, M.C.; Altieri, A.; D’Orazi, C.; Paliotta, F.; Cicchetti, O.; Minieri, M.; et al. Laboratory Automation in Microbiology: Impact on Turnaround Time of Microbiological Samples in COVID Time. Diagnostics 2023, 13, 2243. https://doi.org/10.3390/diagnostics13132243
Fontana C, Favaro M, Pelliccioni M, Minelli S, Bossa MC, Altieri A, D’Orazi C, Paliotta F, Cicchetti O, Minieri M, et al. Laboratory Automation in Microbiology: Impact on Turnaround Time of Microbiological Samples in COVID Time. Diagnostics. 2023; 13(13):2243. https://doi.org/10.3390/diagnostics13132243
Chicago/Turabian StyleFontana, Carla, Marco Favaro, Marco Pelliccioni, Silvia Minelli, Maria Cristina Bossa, Anna Altieri, Carlo D’Orazi, Federico Paliotta, Oriana Cicchetti, Marilena Minieri, and et al. 2023. "Laboratory Automation in Microbiology: Impact on Turnaround Time of Microbiological Samples in COVID Time" Diagnostics 13, no. 13: 2243. https://doi.org/10.3390/diagnostics13132243
APA StyleFontana, C., Favaro, M., Pelliccioni, M., Minelli, S., Bossa, M. C., Altieri, A., D’Orazi, C., Paliotta, F., Cicchetti, O., Minieri, M., Prezioso, C., Limongi, D., & D’agostini, C. (2023). Laboratory Automation in Microbiology: Impact on Turnaround Time of Microbiological Samples in COVID Time. Diagnostics, 13(13), 2243. https://doi.org/10.3390/diagnostics13132243






